1 Department of Biological Sciences, Virtual University of Pakistan, 55150 Lahore, Punjab, Pakistan
Abstract
Vertebrae protein-coding genes exhibit remarkable diversity and are organized into many gene families. These gene families have emerged through various gene duplication events, the most prominent being the two rounds of whole-genome duplication (WGD). The current research project analyzed a unique class of genes called “singletons”. Notably, we introduce the concept of “super-singletons”: genes that stand as the last representatives of their ancestral families and the sole representatives of their genetic makeup with no ortholog in any other species.
We used the Ensembl/Biomart pipeline to identify duplicated and unduplicated protein-coding genes in different vertebrate species and found orthologs of human genes.
We showed the frequency of duplicated genes and singletons, demonstrating that singletons are more vulnerable to evolutionary loss than duplicated genes. Additionally, we found that contractions in vertebrate gene families are more prevalent than expansion.
Our study provides insight into the evolution of gene families and presents a novel scenario where the extinction of species would lead to the extinction of a gene, ultimately shifting the narrative from the impact of genetics on species extinction to the extinction of genes.
Keywords
- genome evolution
- gene duplication
- whole genome duplication
- gene loss
- singletons
Vertebrates are among the most complex and successful living organisms on our planet [1, 2]. According to the Linnaean classification system, vertebrates consist of five major classes of animals: fish, amphibians, reptiles, birds, and mammals, representing a diverse array of animals [3]. A notable characteristic of vertebrates is that their genomes have undergone various rounds of whole genome duplication (WGD) events [4, 5]. For instance, approximately 450 million years ago, our ancestral vertebrate species underwent two rounds of duplication. Thus, the current vertebrate genomes result from these two rounds of duplications, except in fish, which underwent an additional round of duplication events [6, 7]. Consequently, there are four and eight genes in tetrapods and fish, respectively, for each orthologous gene in invertebrates. A vertebrate gene and all its paralogs form a gene family. However, the size of gene families in vertebrates is not always a multiple of their unduplicated ortholog in invertebrates. This divergence in the size of vertebrate gene families is due to the presence of redundant copies of paralogs within a gene family, which leads to a relaxation in functional constraint and results in differential selective pressure. This ultimately leads to a differential rate of evolution of different genes within a gene family [8, 9, 10]. There are two consequences of this differential selective pressure. First, the genetic redundancy of paralogs results in genetic differences between genes, leading to functional diversity through neofunctionalization and subfunctionalization [11, 12, 13]. Secondly, selective pressure in different species leads to gene gain, loss, and retention in a gene family [14, 15]. This gain and loss of genes ultimately affect the physiological capabilities of a gene family in a species and, hence, contribute to shaping the evolution of these species. An important aspect of the loss of genes in vertebrate gene families is that there are certain instances where all the paralogs of a gene are lost during evolution, meaning only one surviving member gene remains—designated as the singleton.
This research project focused on the evolution of vertebrate gene families, specifically on singletons. Moreover, this study identified genes present in only one species that have lost all their orthologs, called super-singletons. This study also detailed why the orphan gene concept does not truly reflect the evolutionary importance of these genes. To our knowledge, this is the most up-to-date and comprehensive study on this topic, using data from approximately 200 vertebrate species and providing some intriguing insights into the evolution of the vertebrate genome.
Data from 200 vertebrate species were utilized in this study, comprising 78 mammals, 20 birds, 12 reptiles, and 78 fish species. To our knowledge, this is the most comprehensive study focusing on the evolution of vertebrate singletons (Supplementary File 1). All genomic data were extracted from Ensembl 111 using BioMart [16, 17, 18]. In our comparative genomics analysis of orthologs, we used humans (Homo sapiens) as the reference genome, with human protein-coding genes in the filter section as the gene type. In the attributes section, we chose different vertebrate species for which we wanted to identify human orthologous genes. Additional attributes for this study included stable gene IDs of ortholog genes, homology category, percent identity, and Gene Order Conservation (GOC) score. For the quality control of the assigned orthology between different genes, we used two Biomart pipelines: the gene order conservation (GOC) score and the percentage identity score. The GOC score defines orthologous relationships based on the two genes upstream and downstream of the target gene being conserved in the reference and query species. In addition, only genes that showed a 1-to-1 homology/orthology type were selected as orthologous to minimize the inclusion of any false positive orthologs [19].
In our article, wherever we use the term ‘gene’, it specifically means protein-coding gene. Data on gene families were gathered by downloading the paralogs of all protein-coding genes from all species in our dataset from the Ensembl/Biomart genome browser. In our study, a gene family consists of the provided gene and all its paralogs. Any gene with a paralog is termed a ‘duplicated gene’ and part of a gene family. Conversely, a gene lacking a paralog is termed a ‘singleton’ and is not associated with any gene family [20]. The conservation status of each species was obtained from the International Union for Conservation of Nature (IUCN) data on endangered species [21, 22]. Boxplots and histograms were created using Microsoft Excel 2019 (Version 2019, Build 16.0.14430.20256 Product ID: 00470-90000-00000-AA855, Microsoft Corporation, Redmond, WA, USA) [23]. Statistical analysis was made by using online statistics tool https://www.socscistatistics.com/tests/ztest/default2.aspx.
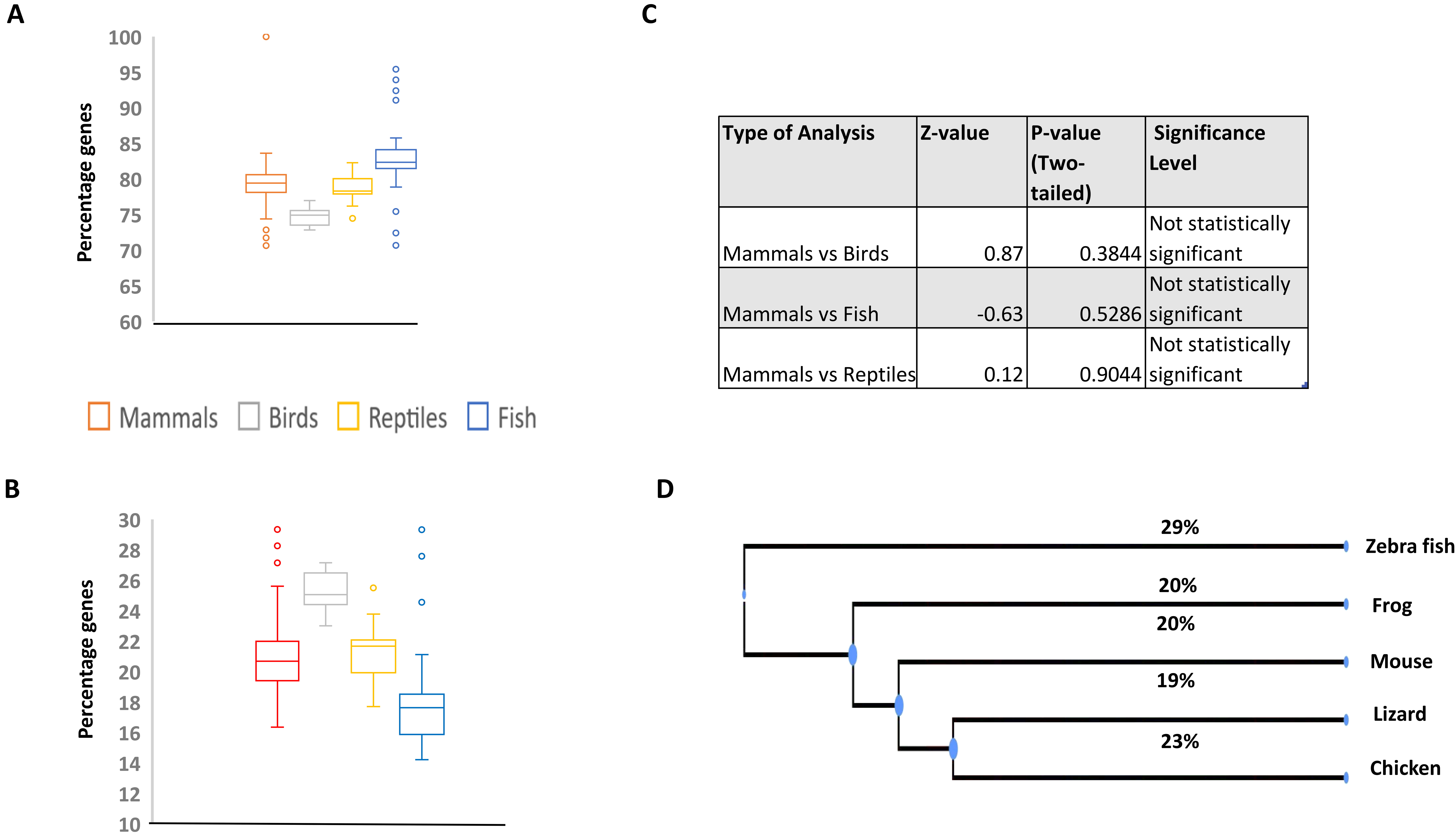
Gene and whole genome duplications have been pivotal in vertebrate genome evolution [24]. Therefore, it is pertinent to explore the frequency of duplicated and unduplicated genes in different vertebrate species to understand the evolution of the vertebrate genome. Thus, we chose a comprehensive set of species from major vertebrate clades. As expected, duplicated genes comprised the predominant part of vertebrate protein-coding genomes, with median values of 79% in mammals, 76% in birds, 78% in reptiles, and 84% in fish (Fig. 1A,D). One reason fish species exhibited the highest duplicated genes is that fish undergo an extra round of genome duplication [6]. Comparatively, fewer unduplicated genes or singletons were found in these vertebrate species, with median values of 21% in mammals, 25% in birds, 22% in reptiles, and 18% in fish (Fig. 1B,D). No significant difference in the frequency of duplicates/singletons was found in any clade (Fig. 1C). There were a few notable exceptions. For instance, in fish, the fewest number of duplicates were found in the lamprey, which is understandable as the lamprey is one of the earliest vertebrates and diverged from other fish species before the third WGD event round [6, 25, 26]
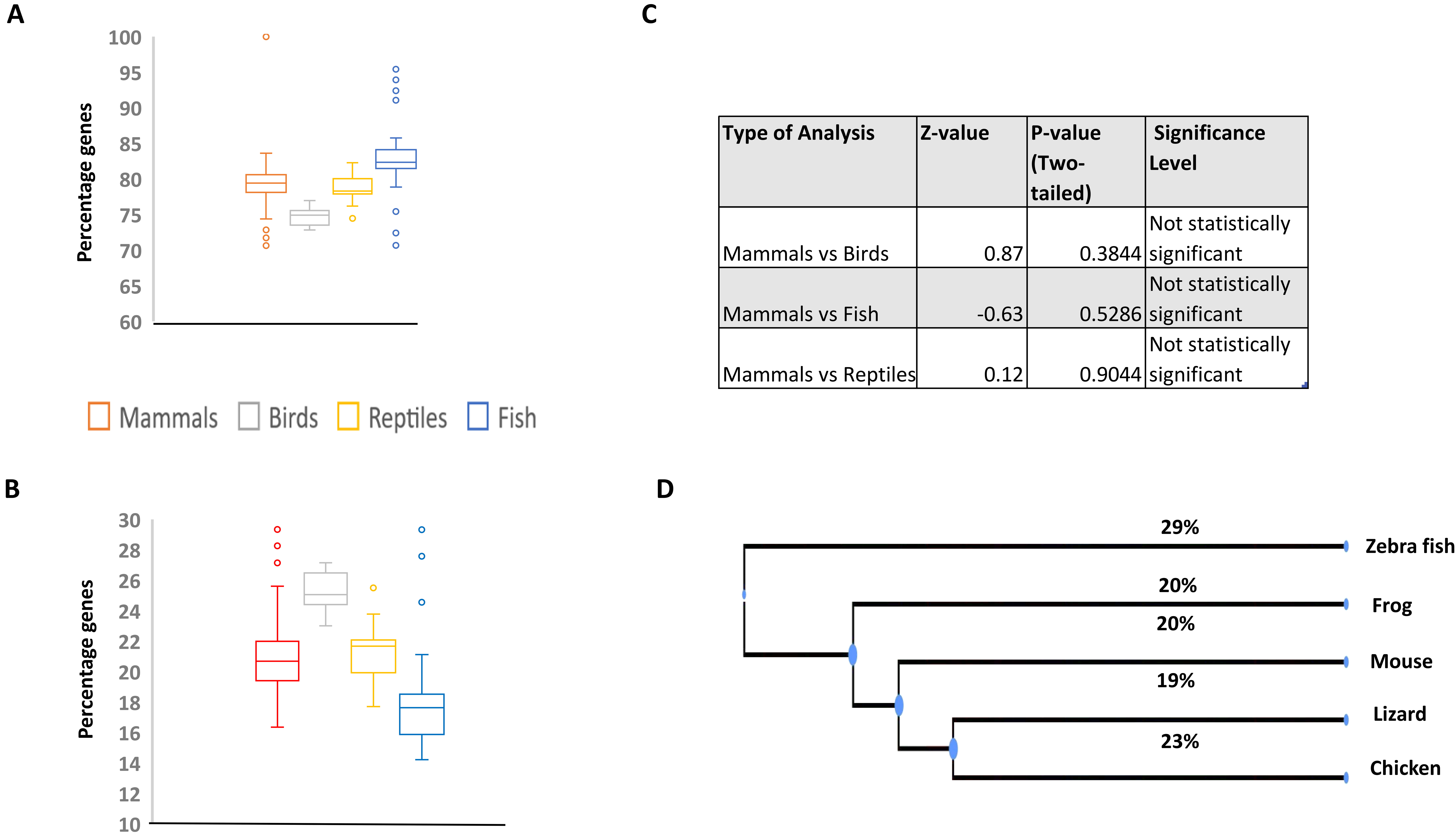 Fig. 1.
Fig. 1.
Duplicates and singletons in vertebrates. (A,B) The frequency of duplicates (A) and singletons (B) in different vertebrate species is represented by boxplots. The bottom and top of the box illustrate the 25th (Q1) and 75th (Q3) percentiles, respectively, while the midline in the box marks the median value. The bottom and top of the whiskers refer to the 10th and 90th percentiles, respectively. Data points beyond the whiskers are considered outliers and are plotted individually as small circles. Outliers represent values that are significantly different from the rest of the data. The scale used for the y-axis is linear, representing values in percentages. The detailed data can be seen in Supplementary File 2. (C) Significance level for difference in duplicated genes. A z-test was applied to estimate the p-values at a significance level of 0.05. (D) The frequency of singletons in the representative species of each of the four vertebrate taxa is shown in a simple cladogram.
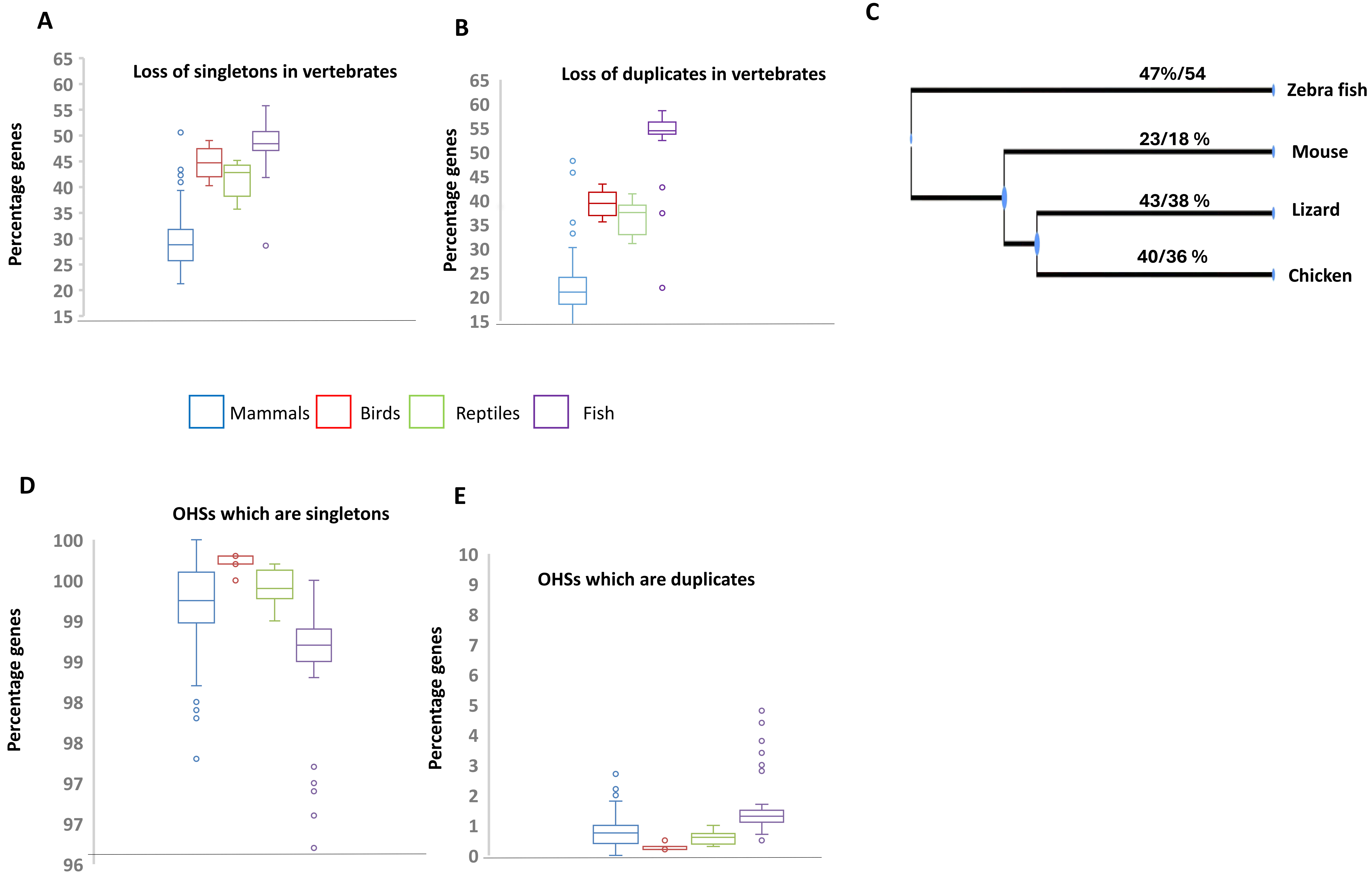
The presence of singletons in a considerable portion of human genes hints towards their evolutionary and physiological importance. The question arises: Are these singletons the evolutionary relics, represented as the sole survivors of an ancestral gene family that has undergone gradual gene losses and eventually ended up as a single gene? Or are singletons, or most of them, representative of convergent evolution, resulting in the formation of new genetic toolkits in the form of novel genes in vertebrate genomes? An evolutionarily surviving singleton will most probably have orthologs. Conversely, it would be difficult for a de novo singleton to present in multiple species as the chances of forming protein-coding genes with similar genetic makeups in various species must be extremely low. In this regard, we attempted to determine the status of orthologs of human singletons (OHSs) in each vertebrate species in our dataset. We found that OHSs are present in numerous species, indicating that these result from divergent evolution and not merely novel innovations. Interestingly, a predominant majority of human singletons seemed lost in other species, with median values for percentage loss of 29%, 45%, 43%, and 48% in mammals, birds, reptiles, and fish species, respectively (Fig. 2A,C and Supplementary File 3). Thus, a substantial fraction of OHSs seems to be lost throughout the vertebrate clade. The significantly low percentage loss value for mammals (with a z-score of –2.36 and p-value of 0.01 at a 5% significance level) is perhaps due to less evolutionary divergence from mammals than other clades in our dataset. Likewise, because fish are most distantly related to mammals, the loss of OHSs is relatively higher compared to different clades. Notably, the median values for loss of orthologs of duplicates were relatively lower than the loss we observed for the OHSs, i.e., 21%, 39%, 38%, and 54% for mammals, birds, reptiles, and fish, respectively (Fig. 2B and Supplementary File 3). These findings contrast the common assertion that singletons evolve under relatively strict functional constraints compared to duplicates [27, 28]. Indeed, certain previous studies have questioned such assertions [9, 29].
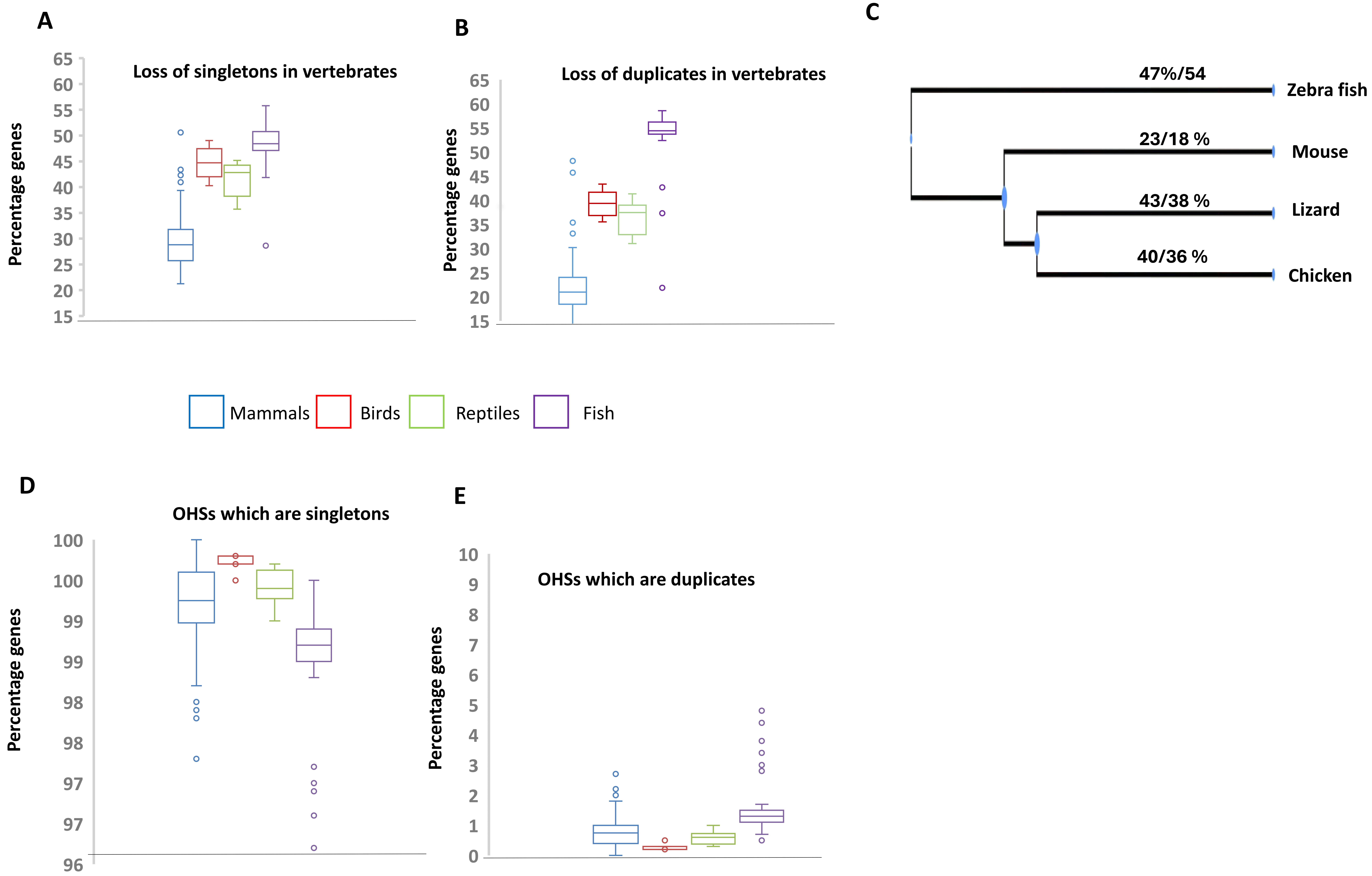 Fig. 2.
Fig. 2.
Singletons, the final survivor in a gene family. (A,B) Loss of orthologs of human singletons and duplicates in vertebrates, respectively. (C) A simple cladogram shows the loss of singletons and duplicates of representative species for each of the four vertebrate taxa. The scores on the left and right sides of the slash represent the percentage loss of orthologs of human singletons and duplicates, respectively. (D,E) The frequency of orthologs of human singletons (OHSs) is for the singletons and duplicates, respectively, in different vertebrate species. For each boxplot, the bottom and top of the box illustrate the 25th (Q1) and 75th (Q3) percentiles, respectively, while the midline in the box marks the median value. The bottom and top of the whiskers refer to the 10th and 90th percentiles, respectively. Data points beyond the whiskers are considered outliers and are plotted individually as small circles. Outliers represent values that are significantly different from the rest of the data. The scale used for the y-axis is linear, representing values in percentages. The detailed data can be seen in Supplementary Files 3,4.
One way of looking at this loss of OHSs is not to see it as a loss of individual singletons but to consider them a part of the ancestral gene family. These singletons would have once been part of a gene family. The loss of their ancestral paralog genes means these gene families were already evolving under relaxed functional constraints. As a result, these ancestral paralogs were lost, leaving behind the singletons as the remnants of their ancestral gene family. Since the ancestral paralogs did not provide a survival advantage, it is intuitive to think that these singletons, or at least some of them, also evolved under relaxed functional constraints and, thus, did not provide a significant survival advantage. Therefore, it was only a matter of time before these singletons faced the same fate as their paralogs. However, the question arises of how a gene family undertakes such a drastic evolutionary route. One answer may be that many gene families are part of a supergene family; hence, they may share potentially redundant protein domains in that superfamily, ultimately promoting the loss of most of the members in that gene family [30]. This potential process also underscores the importance of more rigorous and comprehensive functional and evolutionary studies to evaluate the evolution of singletons. These genes are not unique but also have the potential to provide us with a window into the evolution of the ebbs and flows of gene families. Interestingly, the dominant frequency of the OHSs in different species remained singletons, with a median value of 99.2%, 99.8%, 99.4%, and 98.7% in mammals, birds, reptiles, and fish species, respectively. Consequently, a very negligible frequency of genes, with a median value of 0.8%, 0.2%, 0.6%, and 1.3% in mammals, birds, reptiles, and fish species, respectively, were non-singletons or had paralogs (Fig. 2D,E, and Supplementary File 4). These data show that numerous OHSs are lost (as shown in Fig. 2A), and the tendency of vertebrates to lose singletons is higher than their expansion capacity. Therefore, gene loss is more prevalent than gene gain in vertebrates, irrespective of whether genes are duplicated or singletons. The loss of duplicated genes leads to the compaction in gene family size; sometimes, this compaction is so extensive that all the members of a gene family are lost except one surviving member, known as the singleton.
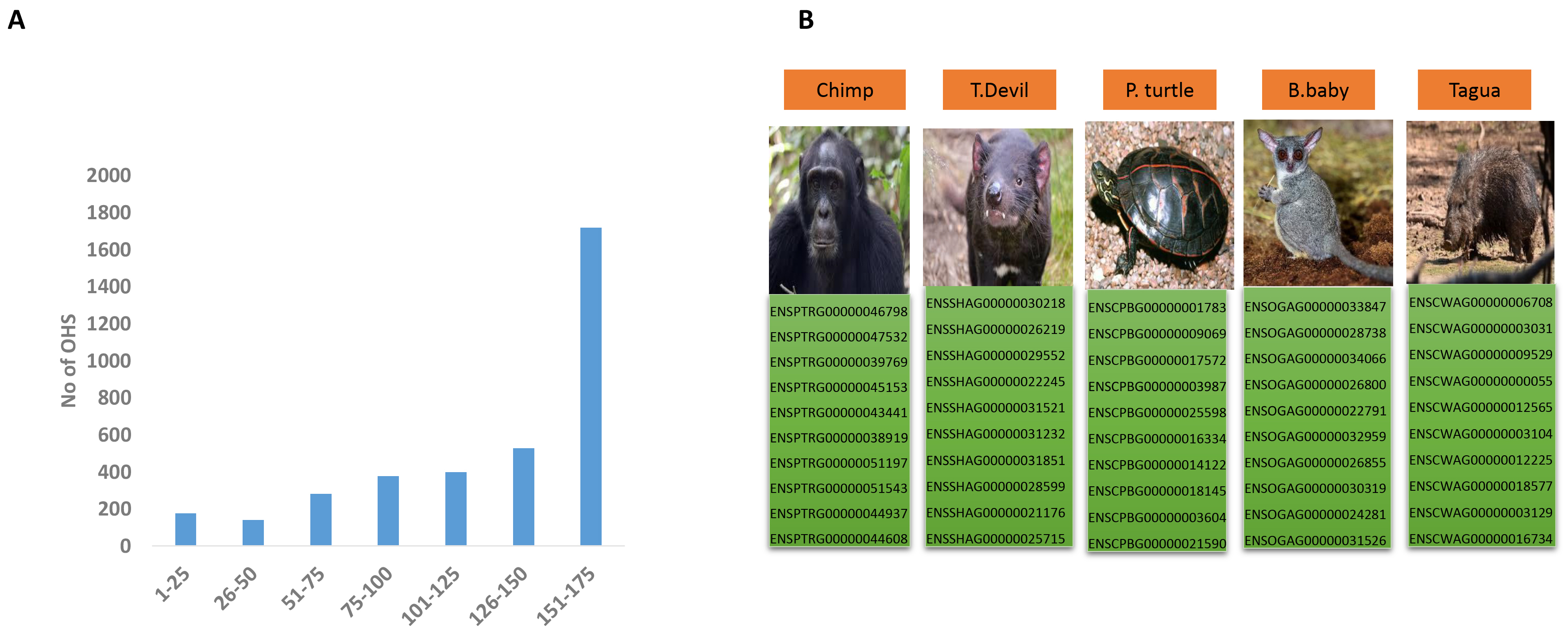
To see to what extent OHSs are present in different species, we determined the
presence or absence of each OHS in individual vertebrate species in our dataset.
Our findings demonstrated that not only do the majority of the human singletons
have orthologs in numerous species, but the largest majority of orthologs were
present in the predominant majority of species in our dataset, with the largest
majority of OHSs (
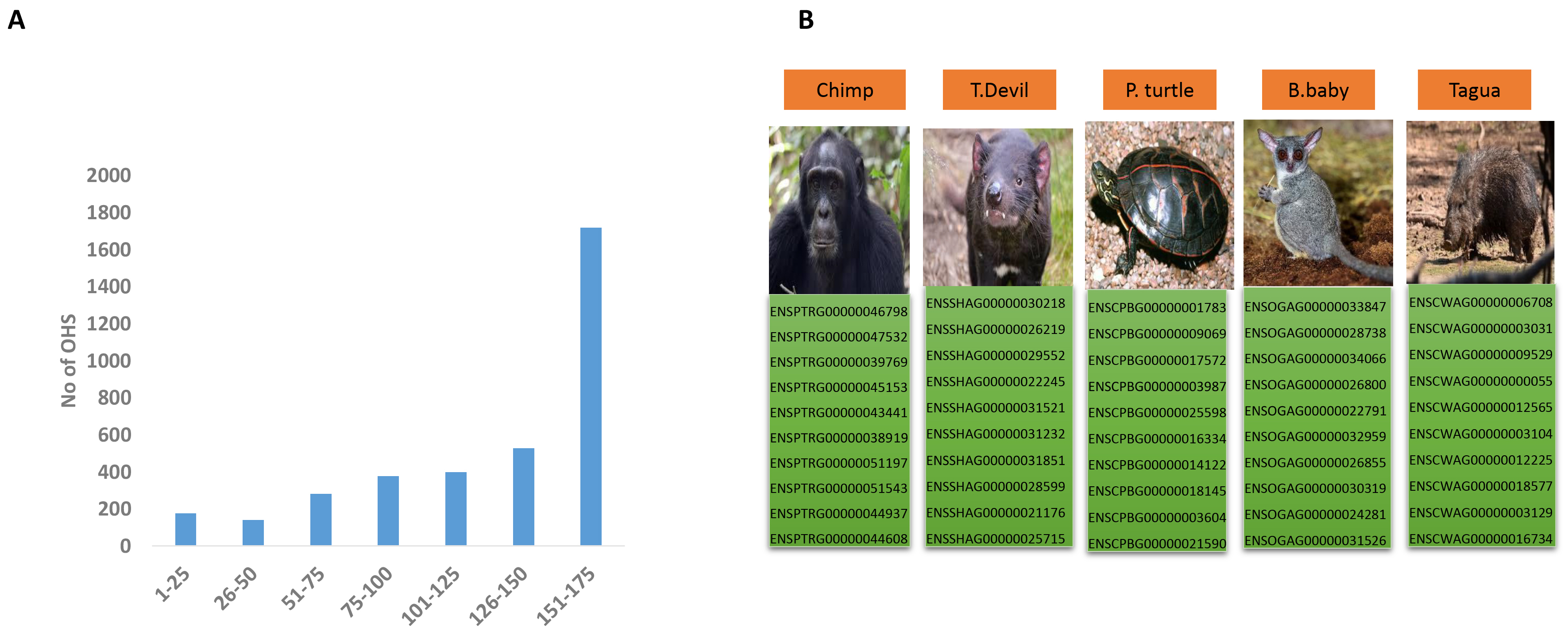 Fig. 3.
Fig. 3.
Super-singletons and gene extinction. (A) The frequency of OHSs is present in a given number of species. Values on the x-axis represent several species, while values on the y-axis represent the number of OHSs present. Please see Supplementary File 5 for details. (B) Super-singletons in endangered species. The figure shows the Gene IDs of 10 super-singletons in five representative endangered species. The images of the species are taken from Wiki Commons (Chimp, T.devil, P.turtle, B.baby, Tagua). For a complete list of super-singletons in all the endangered species in our dataset, please refer to Supplementary File 6. Abbreviations: T. devil, Tasmanian devil; B. baby, bush baby; P. turtle, painted turtle.
Gene duplication events, especially whole-genome duplications (WGDs), and the formation of gene families are crucial in vertebrate evolution. The size and composition of these gene families vary, leading to the formation of singletons. The attrition of gene families resulting in the formation of singletons represents a unique scenario in the evolution of species and their genomes. Super-singletons, which have lost all orthologs across species, represent the case of gene loss across species. Species extinction, particularly those harboring super-singletons, results in the permanent loss of unique genetic makeup from the biosphere. Our study emphasizes the need to identify singletons in every species, creating a catalog of genes at risk of extinction. For decades, we have been associating extinction events with the permanent disappearance of species from the biosphere. Now, species extinction will also be related to the extinction of genes.
Most of the relevant data are included in the supplementary materials. However, for detailed data on each individual species, requests can be made to the corresponding author.
AAK conceived the idea, AAK and AF extracted the data, AAK and AF analyzed the data, AAK wrote the article, AAK supervised the project. Both authors have participated sufficiently in the work to take public responsibility for appropriate portions of the content and agreed to be accountable for all aspects of the work in ensuring that questions related to its accuracy or integrity. Both authors read and approved the final manuscript. Both authors contributed to editorial changes in the manuscript.
Not applicable.
Not applicable.
This research was funded by Higher Education Commission of Pakistan, grant number HEC-NRPU-17093.
The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/j.fbs1604022.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.