1 Department of Neurology, Renmin Hospital of Wuhan University, 430060 Wuhan, Hubei, China
2 Center for Genome Analysis, ABLife BioBigData Institute, 430075 Wuhan, Hubei, China
3 Laboratory of Human Health and Genome Regulation, ABLife BioBigData Institute, 430075 Wuhan, Hubei, China
Abstract
Background: Angiogenesis is essential for tissue development, and therefore its dysregulation can cause various diseases, including cerebrovascular disease. Galectin-1, encoded by the lectin galactoside-binding soluble-1 gene (LGALS1), has critical roles in the regulation of angiogenesis, but the underlying mechanisms need further clarification. Methods: LGALS1 was silenced in human umbilical vein endothelial cells (HUVECs) and whole transcriptome sequencing (RNA-seq) was then performed to investigate potential targets for galectin-1. Galectin-1-interacting RNA data was also integrated to explore how galectin-1 might regulate gene expression and alternative splicing (AS). Results: A total of 1451 differentially expressed genes (DEGs) were found to be regulated by silencing LGALS1 (siLGALS1), comprising 604 up- and 847 down-regulated DEGs. Down-regulated DEGs were primarily enriched in angiogenesis and inflammatory response pathways, and included CCL2, GJA5, CALCRL, ACKR3, HEY1, AQP1, CD34, ECM1, RAMP2, and SELP. These were validated by reverse transcription and quantitative polymerase chain reaction (RT-qPCR) experiments. siLGALS1 was also used to analyze dysregulated AS profiles, such as the promotion of exon skipping (ES) and intron retention, and inhibition of cassette exon events. Interestingly, regulated AS genes (RASGs) were found to be enriched in focal adhesion and in the angiogenesis-associated vascular endothelial growth factor (VEGF) signaling pathway. Furthermore, based on our previously published RNA interactome data for galectin-1, hundreds of RASGs were found to be bound by galectin-1, including those enriched in the angiogenesis pathway. Conclusions: Our results demonstrate that galectin-1 can regulate angiogenesis-related genes at transcriptional and post-transcriptional levels, probably by binding to the transcripts. These findings expand our understanding of the functions of galectin-1 and the molecular mechanisms that underlie angiogenesis. They also indicate that galectin-1 could serve as a therapeutic target for future anti-angiogenic treatments.
Keywords
- galectin-1
- angiogenesis
- transcriptome
- alternative splicing
- HUVECs
Angiogenesis, also known as neovascularization, is critical for the progression of many diseases, including cancers and various other disorders [1]. Anti-angiogenesis could serve as an effective strategy for treating cancers [2] and ischemic diseases [3]. Due to their critical roles in various angiogenesis-related disorders, several key proteins have been reported to participate in angiogenesis [4]. Vascular endothelial growth factors (VEGFs) and their receptors play key roles in normal and pathological angiogenesis [5]. Fibroblast growth factors (FGFs) and their receptors (FGFRs) regulate a broad spectrum of biological functions, including angiogenesis [6]. Transforming growth factor-beta (TGF-beta) is highly expressed in the smooth-muscle cells of patients with angiogenic disorder, and is also involved in the regulation of tumor angiogenesis [7]. A recent study found that the environment surrounding extracellular matrix (ECM) compression could regulate microvascular growth and upstream angiogenic signaling pathways [8]. These results indicate that the expression levels of angiogenic factors may be regulated by other proteins whose functions in the pathogenesis of angiogenic disorders require further investigation.
Galectin-1, encoded by lectin galactoside-binding soluble-1 (LGALS1),
was first reported in 2006 as playing a critical role in tumor angiogenesis [9].
Galectin-1 is a carbohydrate-binding protein with an affinity for
To address this question, LGALS1 was silenced in human umbilical vein endothelial cells (HUVECs) and whole transcriptome sequencing (RNA-seq) was then performed to comprehensively investigate gene expression and AS levels. In addition, galectin-1 RNA interactome data was also integrated to explore the relationship between its bound RNAs and dysregulated RNAs. Our study offers a possible explanation for the previously reported functions of galectin-1 in angiogenesis, which appears to be via modulation of the transcriptome profiles of HUVECs, thus highlighting one of the critical roles of galectin-1.
The small interfering RNA (siRNA) duplexes for LGALS1 transcripts were purchased from Gemma (Suzhou, China). Non-targeting control siRNA (si-Negative) sequence was: 5’-UUCUCCGAACGUGUCACGUTT-3’ (sense), while the siRNA targeting LGALS1 (siLGALS1-3) sequence was: 5’-GCAAAGACAGCAACAACCUTT-3’ (sense). Three biological replicates were prepared for the subsequent experiments in this study.
HUVECs were originally obtained from ScienCell (8000, ScienCell, Carlsbad, CA, USA), and provided by Shanghai Zhong Qiao Xin Zhou Biotechnology Co.,Ltd. (Shanghai, China). The cells were authenticated by immunofluorescence analysis of the positive marker gene of vWF, and tested for the free of mycoplasma contamination through PCR.
HUVECs were cultured at 37 °C with 5% CO
After harvesting LGALS1-silenced (siLGALS1) and control cells,
genomic DNA was removed with RQ1 DNase (Promega, Madison, WI, USA) to obtain total RNA. Qualified
RNA, as measured by absorbance at 260 nm/280 nm (A260/A280) using SmartSpec Plus
(BioRad, Hercules, USA), was used in the following experiments. One
Raw RNA-seq reads were filtered by removing N-containing reads, adaptors and
low-quality bases, and discarding short reads (
The ABLas pipeline was used to predict and quantify alternative splicing events
(ASEs) and galectin-1-regulated ASEs in siLGALS1 and Ctrl groups as described
previously [23, 24]. Based on the splicing reads, 9 types of ASEs were analyzed:
alternative 3’ splice site (A3SS), alternative 5’ splice site (A5SS), cassette
exon, exon skipping (ES), A3SS&ES, A5SS&ES, mutually exclusive exons (MXE),
mutually exclusive 5’UTRs (5pMXE), and mutually exclusive 3’UTRs (3pMXE).
Student’s t-test was used to calculate the statistical significance of
the ASEs’ ratio differences in order to identify regulated ASEs (RASEs). ASEs
with a p-value
Functional cluster analyses were conducted using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases to analyze DEGs and RASGs for enriched functional pathways. Briefly, DEGs and RASGs were mapped to all terms in the GO database to count gene numbers, and the hypergeometric distribution test was used to obtain the significance of each GO term for DEGs and RASGs’ against a background of GO annotation for the whole genome. The analysis method was the same as for the KEGG analysis mentioned above.
The iRIP-seq data for galectin-1 was downloaded from the NCBI GEO database with accession number GSE147565. This was used to investigate how galectin-1 regulates DEGs and RASEs. Galectin-1-bound peaks and genes were re-analyzed according to a previously published method [16]. Analyses of identified DEGs and RASGs were performed after the overlapping peaks and genes were obtained from two iRIP-seq replicates.
Data were analyzed with GraPhPad prism application software (San Diego, USA) and were presented
as the mean
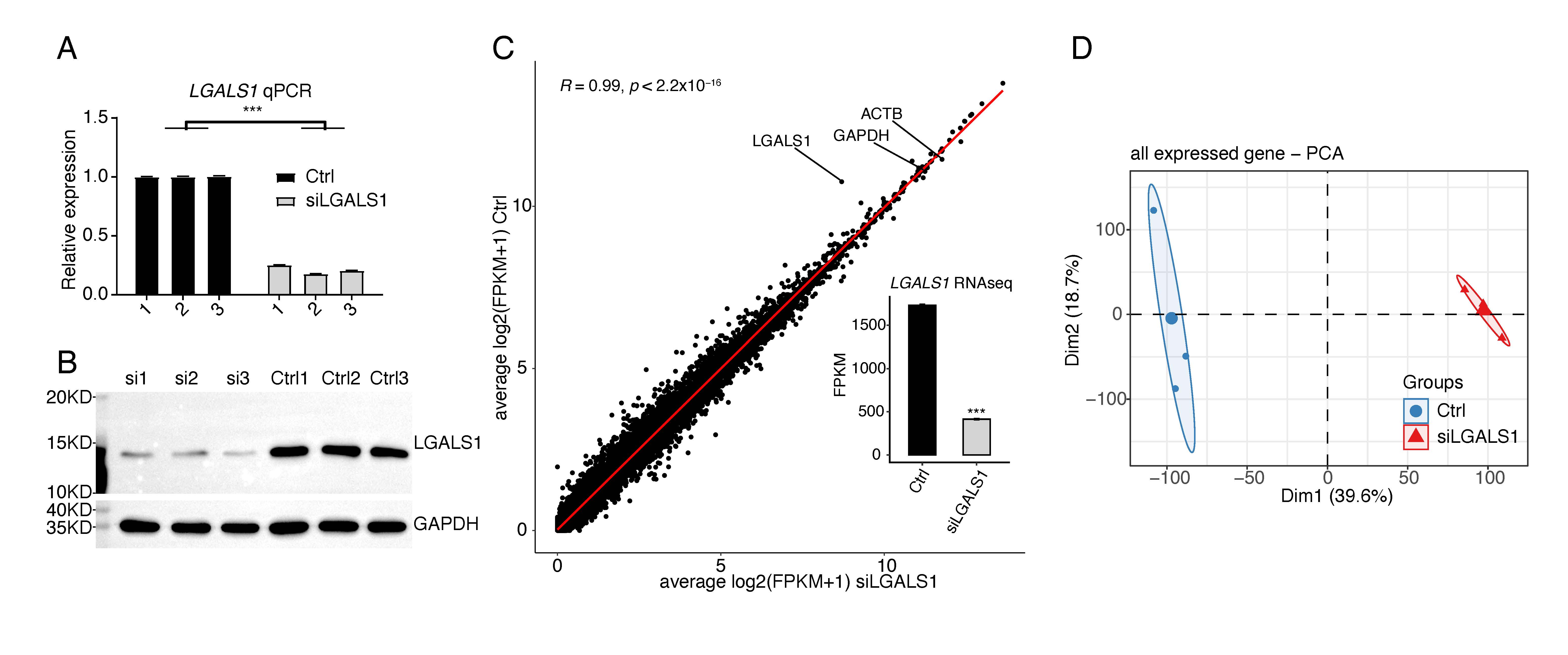
Our previous study demonstrated that silencing of galectin-1 could inhibit capillary tube formation in HUVECs [16]. However, it is not known how galectin-1 regulates angiogenesis, and therefore the LGALS1 knockdown experiment was repeated using siLGALS1. Successful knockdown of LGALS1 was confirmed by RT-qPCR and WB experiments (Fig. 1A,B). RNA-seq was then performed to identify LGALS1-regulated genes at the transcriptional and post-transcriptional levels. Three biological replicates with high-quality sequencing data were obtained for siLGALS1 and negative control (Ctrl) samples (Supplementary Table 2). After aligning quality-filtered reads to the human genome and calculating FPKM for the gene expression level, LGALS1 was found to be highly expressed, with an average FPKM value of 1731. This was only slightly lower than expression of the house-keeping genes ACTB and GAPDH (Fig. 1C). SiLGALS1 samples had a lower FPKM value compared to Ctrl samples (Fig. 1C). Principal component analysis (PCA) for all expressed genes revealed a clear separation between siLGALS1 and Ctrl samples, with a short inner distance for the three biological replicates (Fig. 1D). The sharp contrast indicated the global changes in transcriptome profiles following siLGALS1. These results indicate that galectin-1 is highly expressed in HUVECs and may play a critical role in regulating the transcriptome in these cells.
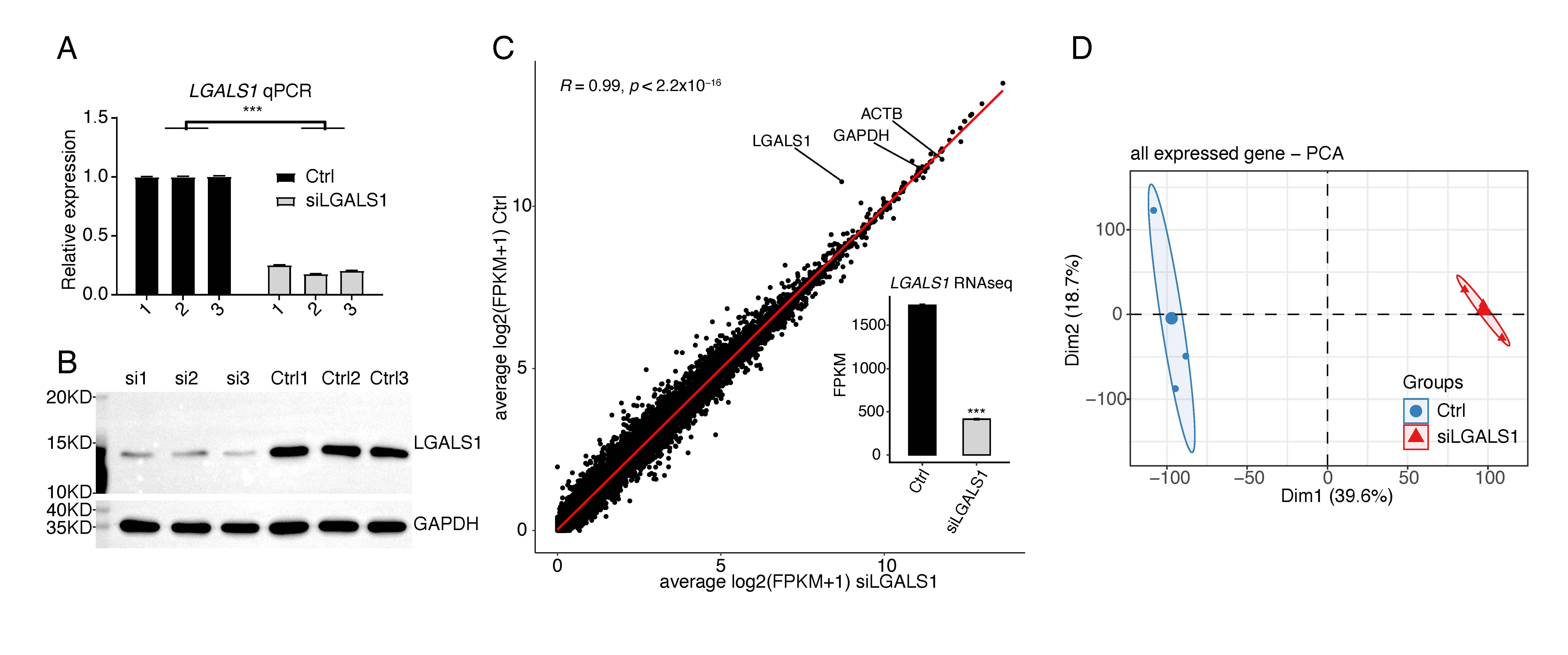 Fig. 1.
Fig. 1.SiLGALS1 has a global influence on the transcriptome profile of
HUVECs. (A) Bar plot showing the relative expression levels of LGALS1 in
siLGALS1 and Ctrl samples, as determined by the RT-qPCR method. N = 3. (B)
Western blot result showing a decreased level of LGALS1 protein in siLGALS1
samples compared with Ctrl samples. N = 3. (C) Scatter plot showing the
correlation between siLGALS1 and Ctrl samples using the FPKM values for all
expressed genes. The FPKM values for LGALS1 are shown in the bar plot.
(D) PCA result showing that siLGALS1 samples are clearly separated from Ctrl
samples at the first principal component. *** p-value
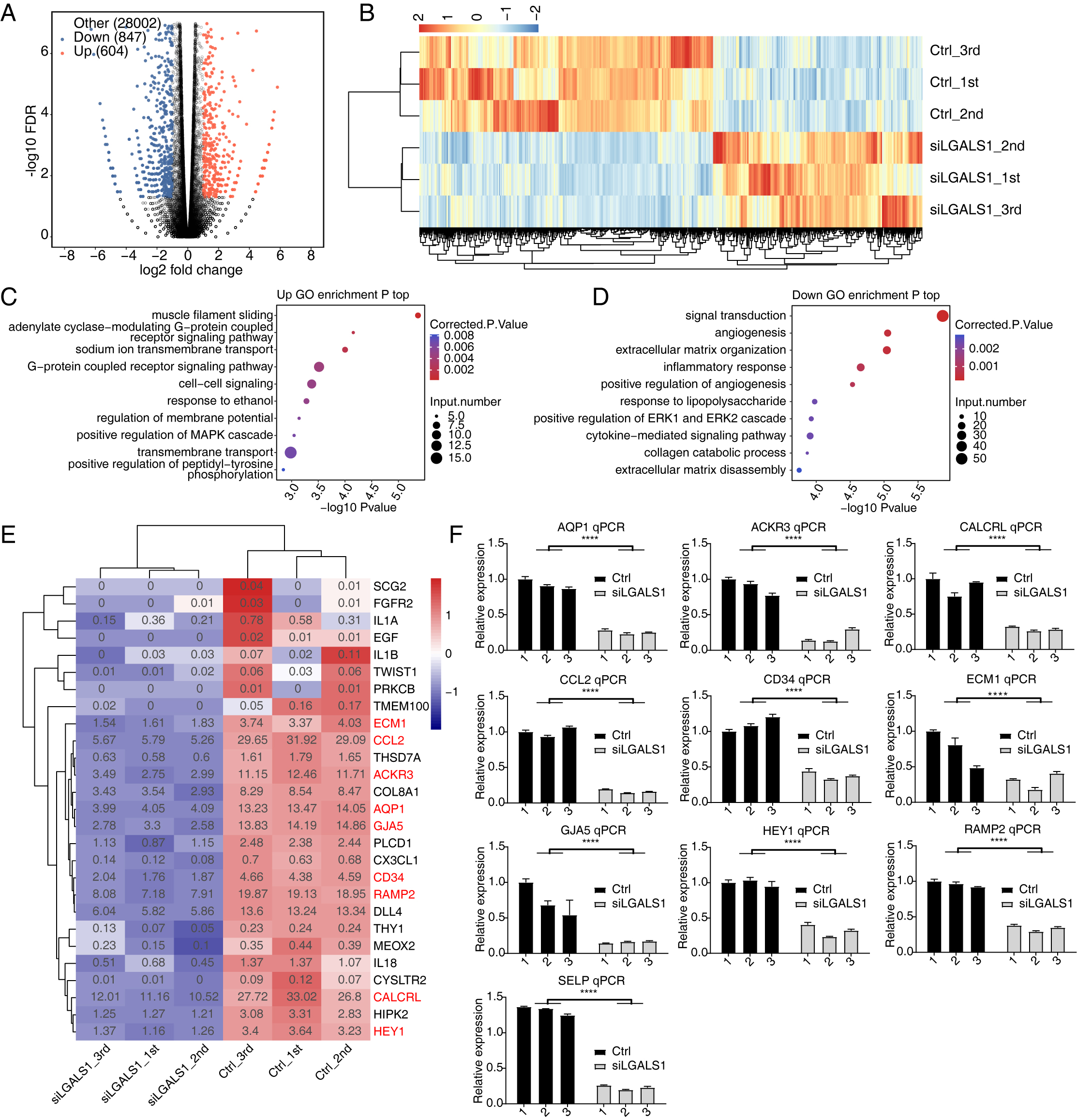
We used siLGALS1 to identify dysregulated genes that could participate in angiogenesis. With the criteria set to a 2-fold change and 0.05 false discovery rate (FDR), 1451 DEGs were identified using the edgeR package [25], comprising 604 up-regulated and 847 down-regulated genes (Fig. 2A, Supplementary Table 3). A hierarchical cluster heatmap of all DEGs showed consistent expression changes for the three replicates (Fig. 2B). Interestingly, more down-DEGs were detected than up-DEGs, suggesting that galectin-1 is likely to promote gene expression. We then investigated the functions of up- and down-DEGs using GO and KEGG databases. The top 5 enriched GO biological process (BP) terms included muscle filament sliding, the adenylate cyclase-modulating G-protein coupled receptor signaling pathway, sodium ion transmembrane transport, the G-protein coupled receptor signaling pathway, and cell-cell signaling (Fig. 2C). We next focused on the galectin-1-promoting genes (down-DEGs). Functional enrichment analysis revealed that the top 5 terms were signal transduction, angiogenesis, ECM organization, inflammatory response, and positive regulation of angiogenesis (Fig. 2D). Down-DEGs were significantly enriched in angiogenesis and in positive regulation of angiogenesis pathways. These two terms included 18 and 12 genes, respectively. A heatmap of the two terms showed that all of the genes were down-regulated in siLGALS1 samples (Fig. 2E). To further validate these genes, 10 were selected from the two terms and RT-qPCR was performed. The results confirmed that all of the genes were down-regulated in siLGALS1 samples (Fig. 2F). These DEG results demonstrate that siLGALS1 decreases the expression of angiogenesis-related genes, which could be associated with the promotion of angiogenesis.
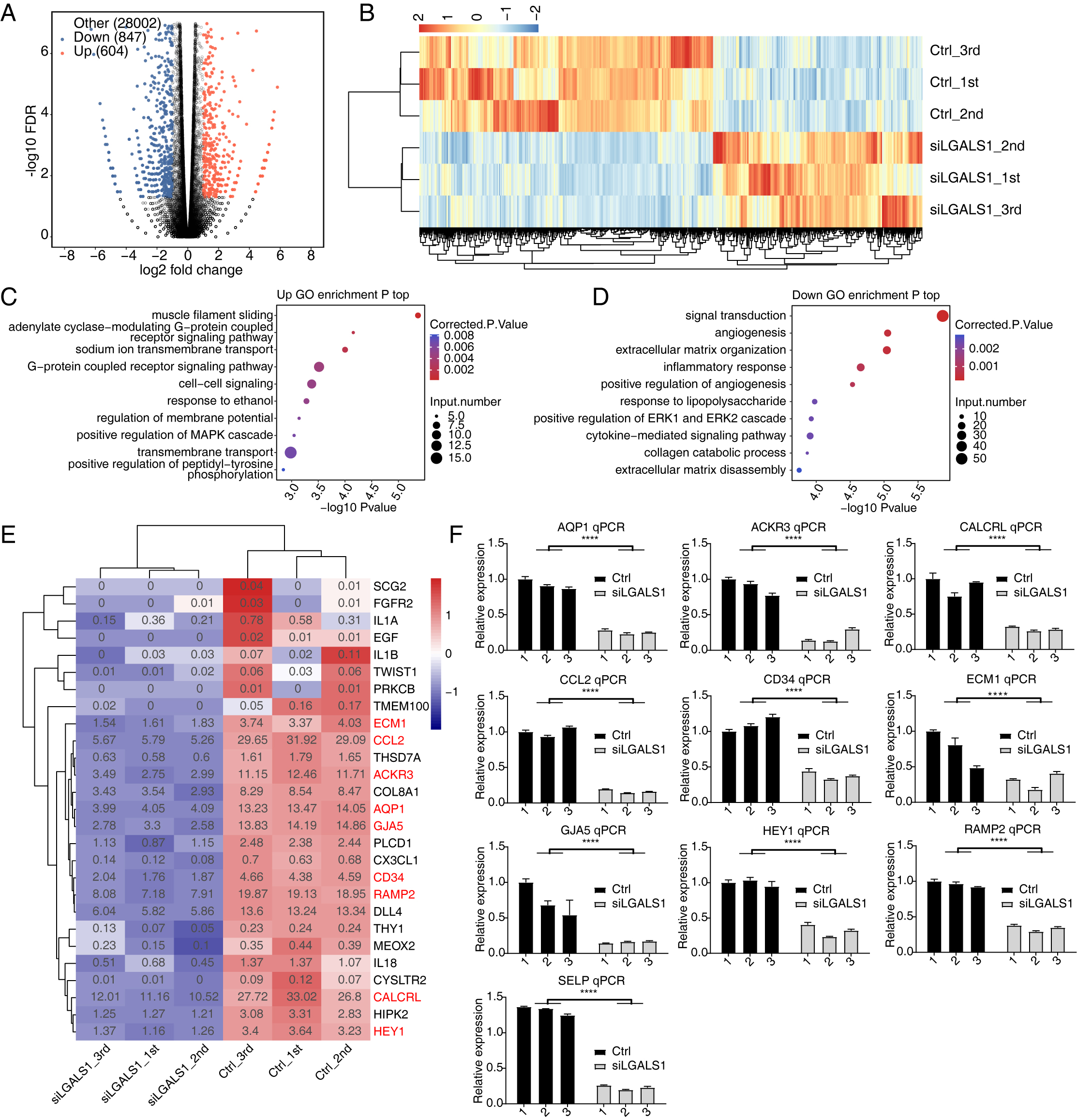 Fig. 2.
Fig. 2.SiLGALS1 reduces the expression of angiogenesis-related genes.
(A) Volcano plot showing the expression pattern of DEGs. Red and blue points
indicate up- and down-regulated DEGs in siLGALS1 samples, respectively. (B)
Hierarchical clustering heatmap showing the expression pattern of all DEGs. (C)
Bubble plot showing the top enriched GO BP pathways of the up-regulated DEGs. (D)
Bubble plot showing the top enriched GO BP pathways of the down-regulated DEGs.
(E) Hierarchical clustering heatmap showing the down-regulated genes selected
from the two angiogenesis pathways. (F) Bar plot showing RT-qPCR results for the
10 genes selected from the two angiogenesis terms. N = 3. **** p-value
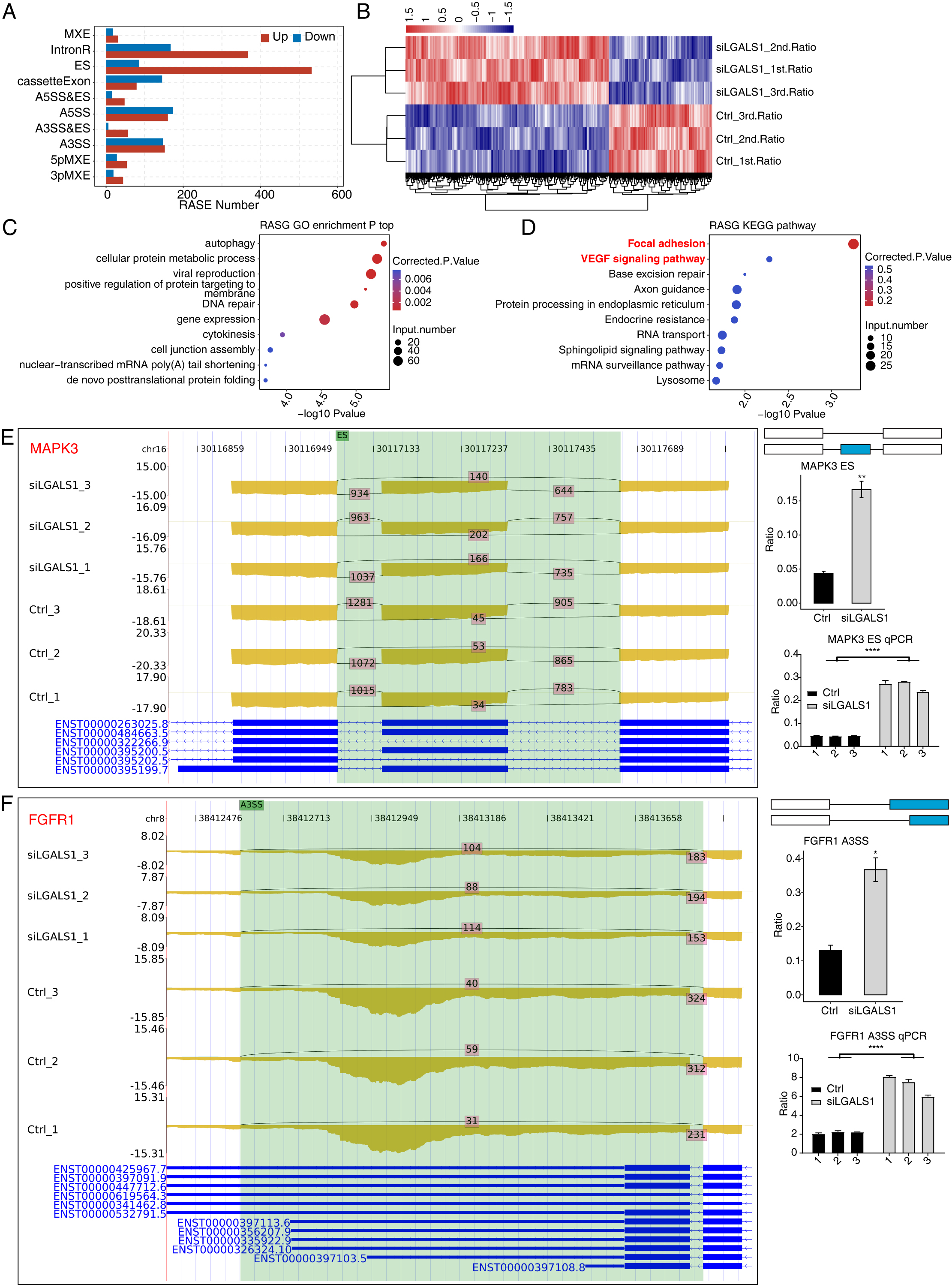
Our previous study found extensive interactions between galectin-1 and thousands of RNA molecules [16], hinting at potential functions for galectin-1 in RNA regulation. AS is an important post-transcriptional regulation mechanism in all eukaryotes. Galectin-1-regulated AS events in HUVECs were systematically analyzed. The ABLas program [24] was applied to identify ASEs and calculate statistical differences between siLGALS1 and Ctrl samples. In total, we identified 2327 RASEs that were significantly different between the two groups. After classifying these RASEs into 10 types, ES and IR events were found to be the most common RASE types (Fig. 3A). More ES (ES, A3SS&ES, and A5SS&ES) and less cassette exon (opposite to ES) events were identified in siLGALS1 samples (Fig. 3A). Therefore, siLGALS1 showed a trend to induce ES events in HUVECs. Moreover, the number of included RASEs (up, 1523) was much higher than the number of excluded RASEs (down, 804), suggesting that galectin-1 has a tendency for AS regulation. A hierarchical clustering heatmap of all RASE ratios demonstrated a clear difference between the siLGALS1 and Ctrl groups (Fig. 3B), suggesting the AS events for these genes were consistently regulated by siLGALS1. We next assessed the enriched functions of these RASGs. The top 5 enriched GO BP terms were found to be autophagy, cellular protein metabolic process, viral reproduction, positive regulation of protein targeting to membrane, and DNA repair (Fig. 3C). KEGG pathway analysis also revealed that RASGs were highly enriched in focal adhesion and VEGF signaling pathways (Fig. 3D). Previous studies have reported that RASGs play important roles in angiogenesis [26, 27]. Of the genes involved in focal adhesion and VEGF signaling pathways, 8 RASEs were selected to confirm the altered AS ratios using RT-qPCR experiments (ADRM1, EGFL7, FGFR1, FLNC, SPHK1, MAPK3, FLT1, and MAPK14). Specific primers for these RASEs (shown in Supplementary Table 1) were designed according to previously published methods [28]. The RNA-seq data revealed that 7 of the 8 RASEs showed consistent ratio changes, with MAPK3, FGFR1, FLNC, and SPHK1 presented as examples in Fig. 3E,F and Supplementary Fig. 1A,B. Together, these results indicate that galectin-1 is a strong regulator of the AS patterns for angiogenesis-associated genes in HUVECs.
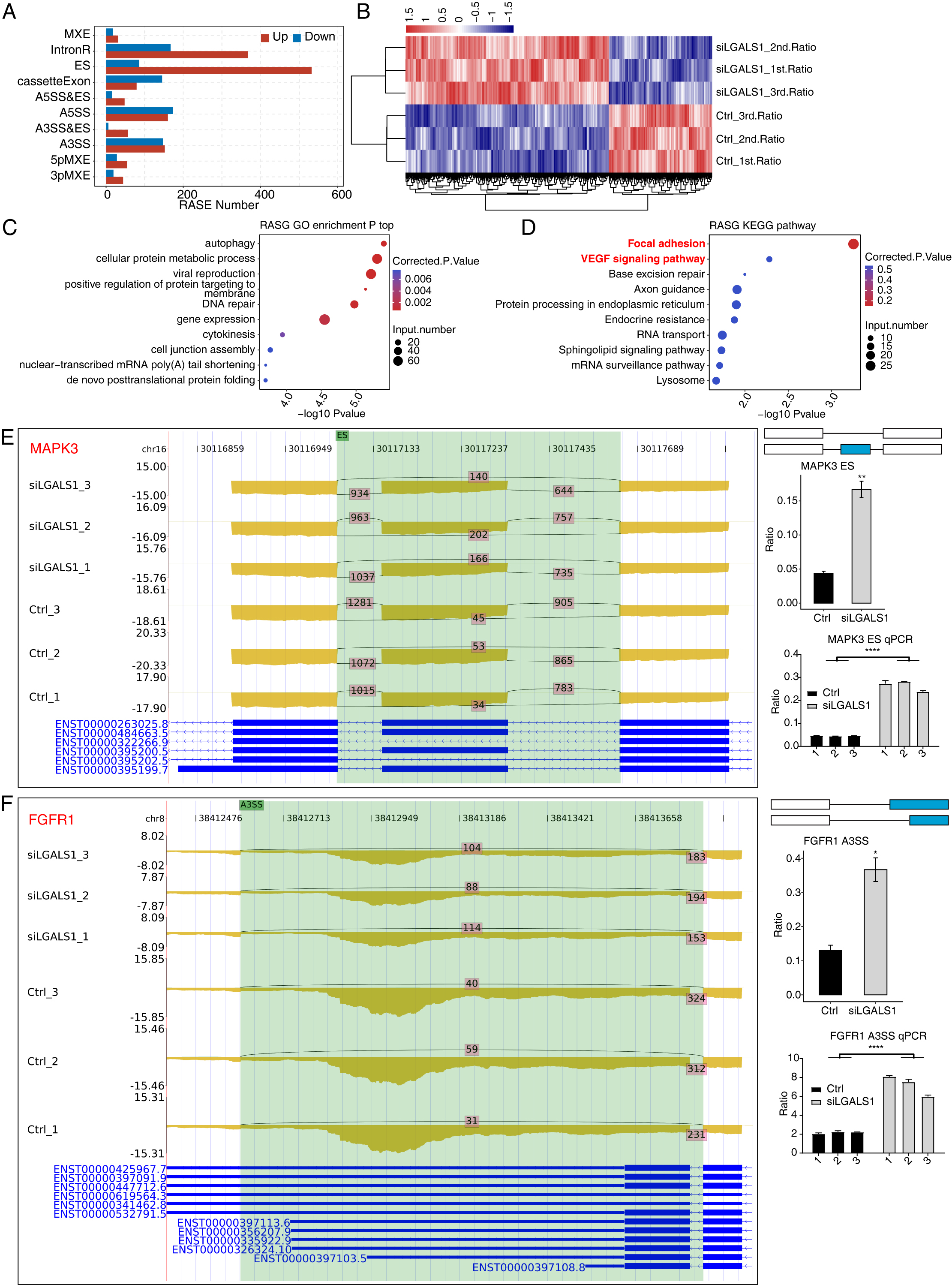 Fig. 3.
Fig. 3. Galectin-1 is a strong regulator of the AS profiles of VEGF
signaling pathway genes in HUVECs. (A) Bar plot showing the number of each RASE
type. Up and down indicates the inclusion and exclusion pattern, respectively, of
each RASE type in siLGALS1 samples. (B) Hierarchical clustering heatmap showing
the ratio patterns of all RASEs. (C) Bubble plot showing the top enriched GO BP
pathways of all RASGs. (D) Bubble plot showing the top enriched KEGG pathways of
all RASGs. (E) Reads plot and RT-qPCR results of ES event for MAPK3 in
the VEGF signaling pathway. N = 3. (F) Reads plot and RT-qPCR results for A3SS
event of FGFR1 in the VEGF signaling pathway. N = 3. * p-value
Based on the above results, we deduced that galectin-1 has significant influence
on both the expression and AS of genes in HUVECs. We next investigated whether
any genes were co-regulated by siLGALS1 at both the expression and alternative
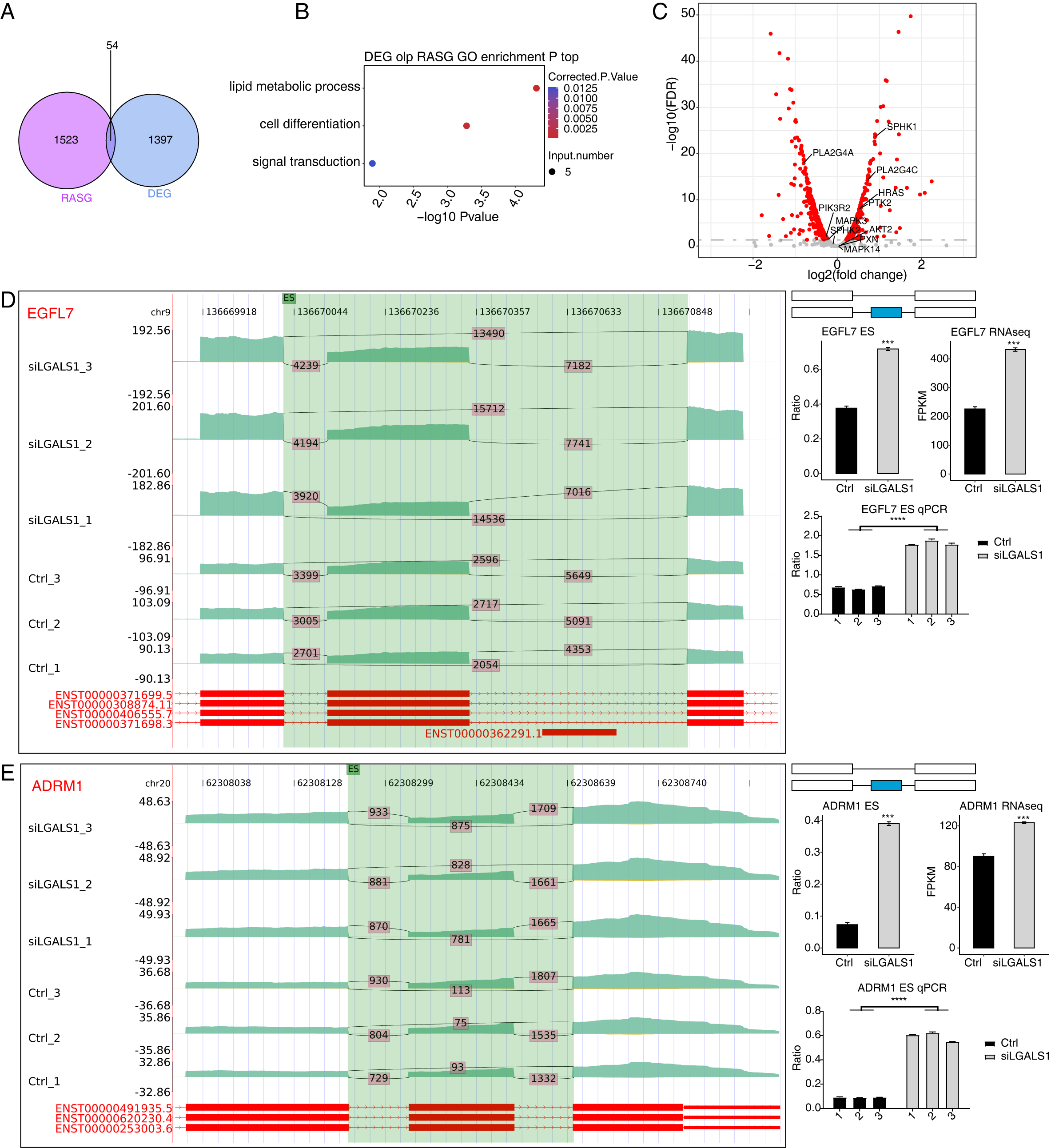
splicing levels. A total of 54 genes were identified both in the 1451 DEGs and in
the 1577 RASGs (Fig. 4A). Functional analyses of these 54 overlapping genes
revealed they were enriched in the sphingolipid metabolism pathway (Fig. 4B,
p-value
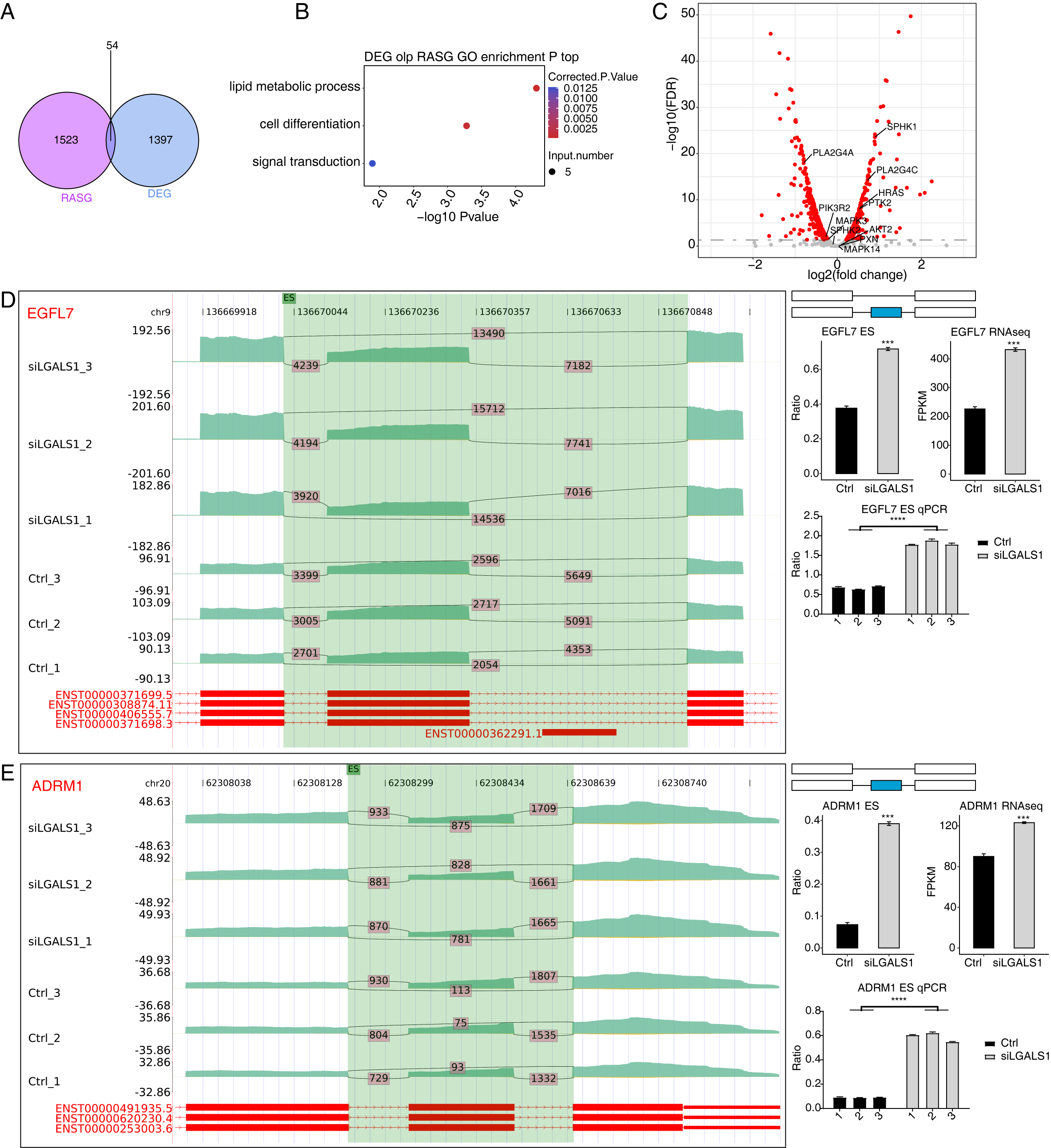 Fig. 4.
Fig. 4.Analysis of the Overlap between DEGs and RASGs. (A) Venn
diagram showing the number of overlapping genes between DEGs and RASGs. (B)
Bubble plot showing the enriched GO BP pathways of the overlapping genes from A.
(C) Volcano plot showing the expression patterns of RASGs. Red points indicate
RASGs with significant FDR (
We previously investigated the RNA binding ability of galectin-1 and its
regulatory roles in angiogenesis using the iRIP-seq method [16]. Here, we focused
on interpreting the relationship between galectin-1-bound RNAs and the
galectin-1-regulated transcriptome. Using the overlapping peaks and genes bound
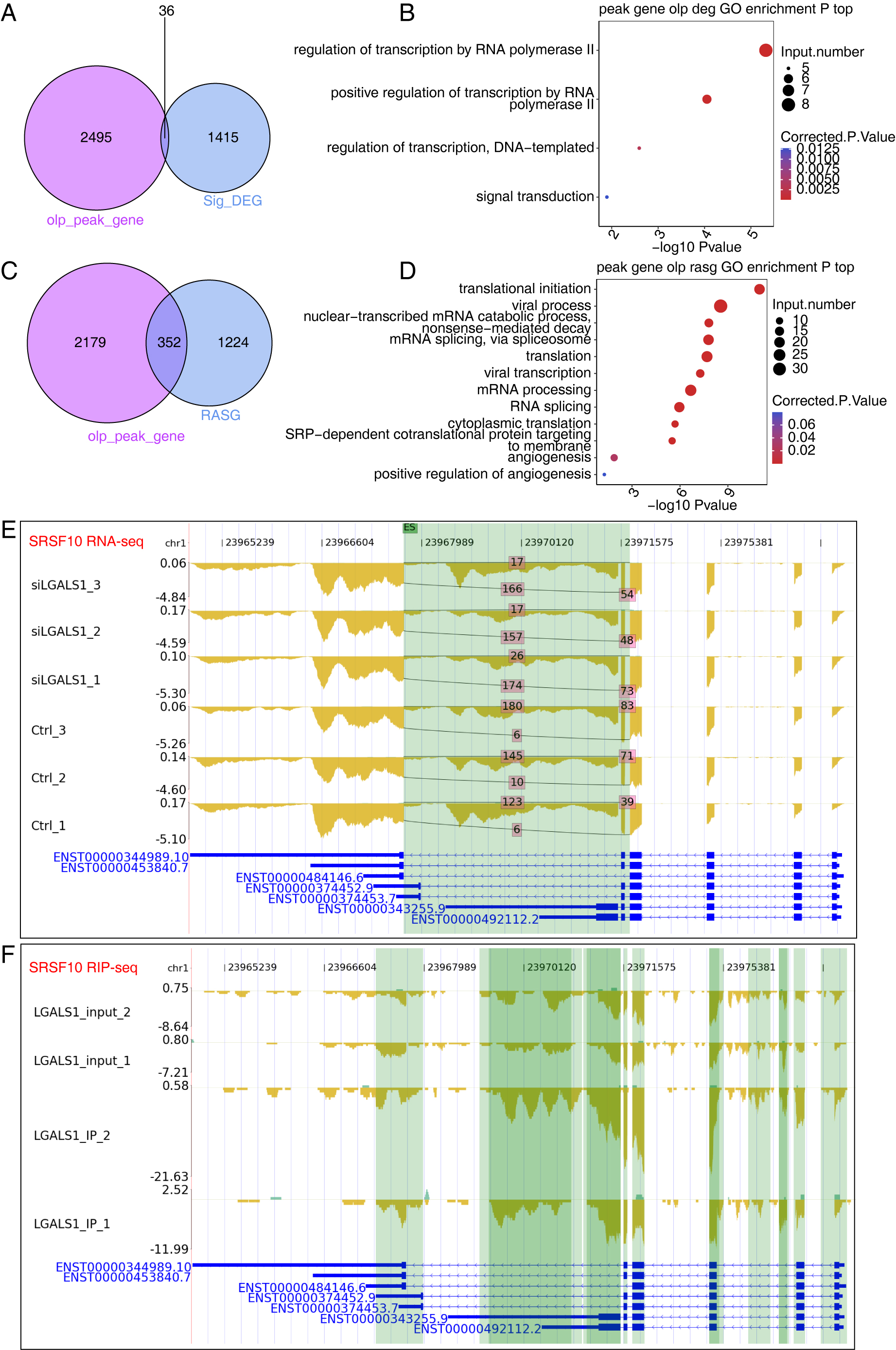
by galectin-1 from two replicates of iRIP-seq, we identified 36 DEGs that were
bound by galectin-1 (Fig. 5A). Although the overlap between DEGs and
galectin-1-bound genes was not significant, 36 genes were found to be enriched in
transcription regulation-associated pathways (Fig. 5B), including HR,
ECM1, AHR, FOSL1, ZNF860, CREBRF,
ATF6B, and SNAI1. These transcription-associated genes play
critical roles in gene expression, indicating that galectin-1 may regulate gene
expression profiles by affecting the expression level of these genes.
Furthermore, galectin-1 may directly influence galectin-1-bound genes by
modulating their AS patterns. The overlap analysis revealed that 352 RASGs were
bound by galectin-1 (Fig. 5C, p-value = 5.646611
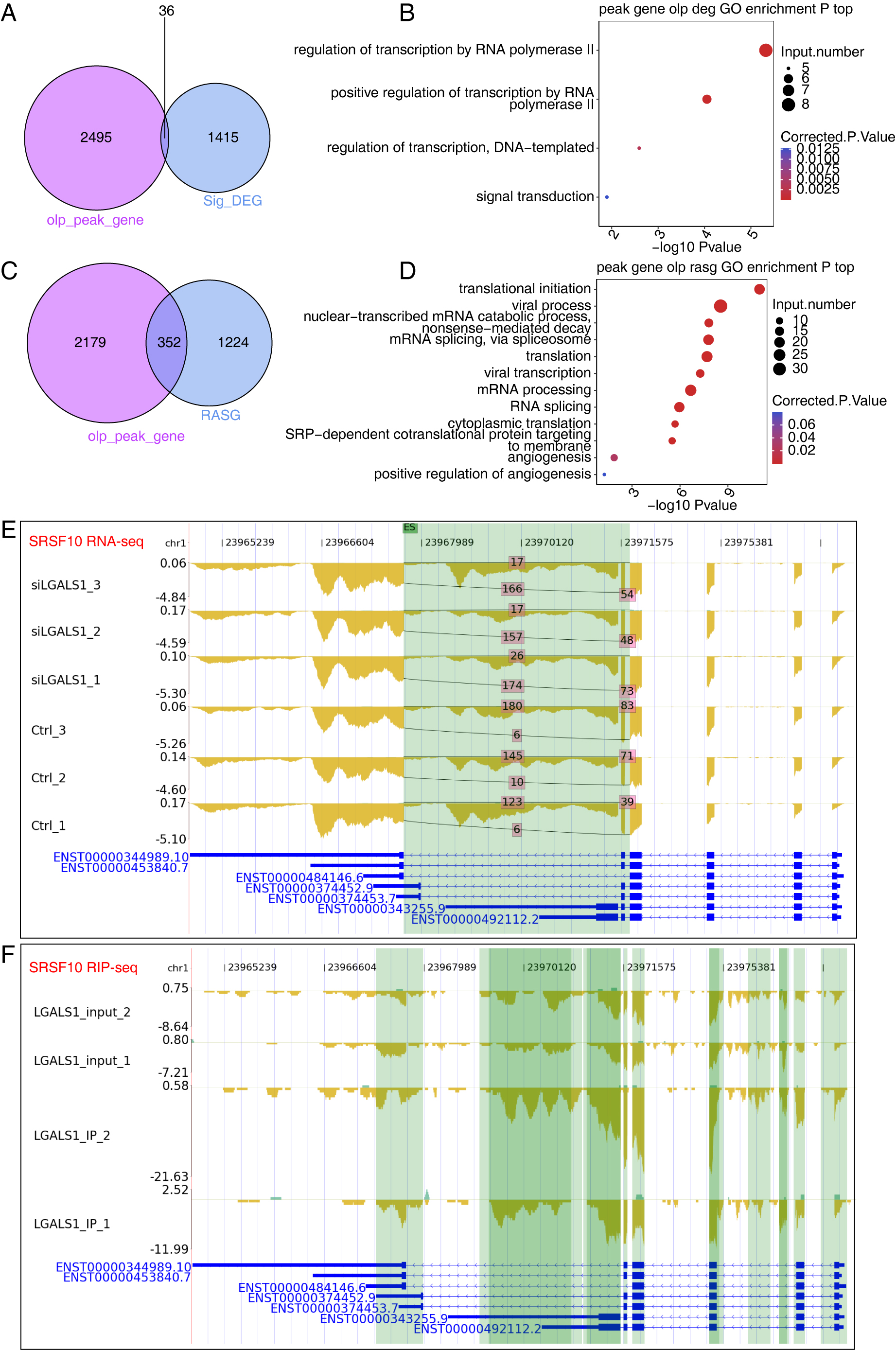 Fig. 5.
Fig. 5.Results of Integration Analysis Between the LGALS1-regulated Transcriptome and RNA Interactome. (A) Venn diagram showing the overlapping genes between DEGs and LGALS1-bound RNAs. (B) Bubble plot showing the functionally enriched BP pathways for the overlapping genes shown in (A). (C) Venn diagram showing the overlapping genes between RASGs and LGALS1-bound RNAs. (D) Bubble plot showing the functionally enriched BP pathways for the overlapping genes in (C). (E) Reads plot showing the AS pattern of SRSF10. The green background indicates the region of the AS event. (F) Reads plot showing the LGALS1-bound profile of SRSF10. The green background indicates the region of LGALS1-bound peaks.
Angiogenesis and neovascularization are often aberrantly regulated in many diseases, including various tumors that are characterized by abnormal angiogenesis [32]. Galectin-1 plays a critical role in tumor angiogenesis by acting as a receptor for the angiogenesis inhibitor anginex [9]. A previous study has discussed the important advances in the potential use of galectin-1 and galectin-3 for clinical therapeutic applications in multiple diseases [33]. To further understand the molecular mechanism that underlies the involvement of galectin-1 in angiogenesis, we systematically investigated how this protein regulates the transcriptome profile in HUVECs. We also investigated how galectin-1 can modulate gene expression and AS by interacting with associated RNAs. Galectin-1-regulated DEGs were found to be strongly associated with angiogenesis and inflammation pathways, and these pathways were consistently repressed after siLGALS1. Galectin-1-RASGs were also enriched in the VEGF signaling pathway. Together, these results indicate that galectin-1 can regulate angiogenesis by altering the global transcriptome profiles in HUVECs. This was partially demonstrated earlier using recombinant galectin-1 to treat pancreatic epithelial cells [34]. Another important finding is that galectin-1 may regulate transcriptome profiles by binding to RNAs. The co-regulated genes identified in both the RNA-seq and iRIP-seq datasets were also highly associated with the functions of galectin-1.
The transcriptome profile is orchestrated by multiple types of proteins, including transcription factors (TFs), epigenetic regulators, and the emerging RBPs [35]. Galectin-1 has been identified as an RBP, and its interaction with RNA was previously investigated by our group [16]. In the present work, we further explored the dysregulated DEGs and RASGs induced by siLGALS1 in HUVECs. The most notable finding was that many angiogenic factors were down-regulated after siLGALS1 treatment, implying that galectin-1 could influence angiogenesis by affecting the expression levels of angiogenic factors. The dysregulation of several of these factors, including AQP1, ACKR3, CALCRL, CCL2, CD34, ECM1, GJA5, HEY1, RAMP2, and SELP, was also validated by RT-qPCR experiments. Extracellular matrix protein 1 (ECM1) is expressed around blood vessels and is a possible angiogenesis trigger in cancer [36]. CCL2 is a chemokine ligand that can accelerate breast cancer metastasis by promoting angiogenesis [37]. The down-regulation of these genes is a hallmark of the angiogenesis process that is inhibited by siLGALS1 [16]. Interestingly, down-regulated DEGs were also enriched in inflammatory response pathways. Angiogenesis and inflammation have been linked and are known to co-occur in many inflammatory disorders [38, 39], suggesting that galectin-1 could also affect angiogenesis and participate in angiogenic disorders by influencing inflammatory response genes. In summary, the present study demonstrates a novel function for galectin-1 in the progression of angiogenesis by modulating the expression levels of associated genes, thus expanding our understanding of high galectin-1 expression in HUVECs.
AS greatly contributes to the proteome diversity of eukaryotes, and its dysregulation is known to cause various diseases [40]. The specific interaction of galectin-1 with thousands of RNAs endows it with the potential to regulate AS patterns. RNA-seq data from the present study confirms this hypothesis and indicates that galectin-1-regulated AS events showed a strong tendency for different AS types. More AS events with inclusion tendency were detected after siLGALS1 treatment, suggesting that galectin-1-regulated AS profiles may alter the functions of translated proteins. Functional analyses of RASGs revealed the VEGF signaling pathway was significantly enriched, including several important genes such as ADRM1, EGFL7, FGFR1, FLNC, SPHK1, MAPK3, FLT1, and MAPK14. Some of these genes act as growth factors that affect moyamoya disease, such as FLT1 (VEGFR1) and FGFR1 [41, 42]. Current evidence suggests that other genes in the VEGF signaling pathway, in addition to FLT1 and FGFR1, are also associated with angiogenesis and could potentially also regulate angiogenic disorders due to their important functions. Different mRNA isoforms of FLT1 (VEGFR1) are observed during gestation and these might be important for regulating VEGF activity [43]. VEGFA could also regulate the AS of VEGFR1 [44]. A previous study reported that AS of FGFR1 in high-grade bladder cancer induced cell proliferation and MAPK pathway activation [45]. These results suggest that altered isoforms of genes in the VEGF signaling pathway may also have altered molecular functions, thus contributing to angiogenesis. Another interesting finding was that the mRNA expression level of several alternatively spliced genes was also altered by siLGALS1. AS can regulate gene expression by affecting mRNA stability or translational efficiency [46]. A recent study demonstrated that antisense oligonucleotide-targeted splicing events could alter mRNA and protein levels [47]. Our findings indicate that galectin-1 may indirectly influence gene expression by modulating the AS patterns of those genes. However, further studies are needed to elucidate the underlying mechanisms, especially for the angiogenic factors in the VEGF signaling pathway.
Finally, we conducted an overlap analysis between the galectin-1-regulated transcriptome and galectin-1-bound RNA interactome. A large number of overlapping DEGs and RASGs were identified. The latter was statistically significant, indicating that galectin-1 may influence gene expression and AS by binding to transcripts and modulating their RNA stability or splicing patterns. The galectin-1-bound RNAs extracted from HeLa cells [16] account for only a small percentage of the galectin-1-regulated RNAs identified in HUVECs. Further experiments are needed to validate the galectin-1-RNA interactions in HUVECs. Overlapping DEGs were found to be enriched in transcription regulation pathways, while overlapping RASGs were enriched in RNA splicing and translation pathways. A previous study reported that EIF2AK2 selectively regulates immune response-related gene expression by affecting TFs [48]. Another study demonstrated that KRT18 regulates AS profiles by modulating the splicing patterns of key splicing factors [49]. Based on the observation that a large number of DEGs and RASGs were not bound by galectin-1, we propose that galectin-1 may indirectly modulate gene expression and AS by regulating TFs and splicing factors. Such an amplified regulatory mechanism could partly explain how galectin-1 regulates changes in the global transcriptome profile of HUVECs, while providing valuable information to better understand the molecular mechanism of galectin-1 action.
In summary, we comprehensively explored the galectin-1-related transcriptome profiles in HUVECs by using siLGALS1. We also used integrated RNA interactome data to investigate the underlying molecular mechanisms of angiogenesis. The study of angiogenic factor expression and splicing patterns revealed a strong association between the galectin-1-regulated transcriptome and angiogenesis. This probably occurs directly via regulation of galectin-1-bound RNAs, or indirectly by modulating angiogenic factors through TFs and splicing factors. Our study sheds light on the molecular functions of galectin-1 as an RBP, and suggests potential therapeutic target genes for angiogenesis and relevant disorders.
LGALS1, Lectin, galactoside-binding, soluble, 1; siLGALS1, Silencing LGALS1; HUVECs, Human umbilical vein endothelial cells; RNA-seq, RNA sequencing; DEGs, Differentially expressed genes; AS, Alternative splicing; RASGs, Regulated alternative splicing genes; VEGFs, Vascular endothelial growth factors; bFGF, Basic fibroblast growth factor; TGFB1, Transforming growth factor-beta1; RNF213, Ring finger protein 213; RBP, RNA binding protein; FDR, False discovery rate; BP, Biological process; GO, Gene ontology; ASEs, Alternative splicing events; RASEs, Regulated alternative splicing events; ES, Exon skipping; A5SS, Alternative 5’ splice site; A3SS, Alternative 3’ splice site; MXE, Mutually exclusive exons; 5pMXE, Mutually exclusive 5’UTRs; 3pMXE, Mutually exclusive 3’UTRs.
The datasets generated and/or analyzed during the current study are available in the NCBI GEO repository with accession number GSE201537.
The experiments were designed by JW and DC. JW and YS collected the samples and conducted the RNA extraction, library preparation, and sequencing. YW performed the bioinformatics analysis; JW wrote the manuscript with essential contributions from DC. All authors revised and approved the final manuscript.
Not applicable.
We would like to express our gratitude to the staff members from ABLife BioBigData Institute for their helpful discussion.
This research received no external funding.
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.