1 Department of Radiation Oncology, Shanghai Ninth People's Hospital, Shanghai Jiaotong University School of Medicine, 200011 Shanghai, China
2 Department of Breast Oncology, Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, 510060 Guangzhou, Guangdong, China
3 Disinfection Supply Center, Shanghai Ninth People's Hospital, Shanghai Jiaotong University School of Medicine, 200011 Shanghai, China
†These authors contributed equally.
Abstract
Background: Breast cancer poses severe threats to human health as radioresistance becomes increasingly prevalent. The mechanisms of radioresistance are hard to expound completely. This study aims to explore proteomic changes of radioresistance, which will help elucidate the potential mechanisms responsible for breast cancer radioresistance and explore potential therapeutic targets. Methods: A radioresistant breast cancer cell line was established by repeated irradiation. Liquid Chromatograph Mass Spectrometer (LC–MS) was used to quantify protein expression. Proteomic changes associated with radioresistance were evaluated by proteomic analysis. Further, cell radioresistance and several identified proteins were verified in in vitro experiments. Results: In the study, more than 3000 proteins were detected, 243 of which were identified as up-regulated proteins and another 633 as down-regulated proteins. Gene Ontology (GO) enrichment analysis indicated that these proteins were mainly expressed in the lysosome and ribosome, associated with coenzyme binding and the structural constituent of the ribosome, involved in mitotic cytokinesis and ribonucleoprotein complex biogenesis. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis indicated that many biological processes were extensively altered, particularly spliceosome and thermogenesis. It is worth noting that the functions and pathways related to ribosomes were significantly enriched, therefore ribosomal proteins (RPL6 and RPS13) were identified through western blot and highly expressed in relatively radiosensitive cells. Additionally, several identified proteins, including S100A4, RanBP9, and ISG15, were also verified to be differentially expressed in different radiosensitive cells. Conclusions: Our results provide a framework for further studies into the mechanisms of radioresistance and serve as a basis to construct a predictive model of radioresistance in breast cancer. Ribosome may participate in the radioresistance of breast cancer, which provides new insights into the proteomic characteristics of the mechanisms of radioresistance.
Keywords
- breast cancer
- radioresistance
- bioinformatics analysis
- proteomic analysis
- ribosome
Breast cancer (BC), the most prevalent female cancer accounting for nearly 1/3 of cancers diagnosed in this population [1], is now widely considered as the leading cause of cancer death among females [2]. The BC mortality rate for females of all ages ranges from 0.4 to 3.4/100 per year due to the differences in the degree of economic development and associated social and lifestyle factors [2, 3], and continues to decrease year-on-year. This decrease has been attributed to the increased use of screening mammography combined with improved BC treatment [4, 5]. Among them, radiotherapy is one of the mainstays in the management of BC, which is either added to mastectomy to eradicate residual subclinical sections or used as a palliative treatment for advanced BC patients without surgical indication [6, 7].
Radiotherapy, a type of ionizing radiation, induces DNA damage directly via ionization or indirectly by the generation of reactive oxygen species (ROS), thereby destroying tumor cells [8, 9]. Radiation dose and frequency of radiotherapy depend principally on the balance between cure and toxicity of treatment [10]. The recommended dose of radiation for each individual treated by radiotherapy alone is 50–70 Gy in 25 to 35 fractions or equivalent [11, 12, 13]. However, a large amount of evidence demonstrated the presence of radioresistance in BC [14, 15, 16, 17, 18]. In addition to the inherent radioresistance of cells, an adaptive response of cancer cells to repeated radiation leads to the development of radioresistance, including alterations in the tumor microenvironment [19], signaling [20], and metabolic [21] pathways. Currently, radioresistance has become a major obstacle to successful cancer therapy, accompanied by high mortality (14.5–22.5%) and recurrence rate (2.4–11.5%) [22, 23, 24]. The majority of previous studies involving radioresistance in BC focused on ROS-DNA damage. DNA damage promotes DNA damage repair by activating various signal pathways, thus resulting in radioresistance. Recently, we established a radioresistant BC cell line by repeated ionizing radiation and found that radioresistant BC cells present elevated expression of ataxia-telangiectasia mutated kinase (ATM) and increased DNA damage repair efficiency, indicating a potential link between ATM, DNA repair pathway, and radioresistance [25]. Additionally, several molecules such as SERPINE1 (serpin family E member 1) [26], YB-1 (Y-box binding protein 1) [27, 28], TTK (TTK protein kinase) [29] and ARID1A (AT-rich interaction domain 1A) [30] have also been reported to contribute to radioresistance.
The majority of previous research has focused at the genomics level, which has inevitable limitations and caveats, including RNA degradation, improper transcription, and inability to analyze the post-translation modification and cellular functions [31, 32]. As the structural and functional elements of cells, protein is regarded as the ultimate executor and effector of genomic biological information [33]. Qualitative and quantitative analysis of proteins will help to explore the basis and conditions for the realization of cell biological functions. Furthermore, a single protein only provides a limited understanding of the biological pathways [34], and systematic large-scale data analysis is essential for expanding our understanding of pathological mechanisms. Therefore, the concept of proteomics was urgently introduced in 1994 by Marc Wilkins [35]. Proteomics is a complete and independent collection involving biological characteristics and activities, covering all the proteins expressed in the genome [36, 37, 38]. Quantitative proteomics provides the difference in protein abundance between healthy samples and tumor samples, as well as relevant information, including protein-protein interactions, signaling pathways, disease prediction, and phenotypic classification [39]. However, due to the limitations of techniques and data analysis methods, the course of proteomics progression in BC seems to be slow and remains in the initial stage [34, 40, 41, 42], and protein detection coverage ranges from hundreds to thousands [43, 44, 45]. Most importantly, it is worth mentioning that previous proteomics studies always adopted the same but imprecise screening criteria for radioresistance-associated proteins, namely the differential proteins between parental cells and radioresistant cells. Thus, it is necessary to conduct a large-scale and systematic study via strict screening criteria to explore the complex radioresistant mechanism.
Herein, using our established radioresistant cells through repeated irradiation [25], and Liquid Chromatograph Mass Spectrometer (LC–MS) to cover more than 3000 proteins, we identified the differential proteins among the radioresistance-related cells lines using a strict triple screening criteria and conducted function and pathway enrichment analysis, thereby facilitating the exploration of radioresistance mechanisms. The identification of novel proteins [46] associated with radioresistance in BC would not only increase our understanding of the molecular mechanisms underlying the therapeutic resistance but also provide tumor molecular characteristics, promote the refinement of BC subtypes, and contribute to the diagnosis, prognosis, and individualized treatment.
All cell lines were authenticated using the short tandem repeat (STR) technique. Additionally, mycoplasma testing was conducted on the cell lines, and the result was negative. Human BC cell lines (MDA-MB-231) were purchased from ATCC, and human pancreatic
cancer cell lines (PANC1 and MIA-PACA2) were obtained from Cell Bank of Type
Culture Collection of Chinese Academy of Sciences (Shanghai, China), grown in
Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% FBS (Gibco,
Loughborough, England) and cultured in a 37 °C incubator containing 95%
O
For the establishment of the radioresistant cell line, as described [25], MDA-MB-231 cells, set to parental BC cell lines, were divided into two groups, PB and PR cells. PR cells were irradiated with 3 Gray (Gy) of X-rays 20 times at a dose rate of 1.43 Gy per minute, with a total dose of 60 Gy over 4 months as radioresistant cell lines [47]. PB cells were treated using the same conditions without irradiation, and used as control cell lines. Compared with PB cells, PR cells had a more stretched and flatter appearance with enhanced anti-apoptotic, migration, and invasion capabilities and presented elevated surviving fraction, reduced percentage of apoptotic cells, and increased DNA damage repair efficiency.
PB cells (hereafter named group A) and PR cells (group B) were collected when
the cells reached approximately 90% in 10 cm culture disks. Additionally, PB and
PR were irradiated with 10 Gy at a dose rate of 1.43 Gy per minute [48, 49] and
collected after 24 hours (group C and group D, respectively). A total of 3
biological replicates were collected. Supplementary Fig. 1 shows the
grouping details. The cells were washed 3 times with ice-cold phosphate buffer
saline (PBS). A total of 1 mL 10M urea was added and incubated for 5 min. Then,
the lysates were sonicated for 5 min. After centrifugation (4 °C, 13,000
g
Proteins were quantified using the modified LC–MS/MS (Agilent 1290, Agilent
Technologies Inc. Santa Clara, CA, USA; QTRAP 4500, AB Sciex Inc. Foster City,
CA, USA) analytical method as described previously [50]. The analysts were
blinded to all the information during testing. The C18 column is balanced with
95% solvent A (0.1% formic acid (FA), H
The scanning parameters were set as follows: DDA acquisition mode, TOP20. The resolution of primary MS was 70,000, the maximum injection time was 50 ms, the fragmentation mode was HCD, and the collision energy of 27% was used for fragmentation; the resolution of secondary MS was 18,000, the maximum injection time was 100 ms, the signal threshold was 1e4 ions/s, and the dynamic exclusion time was 30 s.
Up-regulated proteins were defined based on the following conditions: (1) higher expression in group B than that in group A, (2) increased in group D compared with group B, (3) increased in group C compared with group A. Down-regulated proteins were defined based on the following conditions: (1) lower expression in group B than that in group A, (2) decreased in group D compared with group B, (3) declined in group C than that in group A. A schematic is shown in Supplementary Fig. 2.
Single-cell suspensions were added to 6-well plates in triplicate. After 24 hours, cells were irradiated with 0–6 Gy at a dose rate of 1.43 Gy per minute and then cultured for 14 days. Cell clusters containing over 50 cells were regarded as colonies. The colonies were fixed with 4% paraformaldehyde, stained with crystal violet, and dried naturally. Plating efficiency (PE) = [the mean number of colonies/the number of inoculated cells]. Surviving fraction (SF) was calculated as follows. SF (d) = [the mean PE (d)]/[the mean PE (0)], while d indicates dGy and 0 means 0Gy. The curve was plotted using the linear-quadratic model in Graph Prism 6.01 (GraphPad Software Inc., San Diego, CA, USA) as described previously [51, 52].
Cells were washed by pre-cooling PBS three times, and proteins were extracted in
100 uL lysis buffer containing protease and phosphatase inhibitor cocktail (MCE,
Monmouth Junction, NJ, USA). Then equal amounts of proteins (5 uL) were
size-separated in 10% SDS-PAGE gels and transferred to PVDF membranes.
Immunoreactive proteins were visualized using ECL following the manufacturer’s
instructions (Millipore, Boston, MA, USA). The detailed information for primary
antibodies used in this study was provided as follows: Rabbit polyclonal
antibodies against S100A4 (ABClonal, Wuhan, China, #A19109, 1:1000
dilution), RANBP9 (ABClonal, #A19238, 1:1000 dilution), ISG15
(ABClonal, #A1182, 1:1000 dilution), RPL6 (ABClonal, #A15094, 1:1000
dilution), RPS13 (ABClonal, #A15720, 1:1000 dilution),
Kaplan–Meier Plotter (https://www.kmplot.com) was applied to evaluate the prognosis of BC by selecting the mRNA as a dataset. The relationships between S100A4, RanBP9, ISG15, RPL6, and RPS13 expression with overall survival (OS) of different time points were analyzed with the hazard ratio (HR) with 95% confidence intervals (CIs) and log-rank p-value. “Auto select best cutoff” was set as a method to split patients.
The LC–MS/MS raw files were searched and quantified by MaxQuant (version
1.6.2.0; Max Planck Institute of Biochemistry, Munich, Bavaria Germany) against
UniProt Protein Database (downloaded on 22 April 2021). The enzyme was set by
trypsin with up to two missed cleavages; the main research is 4.5 ppm; The
fragment mass tolerance is 0.02 Da; The peptide and protein FDR is 0.01; Unique
peptides (at least 2 unique peptides) were used for protein quantification. All
data were log2 transformed before analysis. The t-test was employed to
identify significantly differentially expressed proteins between two independent
groups (three biological repeats in each group). According to the
1.5-
In this study, quantitative proteomics of radioresistant cells and
radiosensitive cells enabled the comparison of protein expression profiles in
radiotherapy. Of the 3000+ proteins detected in this study, 876 significant
proteins were identified among four groups. From the distribution of the proteins
as shown by the Venn diagram, 243 up-regulated proteins were screened (Fig. 1A,
Supplementary Table 1), and 633 down-regulated differential proteins
were found (Fig. 1B, Supplementary Table 2) according to the
1.5-
 Fig. 1.
Fig. 1.Protein identification and quantification in three comparison groups. The Venn diagram shows the number of proteins identified in group A vs. C only, group A vs. B only, group B vs. D only, and those shared among three comparison groups. (A) Up-regulated proteins. (B) Down-regulated proteins.
Although the general relationships of proteins from four groups were visualized using the Venn diagram, it was necessary to consider differences at the concentration level graphically. Heat map analyses, constructed from LFQ intensities, gave a significant overall picture of the up-and down-regulated proteins in each of the samples. The rows (identified proteins) were rearranged according to similar profiles. The columns (samples) are sorted by sample type, and each sample type in the data matrix is displayed as a color (purple, green, pink, and blue). As shown in Fig. 2, according to the color distribution of the heat map, it can be inferred that the greatest difference in protein expression was between group D and group A. The difference may be attributed to radiotherapy dose and frequency, namely the presence of radioresistance and radiosensitivity.
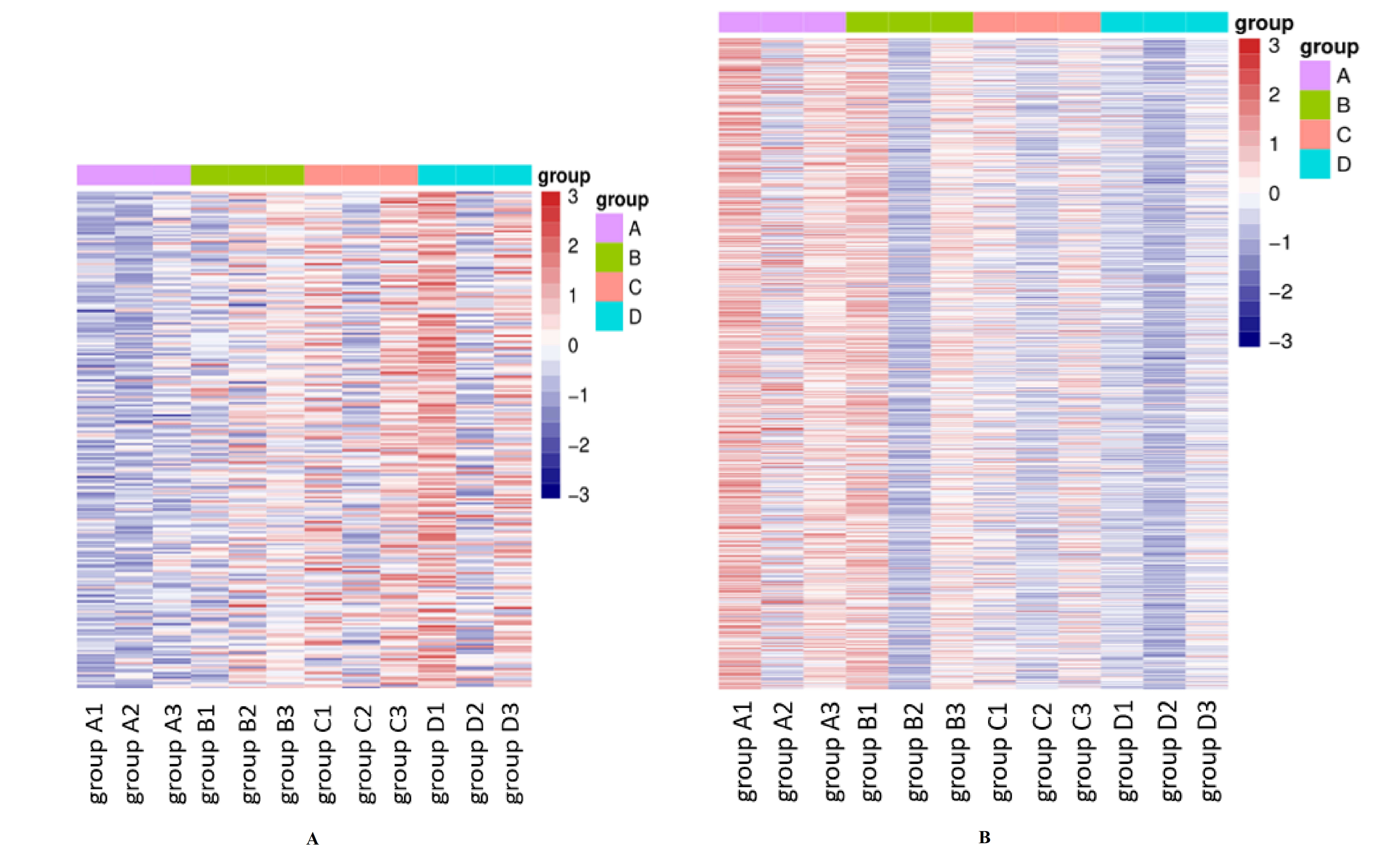 Fig. 2.
Fig. 2.Heatmap visualization of the significantly different proteins among four groups. Columns: samples; Rows: differential proteins. Color key indicates protein expression value: blue, lowest; red, highest. (A) Up-regulated proteins. (B) Down-regulated proteins.
To evaluate the biological and functional associations of up-and down-regulated proteins, we annotated these proteins based on the GO databases. As shown in Fig. 3A, 243 up-regulated proteins were classified by the second level (biological process, cellular component, and molecular function). Biological process analysis showed these significant proteins participate in RNA splicing and mitotic cytokinesis. Cellular component GO terms significantly enriched in the up-regulated proteins were azurophil granule and primary lysosome. The molecule function is cadherin binding and coenzyme binding. Additionally, down-regulated proteins were enriched in the ribosome and mitochondrial matrix, involved in the ribonucleoprotein complex biogenesis and ribosome biogenesis, also associated with a structural constituent of ribosome and translation initiation factor activity (Fig. 3B). Interestingly, GO analysis of down-regulated proteins has revealed a highly significant correlation with the ribosome, including ribosomal subunit, organellar ribosome, ribosome biogenesis, and other ribosomal processes. It has been conceivably supposed that ribosome may be significant in the pathogenesis of radioresistance.
 Fig. 3.
Fig. 3.Gene ontology (GO) classifications of up-regulated proteins (A) and down-regulated proteins (B). The figure shows the top 10 GO terms. Horizontal coordinate: GO enrichment value; vertical coordinate: GO term. BP, biological process; CC, cellular component; MF, molecular function.
Despite the advances in GO annotations, it is necessary to further annotate
proteins more comprehensively and precisely. Thus, the directed acyclic graph
(DAG) showed the hierarchical relationship of significantly enriched GO terms,
including up- and down-regulated proteins, where hierarchy was defined with
respect to the available terms associated with the proteins. As DAG is shown in
Fig. 4, from top to bottom, there were two main paths of GO terms in up-regulated
proteins (the number of GO items
 Fig. 4.
Fig. 4.Pathway analysis of up-regulated proteins using directed acyclic graph (DAG). Boxes were used to indicate the top 10 terms with high salience, and the figure shows the corresponding relationship of each layer. Different colors represent different enrichment salience: the redder color, the higher salience.
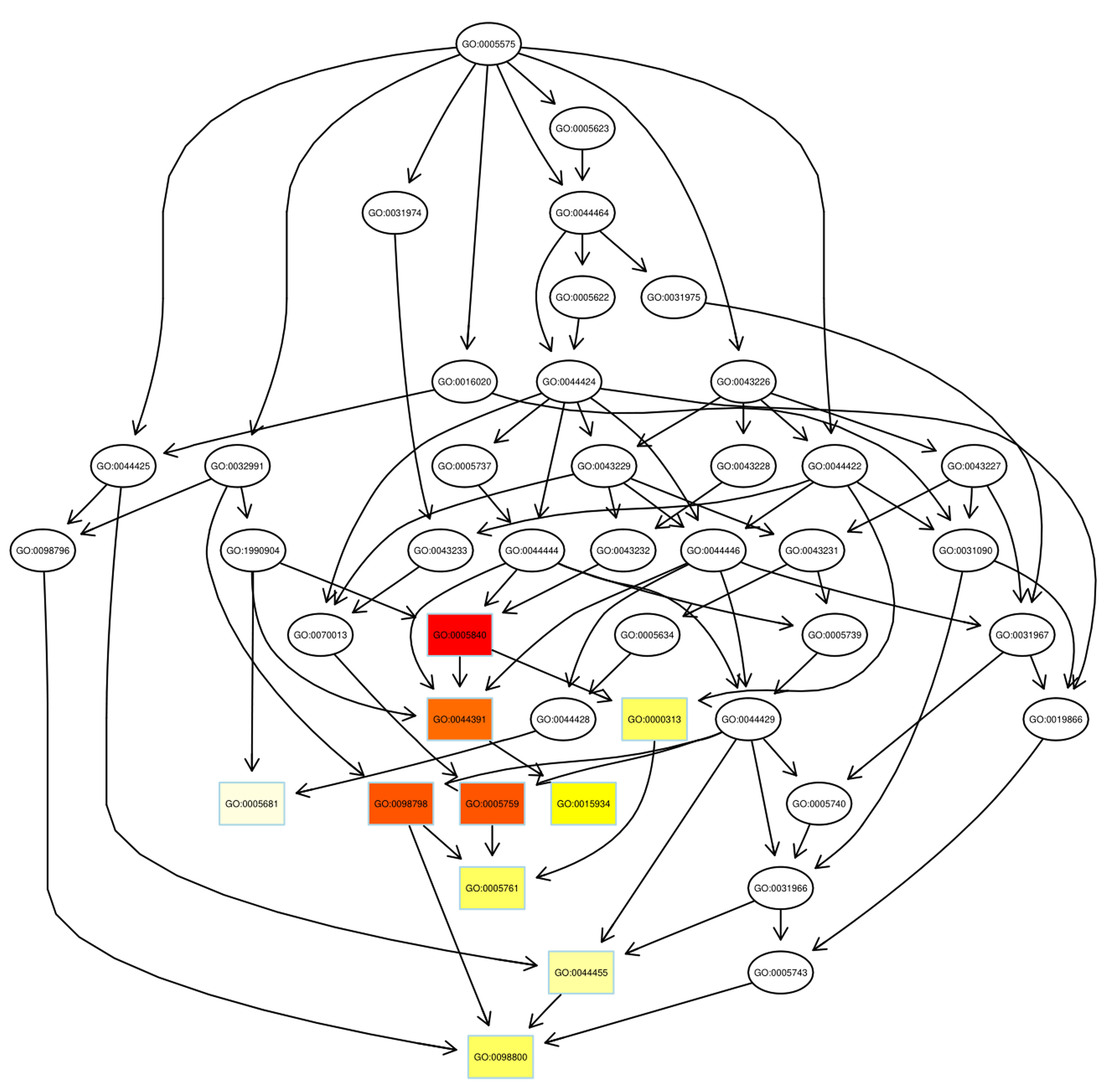 Fig. 5.
Fig. 5.Pathway analysis of down-regulated proteins using DAG. Boxes were used to indicate the top 10 terms with high salience, and the figure shows the corresponding relationship of each layer. Different colors represent different enrichment salience: the redder color, the higher salience.
As illustrated, the azurophil granule membrane (GO:0035577) and inner mitochondrial membrane protein complex (GO: 0098800) were the targets of action of up-regulated proteins and down-regulated proteins, respectively. Similar to GO annotations, ribosome (GO: 0005840) and mitochondrial ribosome (GO: 0005761) were up-regulated in the radiosensitivity of BC.
To further evaluate the biological significance and investigate the disturbed signaling pathways of these proteins, the association network of differentially expressed proteins was constructed. As depicted in Fig. 6A, the most significantly enriched KEGG pathway in up-regulated proteins was the spliceosome. In addition, thermogenesis was the most important metabolic pathway related to down-regulated proteins (Fig. 6B). In the pathway analysis, several significant pathways were highly enriched, including spliceosome, thermogenesis, Parkinson’s disease, and oxidative phosphorylation. Among these pathways, spliceosome [55] and oxidative phosphorylation [56, 57] have been reported to be linked to radioresistance. It is noteworthy that ribosome was strongly enriched among differentially expressed proteins, consistent with GO annotations, further confirming the importance of ribosome for irradiation-induced radiosensitivity/radioresistance.
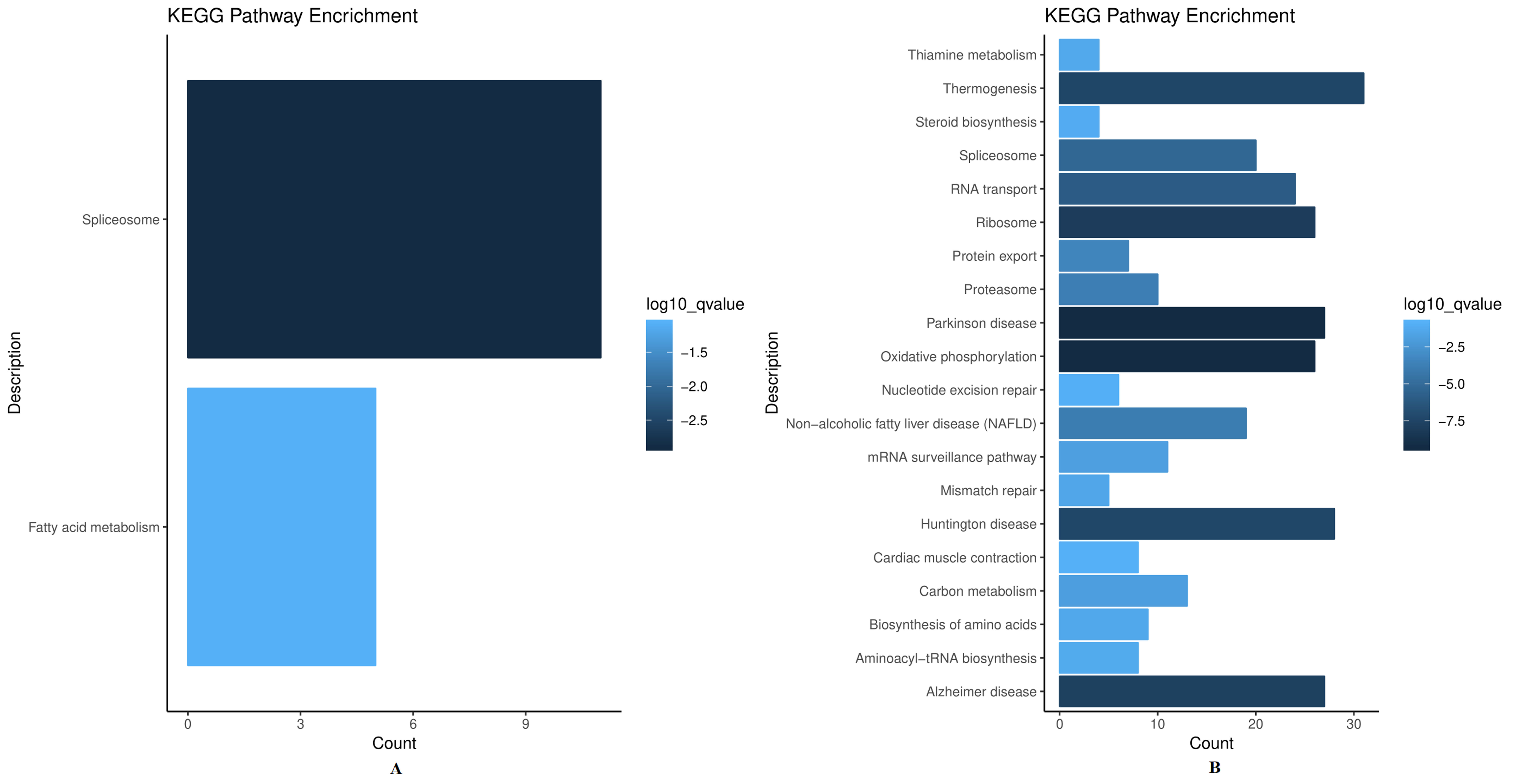 Fig. 6.
Fig. 6.Significantly enriched pathways from Kyoto Encyclopedia of Genes and
Genomes (KEGG). The figure
shows the top 20 KEGG pathways (p
The radioresistance of PB cells and PR cells was compared by surviving fraction (SF). As shown in Fig. 7A, the PR cell line had a higher survival rate than that of PB cells. Next, we randomly selected candidate proteins for validation. S100A4 (S100 calcium-binding protein A4), RanBP9 (RAN binding protein 9), ISG15 (Interferon-induced gene 15), RPL6 (ribosomal protein L6), and RPS13 (ribosomal protein S13), which were found differentially expressed in our study, were verified with immunoblotting. Results of western blotting (Fig. 7B) showed that RanBP9, RPL6, and RPS13 in PR cells were lower than that of PB cells, while S100A4 and ISG15 in PR cells were higher than that in PB cells. What is noteworthy is that RPL6 [58, 59] and RPS13 [60, 61] have been reported to influence multidrug sensitivity in gastric cancer cells, similar to our finding that ribosomal proteins affect radiosensitivity in BC cells.
 Fig. 7.
Fig. 7.Surviving fraction in PB and PR cells and validation of
differential proteins. (A) Surviving fraction of PB and PR cells. Representative
images (left panel) and the quantitative analysis of three independent
experiments (right panel) are shown, respectively. (B) Western blot of
S100A4, RanBP9, ISG15, RPL6, and
RPS13 in PB and PR cells.
Next, the impact of S100A4, RanBP9, ISG15,
RPL6, and RPS13 expression on prognosis in BC patients was
evaluated (Supplementary Fig. 3). BC patients with higher ISG15
or lower RPS13 expression had significantly shorter overall survival
than those with lower ISG15 or higher RPS13 expression
(p
Further, in order to verify the universality of our findings, we tested the correlation between the expression of ISG15 and the radioresistance in human pancreatic cancer cell lines (PANC1 and PACA2) (Supplementary Fig. 4). Similar to the results of PR and PB cells, the expression of ISG15 in PACA2 cells is higher than that in PANC1, whereas the SF of PACA2 is higher than that of PANC1 (Supplementary Fig. 2).
Here, we performed a label-free quantitative proteomics analysis of radiosensitive and radioresistant cells to investigate radioresistance-related proteins.
Among 876 differentially expressed proteins, some proteins have been reported to be related to drug resistance in previous studies, such as S100A4 [62, 63, 64], RanBP9 [65, 66] and ISG15 [67, 68], which are consistent with WB verification in our study. Other differentially expressed proteins, such as LARP4B, CAPRIN1, and PLOD3, need further expression and functional confirmation. In the future, clinically relevant models can be constructed based on these identified proteins to forecast the radioresistance of patients as a precision medicine tool.
In our study, both enrichment analysis and WB results indicated a possible role of ribosomes in the development of radioresisitance. Indeed, evidence has emerged in recent decades regarding the close link between ribosome and therapeutic resistance in cancers.
Ribosome is a complex of ribosomal RNA (rRNA) and ribosomal proteins [69], which synthesize proteins in living cells. Ribosome biogenesis and processing are under surveillance of multiple checkpoints and pathways [70]. As an essential component of the 50S subunit of ribosomes, RPL6 is a highly conserved ribosome protein [71]. Previous studies have shown that RPL6 can up-regulate Bcl-2 and down-regulate Bax [59, 72]; Inhibition of RPL6 inhibits the transition from G1 to S phase [58, 73, 74]; Decreased RPL6 can reduce ubiquitin-dependent peptide presentation and trigger tumor immune escape [75]. Moreover, it has been suggested that RPL6 can be recruited to DNA damage sites and regulate DNA damage response in a poly (ADP-ribose) polymerase-dependent manner [76, 77]. Evidently, RPL6 can inhibit tumor cell apoptosis and increase therapy resistance through the above mechanisms.
RPS13 is a common and stable protein [78], which is primarily distributed in the cytoplasm and associated with rRNA in the large and small subunits of the ribosome. The knock down of RPS13 led to G1 arrest [60]; RPS13 increased the expression level of Bcl-2 and Bcl-2/Bax ratio and induced drug resistance of gastric cancer cells to a certain extent [61].
Under radiation stimulation, DNA is damaged and then activates the DNA damage reaction (DDR) [79]. According to the extent of damage, DDR will develop in the direction of repairing DNA damage to maintain cell survival or activating programmed cell death pathways to accelerate cell death [80]. Among them, the high efficiency of DNA damage repair repairs radiation-induced DNA damage, leading to resistance to apoptosis [81], which is the main cause of radioresistance [82]; in the process, ribosomal proteins (such as RPL6 and RPS13) play an important role in the regulation of cell cycle and apoptosis (mentioned above), thus affecting the repair efficiency. Apart from ribosomal proteins, associated ribosome biogenesis factors, such as Bop1 [83], could increase cell recovery by DNA repair regulation, whereas nucleolin [84] played the opposite role.
Most studies involving ribosomes focus on the correlation between gastrointestinal cancer and drug resistance, and there are very few studies involving radioresistance and BC [14, 85]. In our study, radioresistance-related proteins were screened completely and systematically by proteomic analysis, and the findings emphasized the crucial role of ribosomes in radioresistance. Therefore, follow-up research could focus on the association between ribosome and radioresistance, aiming at providing new therapeutic targets for BC.
In addition to ribosomes, KEGG pathway enrichment showed that spliceosome, thermogenesis, and oxidative phosphorylation have a potential correlation with radioresistance. Previous studies have reported that genes involved in spliceosome assembly were significantly up-regulated in BC [86]. Scholars speculated that the spliceosome may play an important mediation role in the tumor microenvironment via cell signal transduction and gene expression regulation [87]. Thermogenesis and oxidative phosphorylation are highly regarded metabolic pathways. As an important part of the tumor microenvironment, thermogenesis can provide an energy source [88], in turn promoting tumor progress [89, 90]. Oxidative phosphorylation is over-active in many cancers [91] and aggravates the carcinogenic behaviors by the hypoxia pathway [92, 93, 94], thereby maintaining drug resistance and resulting in poor prognosis [95, 96].
The findings of our study not only provide a reliable theoretical basis for exploring the mechanism of radioresistance but also provide a data set of radiosensitivity-related proteins for building an evaluation model of individual radiosensitivity. Each individual’s radiosensitivity is varied and unpredictable [97]. Therefore, it is necessary to establish a model to evaluate the individual’s radiosensitivity so as to formulate a more appropriate radiotherapy strategy and obtain the maximum therapeutic benefit with minimal toxicity. In addition, there are already a variety of BC proteins and transcripts as biomarkers. If the differentially expressed protein screened in the study is used as a new biomarker, it must be tested in validation studies with clinical samples in advance.
It is worth mentioning that strict screening criteria were used in the study. Most mass spectrometry analyses only detected the differentially expressed proteins between susceptible and tolerant cell lines and then defined these proteins as sensitivity or resistance-related proteins. However, during the culture process, the cells inevitably produce radiotherapy-independent proteins. Therefore, we included radiation factors (i.e., irradiated groups C and D) to further screen sensitivity or resistance-related proteins.
However, as the biological functions of ribosomes on radiosensitivity remain largely unexplained, future studies are warranted to confirm our findings. Our study was also restricted to BC cell lines. Therefore, caution is needed if generalizing the findings to other cancers and replication research in diverse cancer cells is warranted.
In conclusion, our study screened potential radioresistance-related proteins, which provide a rationale for further investigation of radioresistance and subserve to develop an appropriate predictive model for the evaluation of radioresistance in BC. Moreover, the interaction of ribosomes with radioresistance may serve as a promising therapeutic strategy. However, a detailed delineation of the mechanism of modulating BC radioresistance awaits future experimentation.
In conclusion, our study screened potential radioresistance-related proteins, which provide a rationale for further investigation of radioresistance and subserve to develop an appropriate predictive model for the evaluation of radioresistance in BC. Moreover, the interaction of ribosomes with radioresistance may serve as a promising therapeutic strategy. However, a detailed delineation of the mechanism of modulating BC radioresistance awaits future experimentation.
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Study design and concept: DL, YXY, LB, YY. Data acquisition: YXY, LB, YLM, MCZ. Data analysis and interpretation: YXY, LB, YLM, MCZ, FB. Manuscript preparation: DL, YXY, YLM, FB, YY. Manuscript review: YXY, DL, FB, YY. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work to take public responsibility for appropriate portions of the content and agreed to be accountable for all aspects of the work in ensuring that questions related to its accuracy or integrity.
Not applicable.
Not applicable.
This work was supported by the National Natural Science Foundation of China under Grant 81970094; The Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning under TP2015022; Shanghai Pujiang Program under Grant 15PJ1404800.
The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/j.fbl2810244.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.







