1 Department of General Medicine, Tongde Hospital of Zhejiang Province, 310000 Hangzhou, Zhejiang, China
2 Department of Acupuncture and Massage, Tongde Hospital of Zhejiang Province, 310000 Hangzhou, Zhejiang, China
Abstract
This Mendelian randomization (MR) study systematically examines the causal links between skin disorders and depression in individuals of East Asian descent.
MR analysis employed summary-level genome-wide association study (GWAS) data from East Asian populations. Exposures included six skin diseases: atopic dermatitis (AD) (n = 168,103), urticaria (n = 172,083), vitiligo (n = 13,327), systemic lupus erythematosus (SLE) (n = 51,009), psoriasis (n = 69,688) and acne (n = 2062). Depression was assessed using major depressive disorder (MDD) data from the Psychiatric Genomics Consortium (n = 194,548). The primary analytical methods were the inverse variance weighting (IVW) and Wald Ratio. Sensitivity analyses were conducted to detect heterogeneity and pleiotropy, incorporating Steiger tests to mitigate reverse causation.
In East Asian ancestries, a significant causal relationship was identified between urticaria and an increased risk of MDD (odds ratio [OR] = 1.220, 95% CI 1.022–1.457, p = 0.028). No significant causal link was found between psoriasis and MDD. Both findings are in stark contrast to those from previous MR studies of European ancestries. No significant causal associations were observed between AD, vitiligo, SLE, acne and MDD, consistent with previous MR studies in European populations. Sensitivity analyses revealed no significant evidence of heterogeneity or pleiotropy, supporting the robustness of the causal evidence.
This study identifies a significant positive causal relationship between urticaria and MDD risk and no significant association between psoriasis and MDD in East Asian populations, contrasting with previous European findings. Results for other skin diseases align with previous studies. These findings highlight the need for ancestry-specific research to inform personalized prevention and intervention strategies.
Keywords
- East Asian
- Mendelian randomization
- skin disease
- depression
1. Identified a causal link between urticaria and depression in East Asian populations.
2. Found no causal relationships between other skin diseases and depression.
3. Employed advanced Mendelian randomization techniques for robust causal inferences.
4. Highlights the importance of ancestry-specific research for personalized health strategies.
5. Advocates for larger, multi-ethnic datasets to validate and expand the findings globally.
Depression is a common mental health disorder, defined by sustained low mood, diminished interest or enjoyment in activities and pervasive fatigue or aversion to engagement. These symptoms often lead to substantial impairments in daily functioning and disruptions in social relationships [1]. Major depressive disorder (MDD), a severe form of depression, is among the most serious mental illnesses globally, with clinical manifestations including persistent sadness and loss of interest lasting at least two weeks [2]. The World Health Organization estimates that the global annual incidence of MDD is approximately 4.4% [3]. The prevalence of depression has consistently increased, especially with the worldwide spread of the Coronavirus Disease 2019 (COVID-19) pandemic. By 2030, MDD is expected to be one of the leading contributors to global disease burden [4]. The impact of depression is profound, being a major contributor to disability worldwide and closely linked to an elevated risk of suicide [5]. Beyond greatly increased psychological and physical distress to individuals, depression also imposes a substantial socio-economic burden, including increased healthcare costs and productivity losses. Despite significant research advances, the exact etiology and pathogenesis of depression remain largely unknown. Currently, the condition is viewed as a multifactorial ailment, shaped by a combination of genetic, neurobiological, psychological and environmental influences [4]. Timely interventions and an emphasis on research into causes are vital for halting increasing disease numbers and enhancement of the quality of life for the many individuals impacted. Identifying and understanding the factors associated with depression are crucial for the development of effective preventive strategies.
Skin diseases are often comorbid with mental health issues [6]. As the most visible organ of the human body, the skin plays a crucial role in self-perception and social interaction. Study have shown that individuals with chronic urticaria (CU) frequently exhibit symptoms of anxiety and depression, with depression occurring at a notably higher rate than in the general population [7]. Additionally, a systematic review of 41 studies reported that up to 62.3% of individuals with vitiligo exhibit depressive symptoms [8]. Another study revealed that patients with atopic dermatitis (AD) are more prone to depression and psychosomatic symptoms when compared to healthy controls [9]. Evidence also indicates a bidirectional relationship between anxiety and depression in individuals with systemic lupus erythematosus (SLE) [10], who demonstrate a depression incidence twice that of the general population, potentially mediated by inflammatory factors [11]. Furthermore, psoriasis patients are reported to be at a higher risk of mental health issues, with depression affecting up to 20% of this population [12]. However, conflicting findings have been presented by other studies [13, 14, 15, 16], leaving the relationship between skin disease and depression controversial. It is important to note that most observational studies examining the association between skin diseases and depression rely heavily on self-reported questionnaires. Such methods are susceptible to measurement bias and are influenced by the subjective emotions of patients. Furthermore, these studies are predominantly observational and are limited by their inability to effectively remove the impact of confounding factors. Although randomized controlled trials are the primary standard for confirming causal relationships, they often face challenges such as high economic cost and ethical constraints, limiting their application in certain fields.
Mendelian randomization (MR) analysis employs genetic variation as an instrumental variable (IVs) to estimate causal relationships between exposure, such as lifestyle factors or biomarkers and disease. This approach inherently avoids confounding factors common in traditional observational studies, as genetic variations are randomly allocated to individuals at conception [17]. By leveraging this “natural experiment”, MR analysis provides “purer” and more reliable causal inferences, offering a principled basis for understanding complex disease mechanisms and the development of effective prevention measures [18]. Interestingly, existing MR studies on the relationship between skin diseases and depression have primarily focused on European ancestries, acknowledging a significant limitation due to the homogeneity of the data [19, 20, 21, 22, 23]. The frequency of single nucleotide polymorphisms (SNPs) and the effect size of associations between SNPs and traits (exposures or outcomes) can vary markedly across different ancestries, which may lead to applicability issues affecting clinical relevance and the formulation of public health policy. This study aims to fill this gap by investigating the causal relationships between different skin diseases and MDD in East Asian populations, offering specific insights for global prevention and intervention strategies.
Research followed the Strengthening the Reporting of Observational Studies in Epidemiology-Mendelian Randomization (STROBE-MR) standards for the documentation of MR studies [24]. For STROBE-MR, please see the Supplementary material-STROBE MR checklist fillable. The choice of SNPs as IVs was directed by the three fundamental principles of MR: (1) The selected genetic variants were associated with the exposure, specifically various skin disease phenotypes. (2) Variants were further evaluated to ensure no association with potential confounding factors. (3) Effects of the genetic variants on the outcome were required to arise exclusively through the exposure, rather than through alternative pathways, to satisfy the core assumptions of MR [25]. Fig. 1 provides a detailed overview of the study design. As a secondary analysis of publicly available data, this study did not require approval from an ethical review committee.
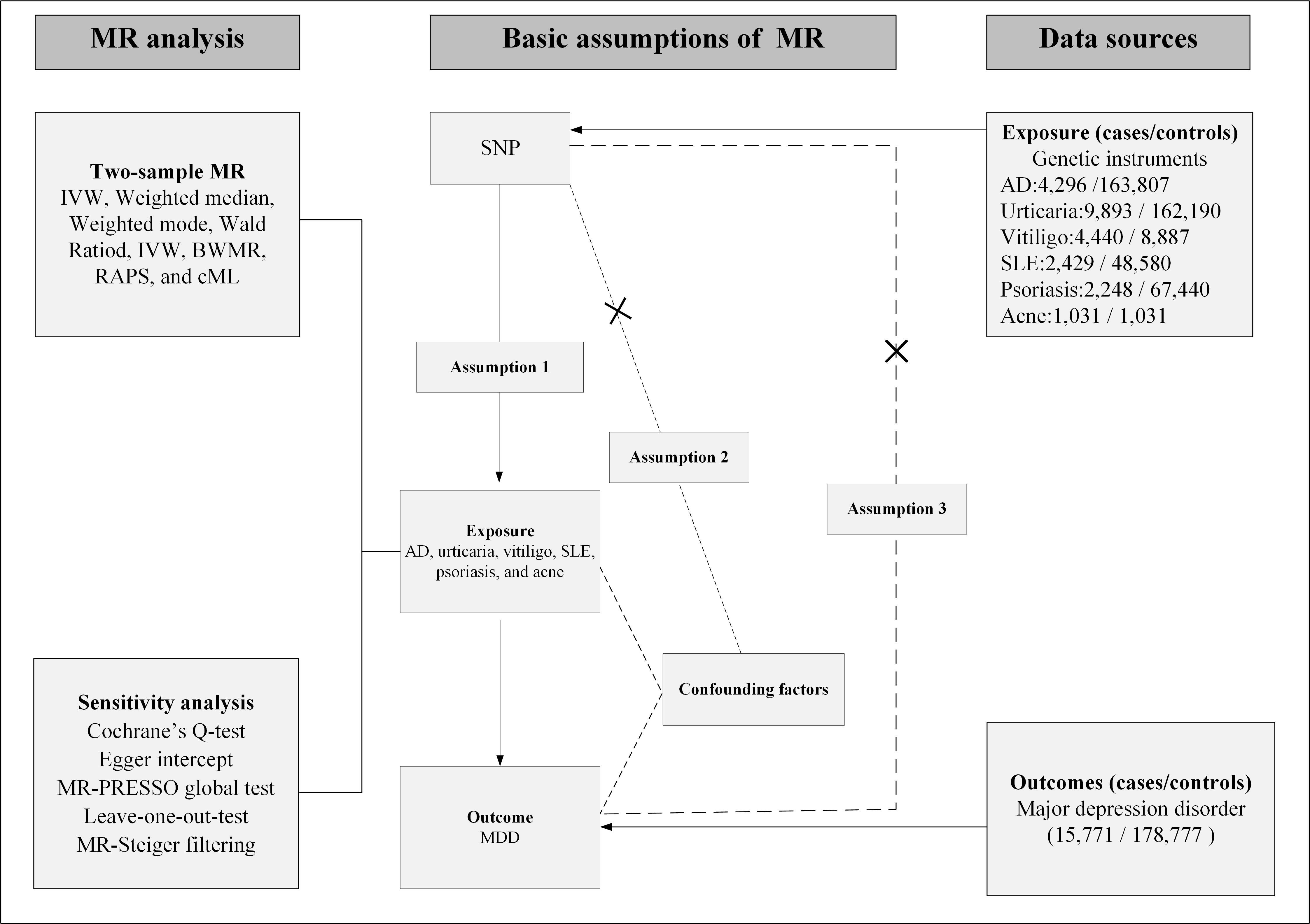 Fig. 1.
Fig. 1. Study design. Solid arrows depict the valid causal chain—SNP → Exposure → Outcome—required by MR. Dashed arrows with “×” mark bias-inducing routes that must be absent: (i) SNP → Confounders → Outcome, breaching Assumption 2; (ii) a direct SNP → Outcome path, i.e., horizontal pleiotropy, breaching Assumption 3. The dashed corner brackets the forbidden direct path for clarity. AD, atopic dermatitis; SLE, systemic lupus erythematosus; MDD, major depressive disorder; MR, Mendelian randomization; MR-PRESSO, MR Pleiotropy Residual Sum and Outlier; cML, constrained maximum likelihood; BWMR, Bayesian weighted Mendelian randomization; RAPS, robust adjusted profile score; IVW, inverse-variance weighted; SNP, single nucleotide polymorphism.
(1) Genetic variants were initially selected based on genome-wide significance (p
Table 1 (Ref. [30, 31, 32, 33, 34, 35]) gives an extensive overview of the data sources employed. Atopic dermatitis and urticaria were obtained from a cross-ancestry meta-analysis by Sakaue et al. [30], which incorporated data from the BioBank Japan project. This study included 220 deeply phenotyped genome-wide association study (GWAS) and integrated meta-analyses with data from the UK Biobank and FinnGen to enhance the resolution of the human genomic atlas. For this research, only East Asian GWAS data were used. AD data consisted of 4296 cases and 163,807 controls, with cases defined by the International Classification of Diseases (ICD)-10-L20, phecode-939, and FinnGen endpoint L12_ATOPIC. Urticaria data included 9893 cases and 162,190 controls, with cases defined by ICD-10-L50, phecode-947, and FinnGen endpoint L12_URTICA_ALLERG.
| Ref | Data source | Ancestry | Participants | |
| AD | 34594039 | Sakaue et al. [30] | EAS | 4296 cases and 163,807 controls |
| Urticaria | 34594039 | Sakaue et al. [30] | EAS | 9893 cases and 162,190 controls |
| Vitiligo | 38286188 | Wang et al. [31] | EAS | 4440 cases and 8887 controls |
| SLE | 38724181 | CMUH. [32] | EAS | 2429 cases and 48,580 controls |
| Psoriasis | 38757301 | CMUH and BBJ. [33] | EAS | 2248 cases and 67,440 controls |
| Acne | 24399259 | He et al. [34] | EAS | 1031 cases and 1031 controls |
| MDD | 34586374 | PGC. [35] | EAS | 15,771 cases and 178,777 controls |
Ref, PMID; CMUH, China Medical University Hospital; BBJ, BioBank Japan; PGC, Psychiatric Genomics Consortium; EAS, East Asianr.
The vitiligo dataset was obtained from Wang et al. [31], who combined data from two independent cohorts (4440 cases and 8887 controls). Diagnoses adhered to the Vitiligo European Task Force criteria and were confirmed by at least two dermatologists. Among the 11 identified susceptibility loci, six were located in intronic regions, while five were intergenic. SLE data were derived from the China Medical University Hospital (CMUH) Biobank [32]. This study included 2429 SLE cases and 48,580 controls. SLE was defined using ICD-9-710.0 and ICD-10-M32 and exclusion criteria included individuals without a history of SLE-specific medication use or diagnosed with other autoimmune diseases. An additive genetic model was applied, with logistic regression analysis adjusting for covariates such as sex, age and principal components. The study identified 20 significant genetic loci, including genes such as general transcription factor II-I (GTF2I), major histocompatibility complex, class II, DQ beta 1 (HLA-DQB1) and signal transducer and activator of transcription 4 (STAT4).
The psoriasis GWAS summary statistics were sourced from Yang et al. [33], who conducted a meta-analysis combining data from the CMUH Taiwan Biobank and BioBank Japan. This dataset included 2248 psoriasis cases and 67,440 controls. Diagnoses were confirmed by dermatologists using ICD-CM codes L40–L41, as well as ICD-9-CM-696. These SNPs were primarily concentrated on chromosomes 5 and 6. Acne data were also sourced from Yang et al. [33], encompassing 1031 cases and 1031 controls of Han Chinese ancestry Severe acne was diagnosed based on grade IV of the Pillsbury system. The study identified two novel susceptibility loci located at 11p11.2 and 1q24.2.
MDD data were sourced from the Psychiatric Genomics Consortium and included in a meta-analysis by Giannakopoulou et al. [35], which combined GWAS data from nine cohorts comprising 15,771 MDD cases and 178,777 controls. The identification of MDD was carried out through various methods, such as structured clinical interviews, examination of medical records, questionnaires focused on symptoms and self-report surveys. Results indicated variability in the heritability of MDD across East Asian populations, ranging from 6.5% to 15%.
The main analytical technique employed was the inverse-variance weighted (IVW) method. The choice between fixed-effect and random-effect models was based on the calculated heterogeneity (I2). If I2
To enhance the robustness and reliability of its primary analytical framework, this MR study employed a diverse range of complementary methods. These techniques were carefully selected to validate the consistency and stability of the findings across different analytical settings. Specifically, the study utilized the debiased inverse-variance weighted (dIVW) approach [38], which addresses potential biases in conventional IVW methods. Additionally, Bayesian weighted Mendelian randomization (BWMR) [39] was applied, leveraging Bayesian principles to account for potential pleiotropy. The robust adjusted profile score (RAPS) [40] method was incorporated to mitigate the impact of outliers and heterogeneity in the instrumental variable estimates. Furthermore, the constrained maximum likelihood (cML) [41] approach was included to ensure robustness under scenarios with complex pleiotropic effects.
Sensitivity analysis began with the use of Cochran’s Q test to assess heterogeneity and calculate the I2 statistic [42]. Horizontal pleiotropy was evaluated using MR-Egger regression [43] and the MR Pleiotropy Residual Sum and Outlier (MR-PRESSO) [44] methods, which also identified and excluded outliers [45]. Finally, a leave-one-out analysis was conducted to examine the robustness of the overall causal effect [46]. To address multiple testing, the Bonferroni correction was utilized, setting significant causal evidence at p
All statistical analyses were conducted using R software 4.2.2 (R Foundation for Statistical Computing, Vienna, Austria), along with the following packages: TwoSampleMR 0.5.6 (MRC Integrative Epidemiology Unit, University of Bristol, Bristol, UK), MR-PRESSO (version 1.0, Ron Do Laboratory, Icahn School of Medicine at Mount Sinai, New York, NY, USA), MRcML (version 0.9, School of Statistics & Division of Biostatistics, University of Minnesota, Minneapolis, MN, USA), mr.raps (version 0.2, Statistical Laboratory, University of Cambridge, Cambridge, UK), BWMR (version 0.1, Department of Mathematics, The Hong Kong University of Science and Technology, Hong Kong SAR, China), and MendelianRandomization (version 0.9.0, MRC Biostatistics Unit, University of Cambridge, Cambridge, UK).
The summarized results for the six different skin diseases and their association with MDD are given in Fig. 2. The number of SNPs included in the study ranged from 1 to 46, all of which passed the Steiger test, with the direction confirmed as “TRUE”, effectively filtering out genetic variants that violated MR assumption (3). This ensured the avoidance of reverse causation. All included SNPs had F-statistics greater than 10, minimizing the risk of bias from weak instrument variables. For SLE, the primary analytical method was the Wald Ratio (SNP = 1). For urticaria, no SNPs met the genome-wide significance threshold, necessitating the use of a less stringent threshold to obtain sufficient SNPs for MR analysis (p
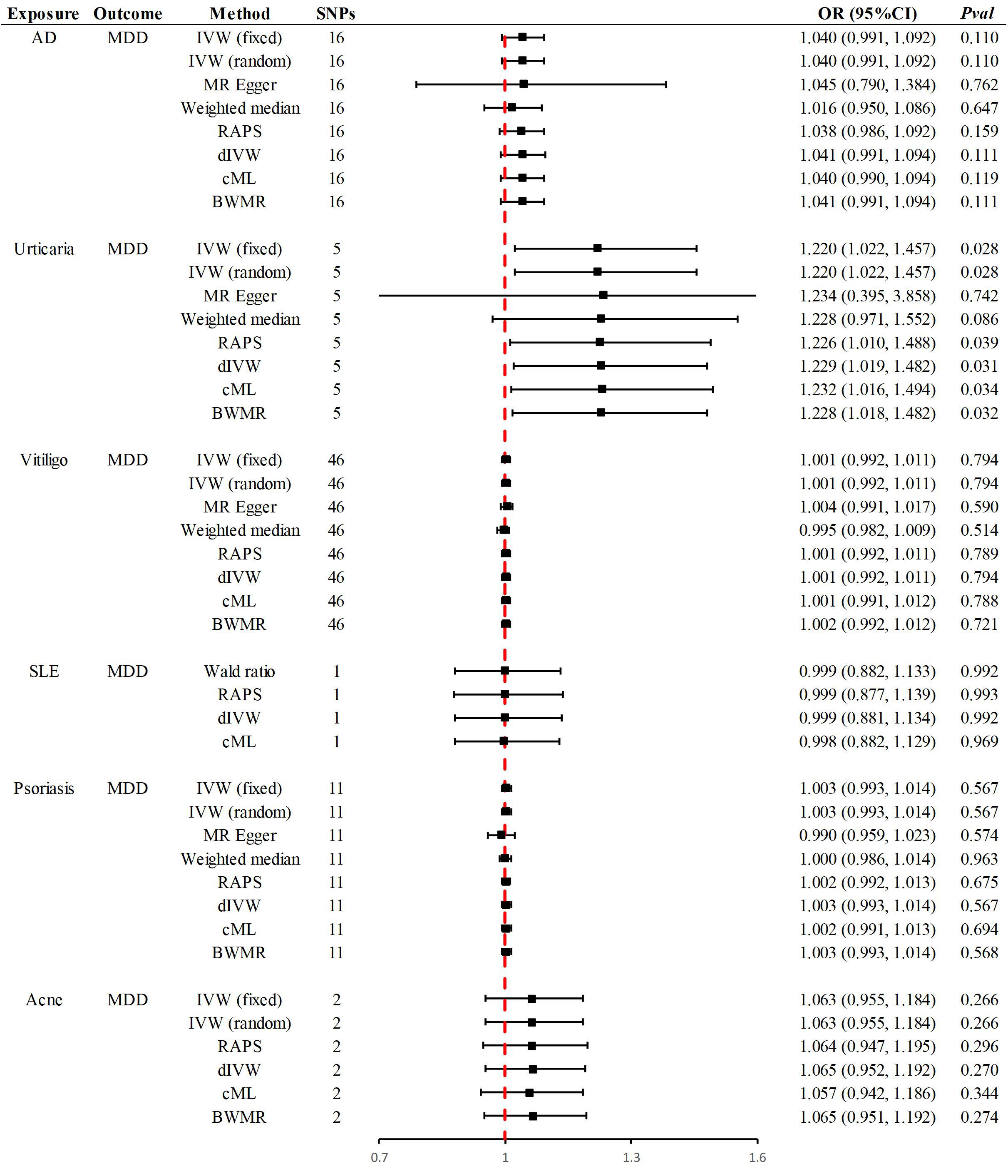 Fig. 2.
Fig. 2. Summary of Mendelian randomization results. OR, odds ratio; dIVW, debiased inverse-variance weighted.
The primary method revealed that a one-standard-deviation increase in genetically predicted urticaria corresponded to a 22% higher risk of MDD [odds ratio (OR) = 1.220, 95% CI 1.022–1.457, p = 0.028]. Supplementary methods, including RAPS (OR = 1.226, 95% CI 1.010–1.488, p = 0.039), dIVW (OR = 1.229, 95% CI 1.019–1.482, p = 0.031), cML (OR = 1.232, 95% CI 1.016–1.494, p = 0.034) and BWMR (OR = 1.228, 95% CI 1.018–1.482, p = 0.032), provided consistent causal evidence (Fig. 2). At an OR of 1.220, the analysis provided 92% statistical power, further strengthening the causal evidence. A forest plot highlighted rs375132703 as the genetic variant with the most significant causal effect. Leave-one-out analysis verified that this association was not influenced by any single SNP. Furthermore, no evidence of heterogeneity or pleiotropy was detected, reinforcing the robustness of the findings (Table 2). However, the significance level did not exceed the Bonferroni corrected p-value, so the relationship between urticaria and MDD is considered a potential causal association.
| Exposure | Outcome | MR-Egger regression | MR-PRESSO | Cochrane’s Q-IVW | Effect model | Steiger_test | ||||||||
| Intercept | SE | pval | RSSobs | pvalglobal test | Outlier | I2 | Q | Q_df | Q_pval | Direction | pval | |||
| AD | MDD | –0.001 | 0.027 | 0.974 | 11.825 | 0.777 | NA | 0 | 10.482 | 15 | 0.788 | Fixed | TRUE | 9.31 × 10 -69 |
| Urticaria | MDD | –0.001 | 0.055 | 0.985 | 5.503 | 0.525 | NA | 0 | 3.563 | 4 | 0.468 | Fixed | TRUE | 2.81 × 10 -11 |
| Vitiligo | MDD | –0.002 | 0.004 | 0.606 | 40.723 | 0.753 | NA | 0 | 38.147 | 45 | 0.755 | Fixed | TRUE | 0 |
| SLE | MDD | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | TRUE | 1.31 × 10 -19 |
| Psoriasis | MDD | 0.016 | 0.020 | 0.438 | 10.995 | 0.547 | NA | 0 | 9.044 | 10 | 0.528 | Fixed | TRUE | 0 |
| Acne | MDD | NA | NA | NA | NA | NA | NA | 0 | 0.756 | 1 | 0.385 | Fixed | TRUE | 1.12 × 10 -15 |
NA, not available.
No significant causal associations were identified between other skin diseases and MDD. Specifically, in East Asian populations, the genetic susceptibility to AD, vitiligo, SLE, psoriasis and acne showed no causal relationship with an increased risk of MDD. Supplementary Methods confirmed the consistency of these causal directions. At ORs of 1.040, 1.001, 0.999, 1.003 and 1.063, the statistical power was 65%, 6%, 5%, 14% and 34%, respectively. The leave-one-out examination verified that none of the individual SNPs affected the identified null associations. Furthermore, there was no indication of heterogeneity or horizontal pleiotropy, which reinforces the reliability and strength of the results (Table 2).
This two-sample MR study is the first systematic exploration of the causal relationships between six distinct skin diseases and MDD within East Asian populations. The analysis revealed a significant causal link between genetic susceptibility to urticaria and a heightened risk of MDD. In contrast, no causal relationship was identified between psoriasis and MDD. Notably, these results diverge significantly from prior causal evidence reported in European populations. Furthermore, the study demonstrated no causal associations between the genetic susceptibilities to AD, SLE, vitiligo, acne and MDD risk, which aligns with MR findings from European populations.
A meta-analysis involving 25 cohorts found that nearly one-third of CU patients have at least one underlying mental disorder, including MDD [48]. Interestingly, this study is the first to identify a causal association between urticaria and an increased risk of MDD in East Asian populations, a finding that contrasts sharply with MR studies conducted in European populations [23]. Previous epidemiological studies have highlighted conflicting evidence regarding the association between urticaria and MDD [49, 50]. Furthermore, research by Tzur Bitan et al. [51] demonstrated that the psychological impact of urticaria varies based on individual characteristics, age and socioeconomic status, complicating the establishment of a causal relationship. It should be acknowledged that this study applied a less stringent threshold for the urticaria phenotype to include sufficient SNPs for MR analysis. Furthermore, the phenotype was defined as a summary phenotype (ICD-10-L50), whereas previous studies have focused more specifically on the association between CU and depression. This broader definition may account for the differing findings on disease associations. Future studies utilizing larger GWAS datasets are needed to validate the findings reported here.
Previous MR studies have extensively investigated the causal relationship between psoriasis and MDD in European populations, but the results remain inconsistent [23, 52, 53]. In European ancestries, MR studies by Wang et al. [53] and Chu et al. [52] supported psoriasis as a risk factor for MDD, whereas the findings of Mo et al. [23] contradicted this conclusion. Due to the lack of a systematic review on this topic, this study did not further explore the relationship. However, the findings of this study align with those of Mo et al. [23], emphasizing the negative association between psoriasis and MDD across both European and East Asian ancestries. In this study, genetic variants were used as IVs, recognizing that significant differences in genetic composition exist across populations of different ancestries. These genetic differences may influence the expression and function of disease susceptibility genes, contributing to the ethnic-specific causal association between psoriasis, urticaria and MDD. Additionally, environmental factors and lifestyle differences between European and East Asian populations must be considered. Psoriasis, urticaria and MDD are likely influenced by complex interactions between genetic and environmental factors, with gene-environment interactions potentially exhibiting distinct patterns in different regions.
Previous observational studies have been influenced by various confounding factors and reverse causation, often resulting in biased findings that reflect correlations rather than causal relationships. This study, using MR analysis, revealed negative causal associations between vitiligo, SLE, AD, acne and MDD in East Asian populations, consistent with MR findings in European populations [19, 20, 21, 22]. First, these skin diseases may primarily develop through pathways unrelated to MDD. For instance, vitiligo and AD are commonly associated with autoimmune responses and skin barrier dysfunction, mechanisms that are unlikely to directly influence the neurobiological processes affecting mental health. Secondly, it is noteworthy that the potential link between depression and skin diseases may partly be mediated through inflammatory pathways. Kouba et al. [54], reported significantly elevated serum levels of pro-inflammatory cytokines in patients with depression. These cytokines are implicated not only in the pathophysiology of depression but also in the onset and progression of various skin disorders. For example, increased levels of interleukin (IL)-6 and tumor necrosis factor (TNF)-
This study underscores the importance of conducting disease association research in diverse populations and regional contexts. The genetic backgrounds and environmental factors of different ethnic groups and regions may significantly influence the pathophysiological mechanisms of diseases. Research should incorporate larger datasets and cross-population evidence (e.g., South Asian, Hispanic, African populations) to further validate these causal associations and explore their biological mechanisms. Additionally, future studies should consider investigating the potential links between urticaria subtypes and mental health to provide more targeted therapeutic strategies. Finally, it is crucial to note that MR studies rely on the quality and suitability of genetic instrumental variables. Therefore, research should include larger cohort studies and evidence-based approaches combined with MR analysis to achieve triangulation, thereby strengthening the causal conclusions.
This study offers several notable strengths. Foremost, it reports the first MR analysis to comprehensively investigate the causal relationships between diverse skin diseases and MDD specifically within East Asian populations. This focus expands the applicability of the findings to populations beyond those of European ancestry. Second, advanced MR methods, including RAPS, cML, dIVW and BWMR, were utilized, providing multidimensional validation and ensuring robust causal evidence. Additionally, the Steiger test was utilized to address potential reverse causation. Nonetheless, this study is not without limitations. The reliance on publicly available summary-level GWAS data meant that MR analysis could not be performed for skin diseases such as rosacea, alopecia areata and xerosis, as no GWAS data currently exists for these conditions in East Asian populations. Research should focus on generating GWAS datasets from these populations and diseases. Moreover, reliance on summary-level data limited the ability to perform subgroup analyses and hindered further exploration of urticaria subtypes. While GWAS data on urticaria subtypes have been published for European populations, such as in the FinnGen cohort, larger GWAS datasets for urticaria in Asian populations are needed to address these gaps.
This MR study, utilizing GWAS data from East Asian populations, identified a causal association between urticaria and an elevated risk of MDD. In contrast, no causal link was observed between psoriasis and the risk of MDD. These findings contrast sharply with previous MR studies conducted in European populations. Further, they underscore the importance of personalized mental health assessments and interventions for patients with skin diseases across different ancestries. In clinical practice, early identification and management of depression risk in urticaria patients are particularly critical. Finally, this study emphasizes the necessity of conducting disease association research across diverse populations and regions, providing valuable insights for the development of global healthcare strategies.
Data for MDD are available for download at the Psychiatric Genomics Consortium (https://pgc.unc.edu/for-researchers/download-results/), and data for AD, Urticaria, Vitiligo, SLE, AD, Urticaria, Vitiligo, SLE, Psoriasis, Acne are available in the GWAS Catalog (https://www.ebi.ac.uk/gwas/) based on their PMIDs, or through the original GWAS at the Data Statement.
SL and XY conceived and designed the study. SL performed data collection, data curation, and statistical analyses, and drafted the initial manuscript. XY supervised the study, verified the analyses, and provided critical revisions. Both authors interpreted the results, contributed to editorial changes, and read and approved the final manuscript. Both authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Not applicable.
We thank all GWAS participants and investigators for publicly making the summary statistics data available.
This study was supported by the Clinical Medicine Special Fund Project of Zhejiang Medical Association (2022ZYC-A127).
The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/AP47646.
We utilized AI tools to help reduce the similarity rate and improve the overall quality of the manuscript. We would like to clarify that AI was used for manuscript refinement and editing purposes. After using this tool, the authors reviewed and edited the content as needed and takes full responsibility for the content of the publication.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.


