1 Cardiovascular Disease Laboratory, The First Affiliated Hospital of Chongqing Medical University, 400016 Chongqing, China
2 Division of Cardiology, The First Affiliated Hospital of Chongqing Medical University, 400016 Chongqing, China
3 Vascular Surgery Department, The First Affiliated Hospital of Chongqing Medical University, 400016 Chongqing, China
4 Health Management Center, The First Affiliated Hospital of Chongqing Medical University, 400016 Chongqing, China
5 Department of Epidemiology and Biostatistics, School of Public Health, Jiamusi University, 154000 Jiamusi, Heilongjiang, China
6 Department of Anesthesiology, The First Affiliated Hospital of Chongqing Medical University, 400016 Chongqing, China
7 Department of Thoracic and Cardiovascular Surgery, The First Affiliated Hospital of Chongqing Medical University, 400016 Chongqing, China
†These authors contributed equally.
Abstract
Aortic dissection (AD) is a high-mortality cardiovascular emergency with unclear pathophysiological mechanisms. This study investigated S100 calcium-binding protein A9 (S100A9) as a therapeutic target for AD and explored its underlying mechanisms.
Proteomic analysis compared aortic tissues from patients with acute type A and matched non-dissected vascular tissues from the same patients. An AD model was induced in wild-type and S100A9 knockout mice via β-aminopropionitrile (BAPN). Survival, aortic diameter, and S100A9 expression were quantified. Furthermore, single-cell RNA sequencing was used to analyze cell populations and mitochondrial pathways in AD mice treated with an S100A9 inhibitor. Finally, the effect of S100A9 on mitochondrial function was investigated in Tohoku Hospital Pediatrics-1 (THP-1) cells.
Proteomics identified that S100A9 is significantly upregulated in AD tissue. Furthermore, S100a9 knockout (S100a9 KO) mice conferred protection against AD-induced mortality and aortic dilation. Single-cell RNA analysis revealed that S100A9 is predominantly expressed within the granulocyte population. S100A9 inhibition activated mitochondrial oxidative phosphorylation pathways and upregulated mtDNA-encoded gene expression. Human tissue mRNA levels confirmed decreased mtDNA in AD. Moreover, recombinant human S100A9 and angiotensin-II treatment in THP-1 cells reduced mitochondrial membrane potential and increased oxidative stress.
S100A9 is a potential contributor to AD pathogenesis. Inhibition of S100A9 might be a promising therapeutic target for AD.
Keywords
- aortic dissection
- proteomic analysis
- single-cell RNA sequencing
- mitochondrial function
- S100A9
Aortic dissection (AD) is a life-threatening cardiovascular disease characterized by high mortality rates and a poor prognosis [1]. AD initiation involves a tear in the aortic intima, causing separation of the aortic wall layers [2]. This process enables blood entry into the aortic media, ultimately resulting in organ ischemia, aortic rupture, and potentially sudden death [1]. Despite therapeutic advances, mortality remains high, primarily due to an incomplete understanding of AD’s underlying pathogenic mechanisms.
The pathophysiology of aortic dissection involves inflammation and oxidative stress. Granulocyte macrophage colony-stimulating factor (GM-CSF), a pro-inflammatory cytokine, contributes critically to AD development [3, 4]. Furthermore, interleukin-6 (IL-6) serves as an essential mediator of angiotensin-II (Ang-II)-induced aortic dissection [5]. Notably, experimental evidence from mouse models demonstrates that increased reactive oxygen species (ROS) levels are associated with aortic dissection [6]. Consequently, targeting inflammatory responses and ROS generation may represent promising therapeutic strategies for AD [7]. Despite this potential, the specific cytokines mediating inflammation and oxidative stress in AD pathogenesis remain poorly characterized.
S100A9, a member of the S100 protein family, functions as a small calcium-binding protein primarily involved in inflammatory processes and immune responses [8]. It is frequently upregulated in response to diverse inflammatory stimuli and has been implicated in various cardiovascular pathologies, including myocardial infarction, heart failure, and age-induced vascular dysfunction [9]. Our prior work has associated elevated S100A9 levels with sepsis-induced acute respiratory injury [10]. Emerging evidence suggests that pharmacological S100A9 inhibition may protect against aortic dissection pathophysiology by mitigating inflammatory responses [11]. However, the precise mechanisms remain incompletely understood.
This study aims to identify novel molecular targets indicative of aortic dissection progression and investigate their functional roles and mechanisms. Leveraging proteomic analysis of human patients with type A aortic dissection tissues, we identified that S100A9 was significantly upregulated in AD. Functioning as an S100 protein family member, S100A9 mediates diverse cellular processes such as inflammation and immune responses [12]. Our findings demonstrate that S100A9 inhibition improves survival in experimental AD mouse models and ameliorates AD-related outcomes, potentially through enhanced mitochondrial function and mitochondrial DNA expression. Collectively, these findings position S100A9 as a potential contributor to AD pathogenesis, establishing its inhibition as a possible therapeutic strategy.
Human aortic dissection specimens and non-dissected vascular tissues (harvested distant from the dissection site) were obtained from patients undergoing surgery for type A aortic dissection. Samples were collected during aortic root and ascending aorta replacement surgeries at either the Department of Thoracic and Cardiovascular Surgery of Chongqing University Central Hospital and the First Affiliated Hospital of Chongqing Medical University from April 2024 to June 2024. All procedures received prior approval from the hospital’s ethics committee and for this retrospective analysis of anonymized clinical data, the requirement for written informed consent was waived by the Ethics Committee (2024-173-02) and all procedures conducted in accordance with the Declaration of Helsinki. Protein was extracted from aortic tissue samples and digested with trypsin. The resulting peptides underwent liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis using high-resolution mass spectrometer. The acquired spectra were processed using a database to identify and quantify the proteins. Label-free quantification (LFQ) was used to analyze differential protein expression between dissection and control tissues.
Male C57BL/6J mice and global S100a9 knockout (KO) mice were purchased from Cyagen Biosciences Inc (Suzhou, China) and housed under specific pathogen-free conditions at the Chongqing Medical University Animal Research Center. Aortic dissection was induced in both wild-type (WT) and S100a9 KO mice. Starting from week 6, all mice were received daily
Protein lysates were resolved by SDS-PAGE (Cat: PG212, EpiZyme, Shanghai, China) at 80–120 V constant voltage. Separated proteins were electrophoretically transferred onto PVDF membranes (Cat: ISEQ00010, Millipore, Billerica, MA, USA) using a rapid transfer solution at 400 mA for 30 minutes. The membranes were blocked with 5% skim milk (Cat: D8340, Solarbio, Beijing, China) for 1.5 hours and washed three times with TBST (Cat: G0001-2L, Servicebio, Wuhan, Hubei, China) for 3 minutes each. Primary antibodies S100A9 (1:1000, Cat: ab288715, Abcam, Cambridge, UK), GAPDH (1:10,000, Cat: GB11002, Servicebio, Wuhan, Hubei, China) were incubated overnight at 4 °C, followed by incubation with secondary antibodies (1:7000, Cat: AB-228395, Invitrogen, Carlsbad, CA, USA) the next day. The signal was visualized by enhanced chemiluminescence (Cat: BL520B, Biosharp, Beijing, China).
Total RNA was extracted from tissue samples using Trizol (Takara, Beijing, China). The RNA was then reverse-transcribed into cDNA using a reverse transcription kit (Cat: RK20433, ABclonal, Wuhan, Hubei, China). Quantitative PCR was performed using SYBR Green Master Mix with specific primers (Cat: RK21203, ABclonal, Wuhan, Hubei, China) on a CFX96™ Real-Time system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The mRNA expression levels were normalized to
GO enrichment analysis and visualization were performed using the ClusterProfiler (v4.10.1) in R studio (v4.3.2) [13]. GSEA enrichment analysis was conducted using the GSEABase (v1.64.0, Bioconductor Core Team, Seattle, WA, USA) [14]. All visualization results presented in the main text had Benjamini-Hochberg adjustment for multiple comparisons (p.adj
We constructed a PPI network by STRING (Search Tool for the Retrieval of Interacting Genes/proteins; v12.0, https://string-db.org/). We then visualized PPI and identified hub genes within this network using the Cytoscape (v3.10.2, University of California, San Diego (UCSD), San Diego, CA, USA) and the MCODE plugin (v2.0.3).
GOSemSim analysis was conducted in R studio using the GOSemSim package (v2.28.1) to comprehensively score candidate proteins from the aspects of Biological process, Molecular Function, and Cellular Component [15]. The analysis results were visualized using ggplot2 (v3.5.2), results had Benjamini-Hochberg adjustment for multiple comparisons (p.adj
GSE247260 raw data was downloaded from GEO database. Single cells were extracted under the criteria: nFeature_RNA
The human acute monocytic cell line Tohoku Hospital Pediatrics-1 (THP-1) was purchased from the Procell system (CL-0233, Wuhan, China), authenticated by STR profiling with no evidence of cross-contamination and tested negative for mycoplasma contamination. The cells were cultured in Complete Medium For Cell Culture Tested (Cat: TCH-G361-C, HyCyte, Suzhou, China) and supplemented with 10% fetal Bovine serum (FBS) (Cat: FBP-S005, HyCyte, Suzhou, China) in a humidified atmosphere at 37 °C under 5% CO2. A working cell bank was produced from one of the master cell banks and the cells in the second or third passage were used for experiments. The cells were diluted to 2
Data were analyzed using GraphPad Prism software (v8.0.2, GraphPad Software, LLC, San Diego, CA, USA). Results are presented as mean
Paired aortic tissue specimens (five dissected and five matched non-dissected vascular samples) were obtained from patients undergoing surgical repair for acute Type A aortic dissection. All ten samples underwent proteomic profiling via LC-MS/MS. Following stringent quality control, 4608 proteins were confidently identified. Principal component analysis (PCA) validated dataset integrity, showing a significant separation between dissection and control groups (Fig. 1A). Differential expression analysis identified 190 significantly altered proteins (
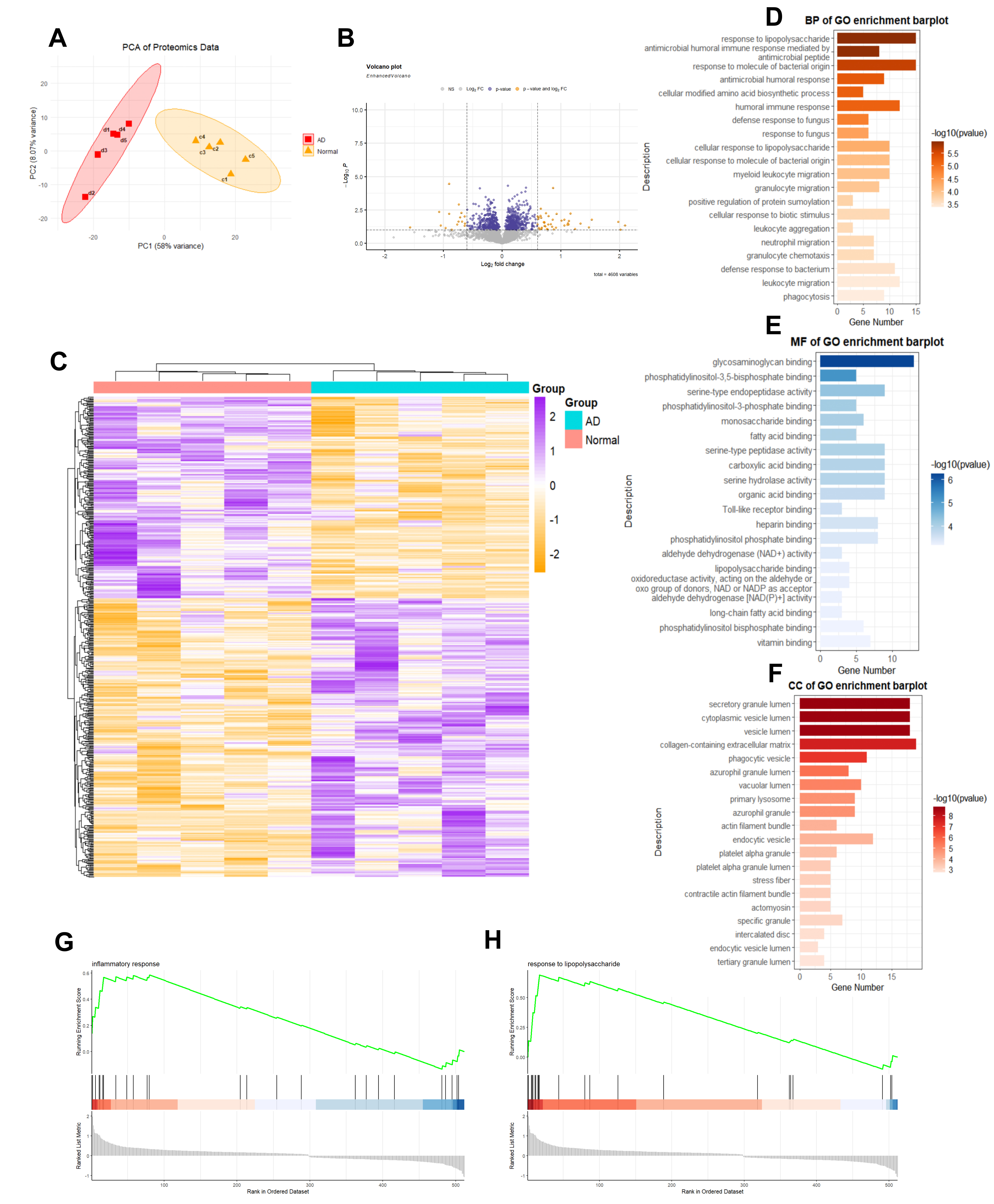 Fig. 1.
Fig. 1. Proteomic sequencing and analysis of type A aortic dissection (n = 5 for normal and dissection vascular samples). (A) Principal Component Analysis (PCA) of the proteomic data. (B) Volcano plot illustrating variable proteins (total proteins analyzed = 4608). (C) Heatmap depicting identified differential proteins. (D–F) Gene Ontology (GO) enrichment analysis of the differential proteins identified in proteomics (p.adj
To characterize functional interactions among differentially expressed proteins, we conducted a PPI network using the STRING database (v12.0). Among the 190 input proteins, 52 lacked predicted interactions, while five formed disconnected singleton pairs (Fig. 2A, Supplementary Table 3). After filtering out these non-networked proteins, the remaining 133 proteins were ranked by topological importance in Cytoscape (v3.10.2), with the highest-scoring hubs visualized at the network core (Fig. 2B, Supplementary Table 4). Topological analysis revealed high-scoring hub proteins concentrated at the network core, indicating functional centrality. Subsequent MCODE clustering identified 17 potential hub proteins from densely interconnected modules (Fig. 2C, Supplementary Table 5). To pinpoint the most significant hub protein, we calculated semantic similarity (GOSemSim v3.8, Bioconductor Core Team, Guangzhou, Guangdong, China) across three GO domains: biological process, molecular functions, and cellular components. S100A9 demonstrated the highest functional coherence score (Fig. 2D). Correlation heatmap analysis showed that S100A9 was closely associated with S100A8 and exhibited interactions with other genes (Fig. 2E). Visualization through heatmap and volcano plot confirmed significant upregulation of S100A9 in the aortic dissection group (Fig. 2F,G). These findings position S100A9 as a central regulator in aortic dissection.
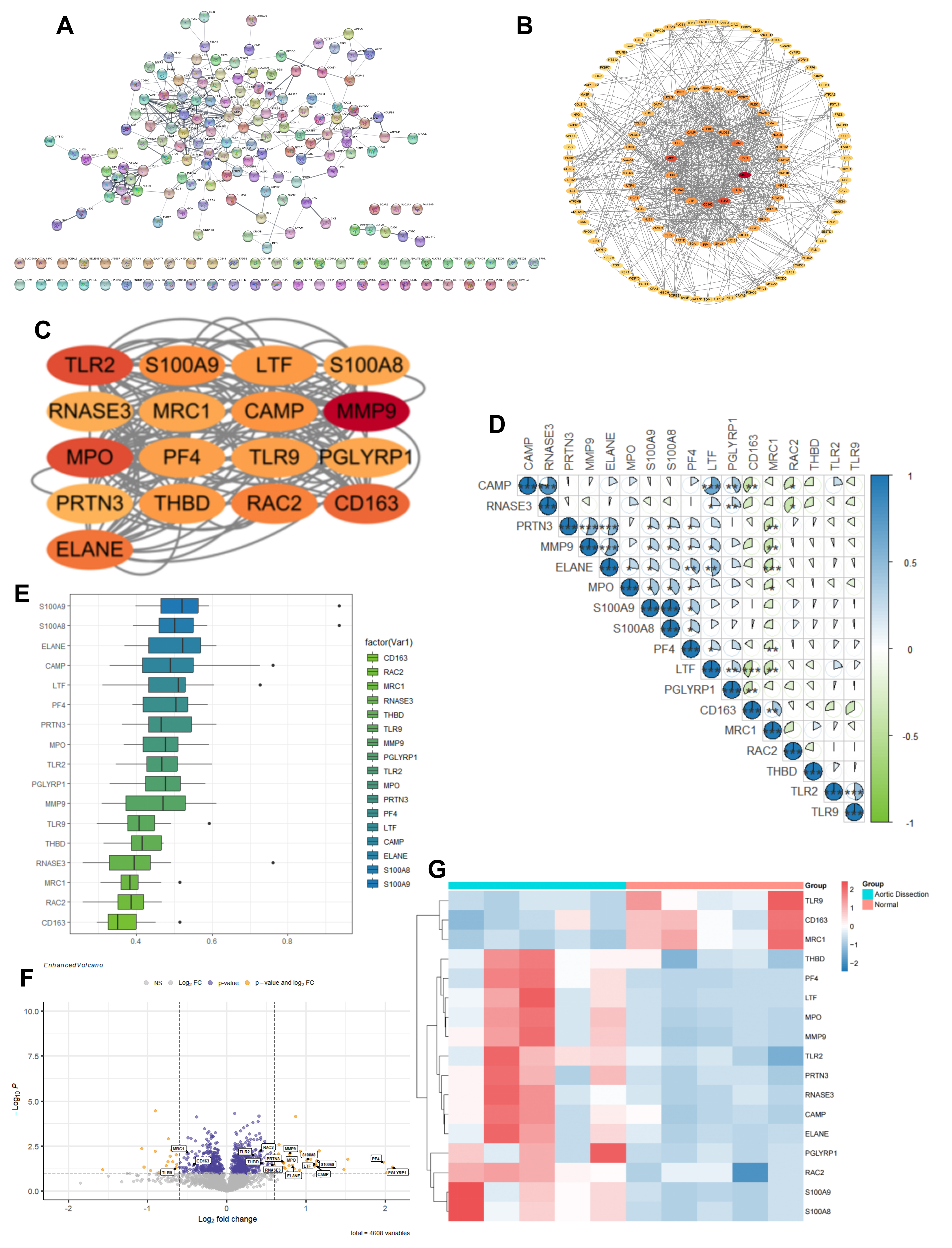 Fig. 2.
Fig. 2. Hub protein screening for type A aortic dissection. (A) Interaction network of differential proteins analyzed using the Search Tool for the Retrieval of Interacting Genes/proteins (STRING) database. (B) Importing protein interaction results into Cytoscape and ranking the proteins. (C) Identification of 17 hub proteins from the analysis of 138 proteins using the MCODE plugin. (D,E) Box plot and correlation heatmap of the final ranking for the 17 hub proteins from GOsensim analysis. (F) Volcano plot of the 17 hub proteins. (G) Heatmap of the 17 hub proteins, displaying each group separately. *p
Following the identification of S100A9 as a hub protein, we validated its expression in dissection vessels. Tissue proteins were extracted from both aortic dissection samples and normal vascular tissues. Western blot analysis revealed a significant increase in S100A9 expression in the aortic dissection group, consistent with proteomic data (Fig. 3A,B). Quantitative PCR further confirmed a marked upregulation of S100a9 mRNA levels (Fig. 3C).
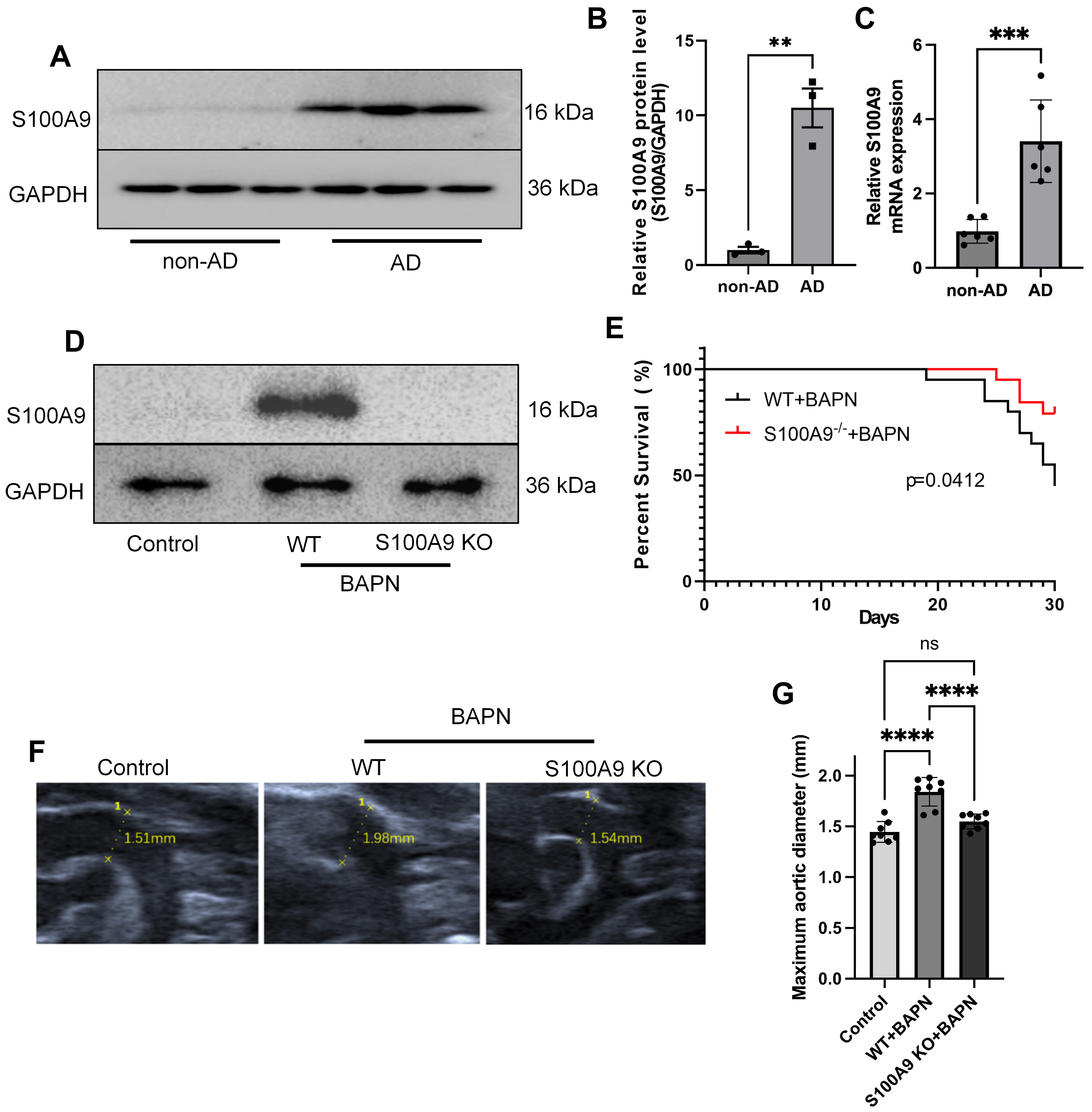 Fig. 3.
Fig. 3. Role of S100A9 in aortic dissection animal models. (A,B) Protein expression of S100A9 in aortic tissues from AD patients evaluated through Western blotting, with statistical analysis presented in (B) (n = 3). (C) S100a9 mRNA expression in vascular tissues (n = 6). (D) Protein expression of S100A9 in aortic tissues evaluated through Western blotting (n = 3). (E) Survival analysis of WT and S100a9 KO AD mice (45% vs 80%; p value = 0.0412). (F,G) Aortic diameter of mice across groups and the statistical result (p value
To explore the effects of S100A9 on aortic dissection development, we utilized wild-type mice and S100a9 KO mice to induce aortic dissection model. Western blot analysis revealed that S100A9 levels were significantly elevated in the aortic vessels of BAPN-treated WT mice, while expression was nearly undetectable in S100a9 KO mice treated with BAPN (Fig. 3D). S100a9 KO significantly increased survival rates of AD mice (Hazard ratio S100a9-/-+BAPN/WT+BAPN: 0.3236; 95% CI of ratio: 0.1143–0.9155, p value = 0.0412; Fig. 3E). Doppler ultrasound examinations showed that S100a9 deficiency also decreased the aortic diameter in AD mice (p value
To further investigate the mechanisms by which S100A9 influences aortic dissection, we analyzed single-cell sequencing data of aortic tissues from both aortic dissection models and ABR (paquinimod)-treated dissection models (ABR, an S100A9-specific inhibitor, intragastric administration, 10 mg/kg/d, single-cell dataset GEO247260) [11]. Using the Seurat package, we performed rigorous quality control, dimensionality reduction, and unsupervised clustering, identifying 15 distinct clusters (Fig. 4A). Through manual cell type annotation based on literature and marker databases (Fig. 4B,C). The results indicated that the proportions of B cells, T cells, Granulocyte, Macrophage, and Endothelial cells were significantly elevated in the dissection group compared to the treated group, while Fibrocyte and Smooth muscle cells (SMC) were more prevalent in the treated group (Fig. 4D). This suggests that inhibiting S100A9 expression leads to an increase in cell populations involved in vascular structure while reducing the numbers of immune and inflammatory cells. These findings align with our observation that deletion of S100a9 improved the aortic dissection phenotype in BAPN-treated mice.
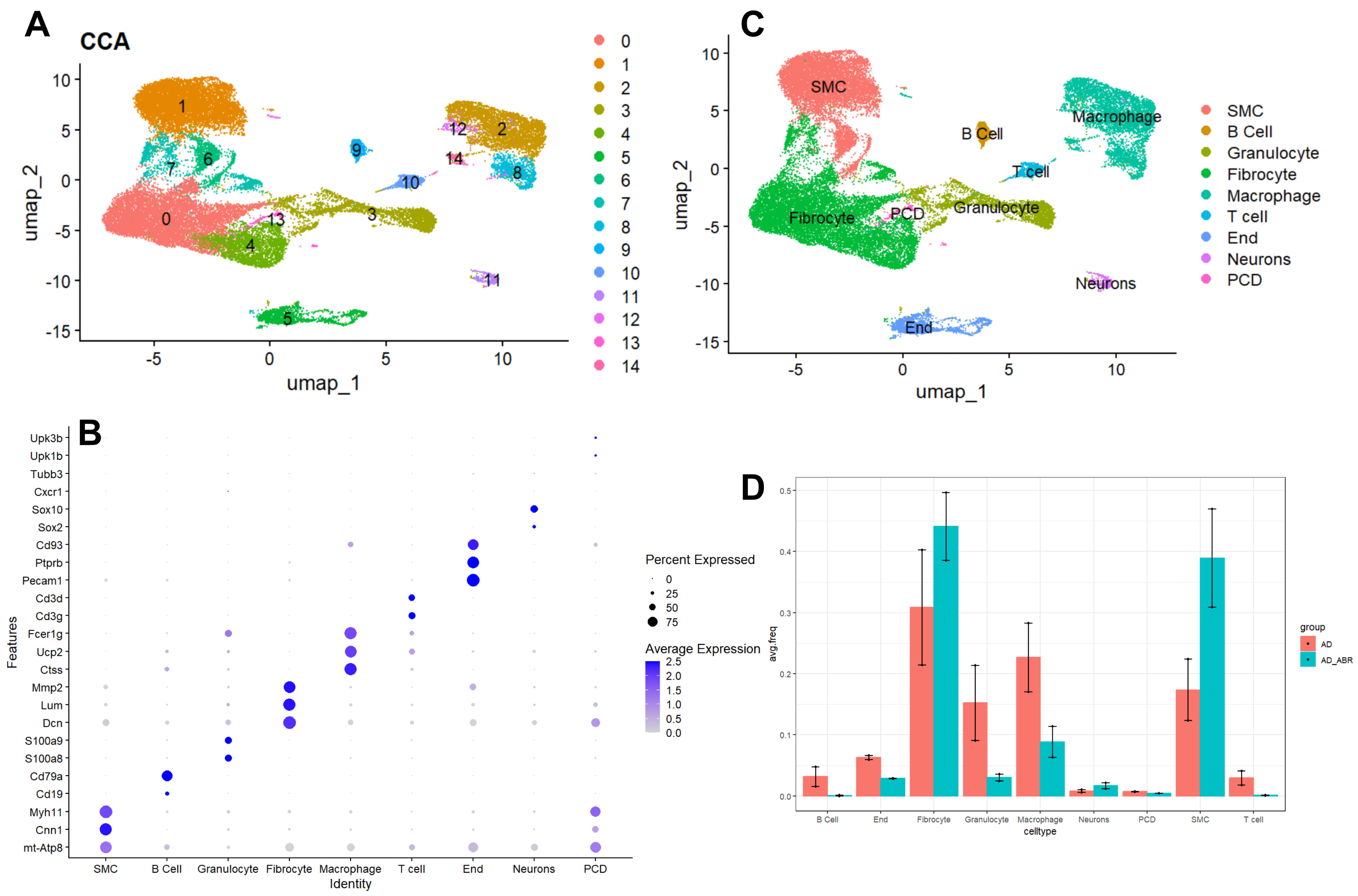 Fig. 4.
Fig. 4. Single-cell transcriptomic analysis of aortic dissection animal models with S100A9 intervention. (A) UMAP plot showing dimensionality reduction and clustering analysis of the GSE247260 dataset. (B) Bubble plot displaying the markers for each cluster. (C) UMAP plot illustrating the clustering based on marker expression. (D) Bar graph comparing the cell populations between the AD and AD+ABR groups (ABR, S100A9 inhibitor; 10 mg/kg/d).
Our analysis confirmed that S100a9 is predominantly expressed in granulocyte (Fig. 5A). To elucidate the mechanisms involved, we performed single-cell Gene Set Variation Analysis (GSVA; v1.50.2, https://bioconductor.org/packages/GSVA/) specifically on granulocyte. We found that the oxidative phosphorylation pathway was significantly downregulated in granulocyte from the aortic dissection group compared to the treated group, indicating that inhibiting S100A9 expression enhances mitochondrial function (Fig. 5B). GO enrichment analysis for the granulocyte population revealed significant enrichment of the reactive oxygen species (ROS) pathway in the dissection group (p.adj
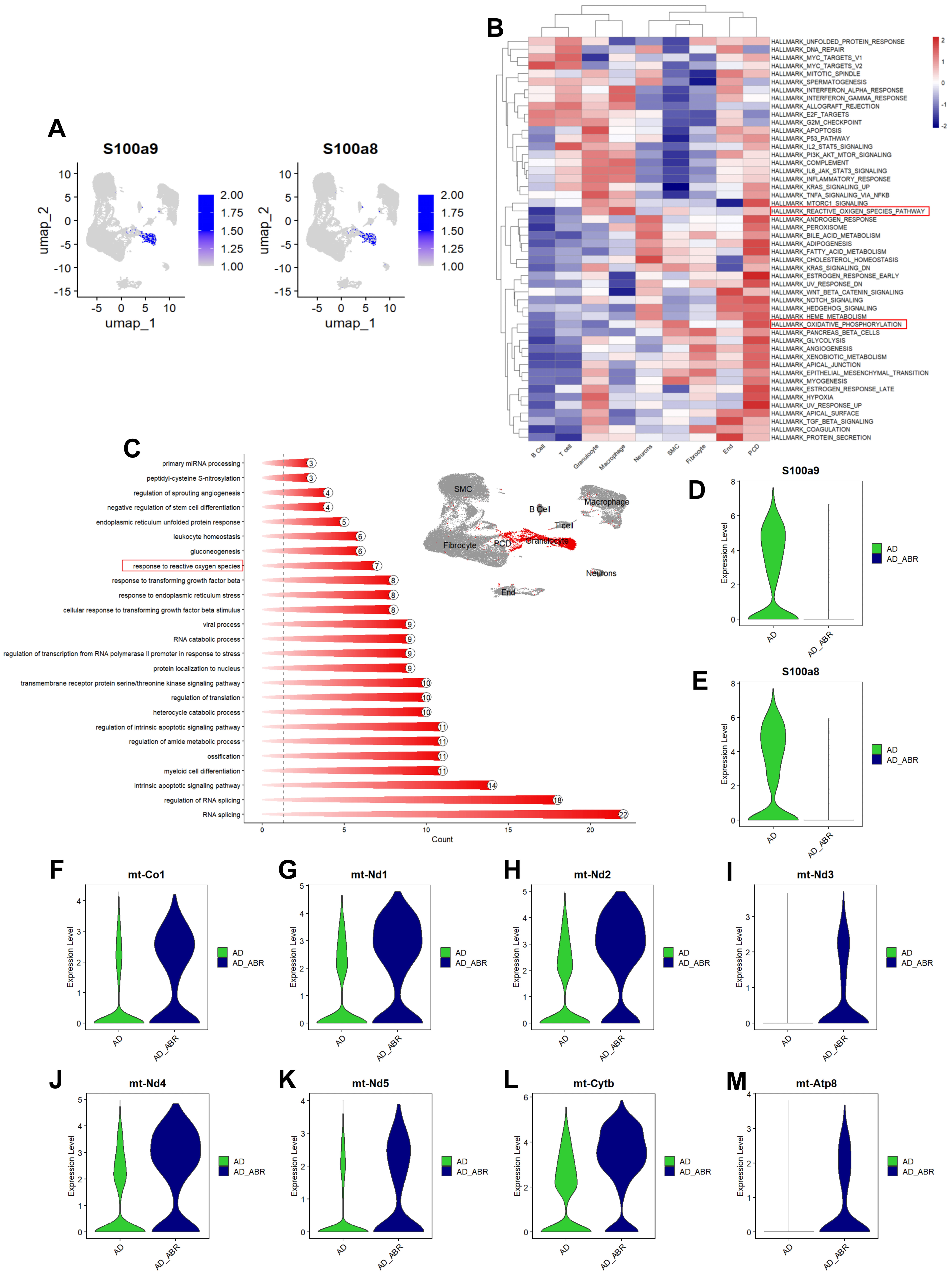 Fig. 5.
Fig. 5. Inhibition of S100a9 mitochondrial quality in granulocytes. (A) UMAP plots displaying the separate visualization of S100A9 and S100A8. (B) GSVA on granulocyte, with the visualized heatmap was reflecting changes in AD group (p.adj
To validate the single-cell analysis, we measured mRNA levels of key mitochondrial genes. MT-CO1, MT-ND1, MT-ND2, MT-ND3, MT-ND4, MT-ND5, MT-CYB, and MT-ATP8 were significantly downregulated in human AD tissues (Fig. 6A–H). Using THP-1 cells, we further assessed the role of S100A9 in mitochondrial function. JC-1 staining showed reduced mitochondrial membrane potential, and MitoSOX staining revealed increased oxidative stress after treatment with Ang-II (Fig. 6I) and recombinant human S100A9 (rhS100A9) (Fig. 6J). These findings indicate that S100A9 deteriorate mitochondrial quality, targeting S100A9 to improve mitochondrial quality may offer a therapeutic strategy for aortic dissection.
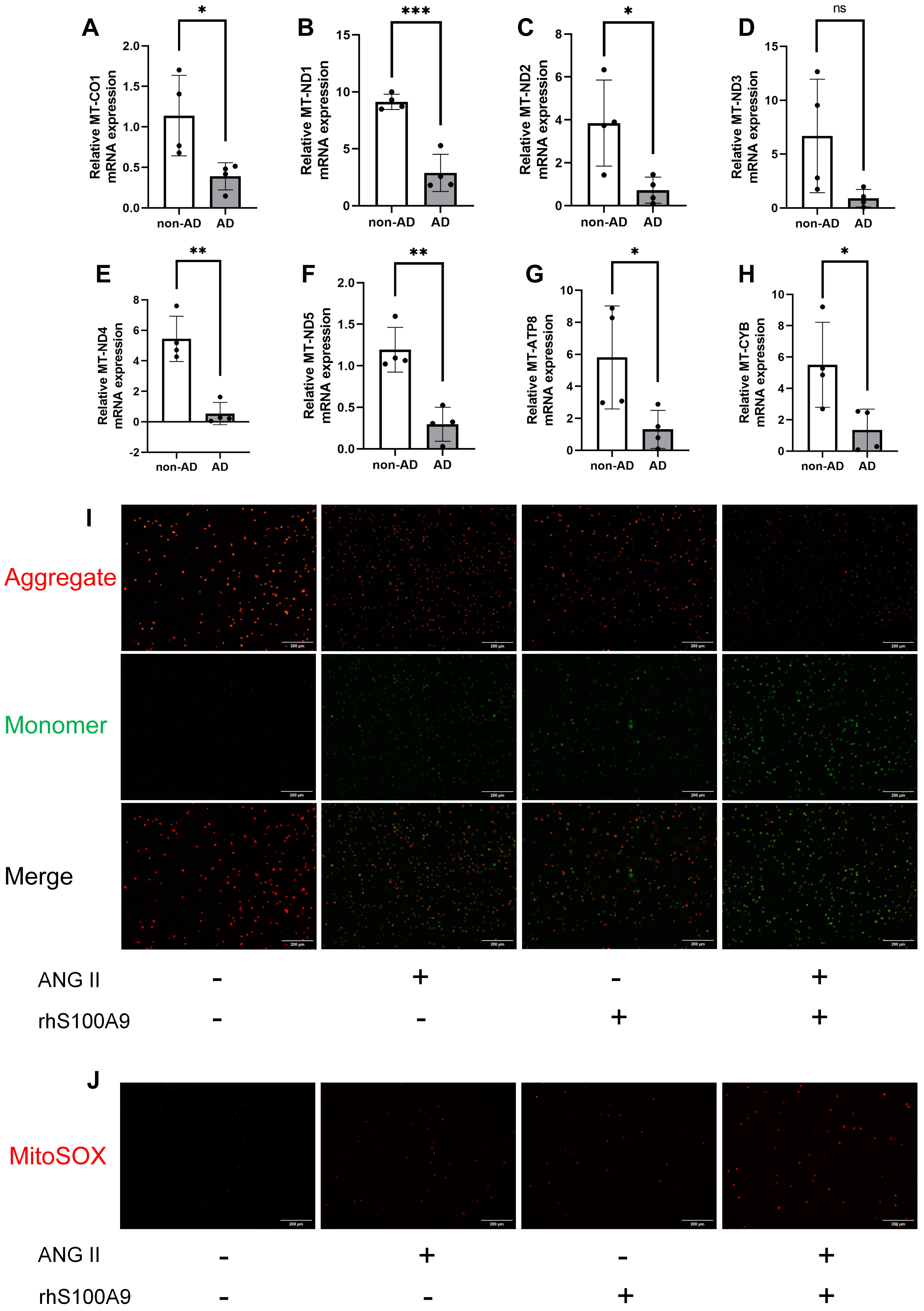 Fig. 6.
Fig. 6. S100A9 deteriorate mitochondrial quality. (A–H) The core mitochondrial genes mRNA expression in vascular tissues (n = 4). (I) JC-1 fluorescence images of THP-1 cells treated with Ang-II or recombinant human S100A9 (rhS100A9) for 24 hours (scale bar: 200 µm). (J) MitoSOX fluorescence images of THP-1 cells treated with Ang-II or rhS100A9 for 24 hours (scale bar: 200 µm). The data are presented as the mean
Aortic dissection is a complex and life-threatening condition characterized by the separation of the layers of the aortic wall [5]. Our study highlights the role of S100A9 as a central player in the pathology of aortic dissection. Our proteomic analysis identified S100A9 as a hub protein, demonstrating its significant upregulation in aortic tissue from patients with type A dissection compared to healthy controls, and that is validated by experiments. This finding aligns with previous research suggesting that S100A9 is involved in inflammatory responses, underscores its relevance in the pathogenesis of aortic dissection [8].
S100A9, a member of the S100 protein family, consists of small calcium-binding proteins that play crucial roles in immune response and inflammation [8]. Some S100A9 inhibitor, such as ABR-215757 and related quinoline-3-carboxamides, have shown promise in preclinical cardiovascular and autoimmune disease models [17]. And another S100A9 inhibitor, Paquinimod, has established safety and pharmacokinetics in early human trials and demonstrates therapeutic efficacy in animal models of fibrosis [18]. Our team also find that S100A9 promotes the progress of sepsis by participating in pulmonary microvascular leakage [10]. S100A9 is often upregulated in response to inflammatory stimuli, participating in the recruitment and activation of immune cells at sites of injury or infection [19]. Elevated levels of S100A9 have been associated with chronic inflammatory diseases, autoimmune disorders, and certain types of cancer [20]. S100A9 also correlates with plaque instability and vascular calcification. Both inflammation, plaque and vascular calcification is known to significantly impact the pathophysiology of aortic dissection through various mechanisms [21]. Thus, we further investigated the effects of S100A9 on aortic dissection using S100A9 KO mice. We observed that S100A9 was markedly increased in the aortic tissues of wild-type mice with induced dissection, while S100A9 knockout mice showed significantly improved outcomes. The enhanced survival rates and reduced aortic diameter in S100A9 knockout mice suggest that targeting this protein may offer a promising therapeutic strategy, which is consistent with a recent study using S100A9 inhibitor [11]. This highlights the potential of S100A9 as a therapeutic target for managing aortic dissection.
Our single-cell analysis provided additional insights into the cellular mechanisms by which S100A9 influences aortic dissection. The inhibition of S100A9 altered the composition of cell populations within the aortic tissue, favoring an increase in fibroblasts and smooth muscle cells, which are essential for vascular integrity. Concurrently, there was a reduction in immune and inflammatory cells, indicating that S100A9 plays a role in modulating the inflammatory environment associated with aortic dissection. This shift in cellular dynamics underscores the importance of S100A9 in maintaining vascular homeostasis.
Previous research has indicated that the elevation of nuclear factor kappa B (NF-
Emerging evidence suggests that S100A9 may regulate mitochondrial function indirectly through pattern recognition receptors such as Toll-like receptor 4 (TLR4) and receptor for advanced glycation end products (RAGE) [25, 26]. Engagement of these receptors activates NF-
Although the protective effects of S100A9 deficiency on AD, therapeutic inhibition of S100A9 should be cautiously considered, as its blockade may induce immunosuppressive effects, given its essential role in innate immunity. Therefore, while targeting S100A9 represents a promising strategy for preventing AD progression, careful evaluation of its safety profile, particularly the risk of immunosuppression, is warranted in future preclinical and clinical studies.
At last, a limitation needs to be pointed out. We used small human cohort size for proteomic analysis, so the future validation in larger, independent patient cohorts would strengthen our results.
In conclusion, our study elucidates the critical role of S100A9 in the development of aortic dissection, suggesting S100A9 inhibition may be a novel approach for preventing aortic dissection.
All data and methods are available from the corresponding author on a reasonable request.
KZ and LL interpreted data, prepared the figures and wrote the main manuscript text. KZ and YZ performed the main analysis and LL helped in bioinformatic analysis. KZ and KG performed the animal experiments. ZZ and MW helped with analysis and editing. XW, YZ and YG designed the study, edited the manuscript and earned the funds. XW and YZ were responsible for the clinical samples collection and supervised the project. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
The study involving human samples was conducted in accordance with the principles of the Declaration of Helsinki. Ethical approval was granted by the local ethics committee of the First Affiliated Hospital of Chongqing Medical University (approval number: 2024-173-02). The requirement for informed consent was waived by the committee due to the retrospective nature of the study and the use of anonymized data. All animal experiments were conducted in accordance with the National Institutes of Health Guidelines for the Use of Laboratory Animals and all animal procedures were approved by the Animal Care and Use Committee of Chongqing Medical University (approval number: IACUC-CQMU-2024-04098).
Not applicable.
This work was supported by the Chongqing education committee (KJQN202300480), the doctor “Through Train” project funding of Chongqing Wanzhou district (WZSTC-20220133), Program for Youth Innovation in Future Medicine, Chongqing Medical University (W0168).
The authors declare no conflict of interest.
During the preparation of this work the authors used ChatGpt-4 in order to check spell and grammar. After using this tool, the authors reviewed and edited the content as needed and takes full responsibility for the content of the publication.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/FBL44666.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.






