1 Institute of Genomic Medicine Sciences, King Abdulaziz University, 21589 Jeddah, Saudi Arabia
§Retired.
Abstract
The serine protease 23 (PRSS23) is a highly conserved member of trypsin-like serine proteases, which are associated with numerous essential processes, including digestion, blood coagulation, fibrinolysis, development, fertilization, apoptosis, and immunity. Original reports on PRSS23 unfolded not earlier than 2006 when a molecular biology study characterized and described PRSS23 as an ovarian protease. Then, in 2012, another important study was published linking PRSS23 with proliferation of breast cancer cells by an estrogen receptor 1 (ESR1)-dependent transcriptional activation of the serine protease. Thereafter, a developmental study in zebrafish reported the implication of PRSS23 in endothelial-to-mesenchymal transition (EndMT) during cardiac valve formation. Although these early studies on PRSS23 have revealed its involvement in some critical or fundamental processes, only in recent years an increasing number of studies have evolved describing the expression and functions of PRSS23 in various normal physiological conditions, diseases, and experimental configurations. Besides breast cancer, PRSS23 has been shown to be involved in different types of malignancies, e.g., in gastric cancer, where drug screening found that the protease inhibitor tipranavir impedes cancer-promoting PRSS23 expression. New innovative techniques such as single cell RNA-sequencing (scRNA-seq) and bioinformatics studies accelerated the discovery of gene expression changes in smaller cell populations, which, e.g., led to the identification of marked PRSS23 expression in a myofibroblast-like subpopulation in localized scleroderma. This review compiles major and significant research results that have contributed to our current knowledge of PRSS23 and briefly discusses where prospective studies could add to our understanding of this versatile serine protease.
Keywords
- PRSS23
- serine proteases
- estrogen receptor 1
- single cell RNA-sequencing
- expression analysis
- pathway analysis
- development
- endothelial-to-mesenchymal transition
- epithelial-to-mesenchymal transition
Serine proteases constitute the largest group of proteolytic enzymes. Well-known members of trypsin- and chymotrypsin-like serine proteases are trypsin, chymotrypsin, and elastase, which have been described and characterized by far earlier than serine protease 23 (PRSS23). The only paralogue of PRSS23 in human is PRSS35. Both proteases share some overlapping features.
A molecular biology study has initially described and characterized PRSS23 as an ovarian protease [1]. Other early but important studies include research on the involvement of PRSS23 in breast cancer and a developmental study in zebrafish reporting its involvement in cardiac valve formation, specifically its implication in endothelial-to-mesenchymal transition (EndMT) [2, 3].
EndMT is a biological process through which endothelial cells acquire a mesenchymal phenotype [4]. This process is similar to epithelial-to-mesenchymal transition (EMT), where the transition originates from epithelial cells. EndMT is critical for certain developmental and physiological processes but may also occur under certain pathological conditions, e.g., in cardiovascular disease [5]. In cancer, EndMT can lead to the development of cancer-associated fibroblasts (CAFs). Of notice, some herein reviewed studies have reported the implication of PRSS23 in either of these transition processes.
Given the fact that PRSS23 is highly conserved in vertebrates, it can be assumed that this serine protease is involved in some essential biological processes, and an accumulating number of reports have provided evidence for its versatile biological functions, positioning a framework for reviewing our current scientific knowledge of PRSS23. Therefore, the purpose of this review is to provide a comprehensive overview of the state-of-the-art research on PRSS23 and to address current gaps in our knowledge of PRSS23 that could be the subject of further studies.
PRSS23 (aliases SIG13, ZSIG13, or SPUVE) has been originally identified and characterized as an ovarian protease in 2006 by Miyakoshi et al. [1]. The PRSS23 gene is located in human on chromosome 11q14.2 [1]. The gene covers approximately 162 kb of genomic region and is transcribed into several mRNA variants. Transcript variant 1, human genome assembly (HG) 38 RefSeq NM_007173, is regarded as the default one [6]. This variant is composed of 3713 nucleotides and is the mature transcript of two exons. Exon 1 contains most of the 5′-untranslated region (5′-UTR), whereas exon 2 contains the 5′-UTR in the immediate proximity of the AUG start codon, the coding sequence, and the 3′-UTR. The coding sequence translates for a PRSS23 protein of 383 amino acids with a molecular weight of 43 kDa and a basal isoelectric point of 9.48.
PRSS23 is a member of the trypsin-like family of serine proteases and is highly conserved in vertebrates [1]. In serine proteases, three specific amino acids, i.e., serine, which contributed to the name serine proteases, as well as aspartic acid, and histidine are arranged in a defined catalytic triad and are functionally indispensable for the cleavage capacity of the active site. Trypsin-like proteases in general cleave peptide bonds at the C-terminal side of a positively charged amino acid. A negatively charged amino acid, located at the base of the substrate-binding pocket, is essential for high-level catalysis [7]. The catalysis is a hydrolytic reaction, where a water molecule acts as a nucleophile to break a chemical bond. Serine proteases exert essential biological functions, e.g., in digestion, blood coagulation, wound healing, and immunity. Eight surface loops, encircling the active site, regulate the substrate specificity and activity of trypsin-like serine proteases [8]. PRSS23 belongs to the S1A subfamily of serine proteases (EC: 3.4.21.-), which are synthesized as inactive precursors [9]. The signal peptide of PRSS23 comprises amino acids 1–19. Post-translational modifications of PRSS23 include phosphorylation, glycosylation, and ubiquitination. So far, experimental studies have detected PRSS23 in the nucleus and exosomes [2, 10]. PRSS23 is curated as a secretome protein [11].
In antisense orientation to the PRSS23 gene, the PRSS23-antisense RNA 1 (PRSS23-AS1) gene transcribes two long intergenic non-coding (linc) RNAs. PRSS23-AS1 has its lowest expression in the duodenum and bone marrow and highest expression in adipose tissue [12].
To assess the promoter regulation of the PRSS23 gene, first, the 2022 database of chromatin immunoprecipitation (ChIP)-X enrichment analysis 3 (ChEA3) for transcription factor (TF) targets was interrogated and second, the database of gene expression profiles for cell lines and tissues following TF knockdown or knockout was interrogated [13, 14, 15]. Both databases curate experimental datasets generated from various human and mouse tissues and cell types. Tissues and cell types include, e.g., embryonic stem cells (SCs), trophoblast SCs, placenta, uterus, macrophages, and cancer types include, e.g., breast, prostate, pancreatic, colon, and lung cancer, as well as leukemia and neuroblastoma. From the first dataset those TFs (n = 65) were extracted that were identified in low- and high-throughput functional studies as factors binding in proximity to the PRSS23 gene. From the second dataset those TFs (n = 21) were extracted that changed expression of PRSS23. Only three TFs, i.e., estrogen receptor 1 (ESR1), Fli-1 proto-oncogene/ETS transcription factor (FLI1), and POU class 5 homeobox 1 (POU5F1) were shared by both datasets.
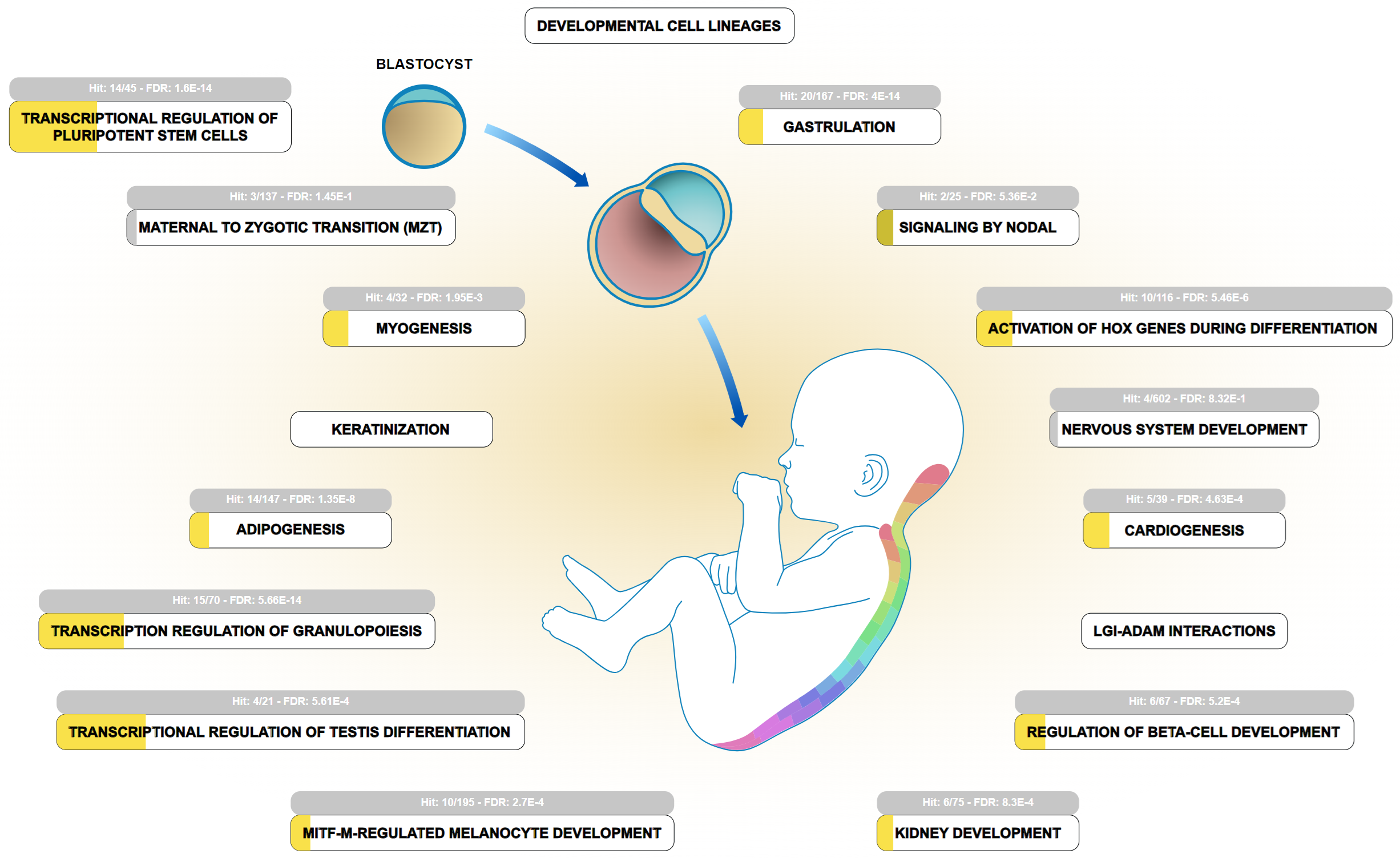
The 83 genes of the two combined datasets, and PRSS23, were analyzed using the statistical overrepresentation test in Reactome v. 90 [16, 17]. The most significantly overrepresented pathways are pluripotent SCs, RNA polymerase II transcription, generic transcription pathway, gene expression (transcription), and development biology (Fig. 1, Ref. [16, 17]) (Table 1) [17]. The developmental biology pathway includes annotation of early embryo processes, e.g., transcriptional regulation of pluripotent SCs and gastrulation, as well as annotation of later, more specialized processes, e.g., transcriptional regulation of granulopoiesis, and adipogenesis. The transcriptional regulation of PRSS23 through TFs that are overrepresented in these processes raises the hypothesis that PRSS23 may exert some proteolytic activities during developmental transition processes. In this regard, a recent single cell RNA-sequencing (scRNA-seq) study detected overexpression of PRSS23 in a defined cell cluster in a late differentiation stage during the transition from human induced pluripotent stem cells (hiPSCs) to trophoblast SCs [18].
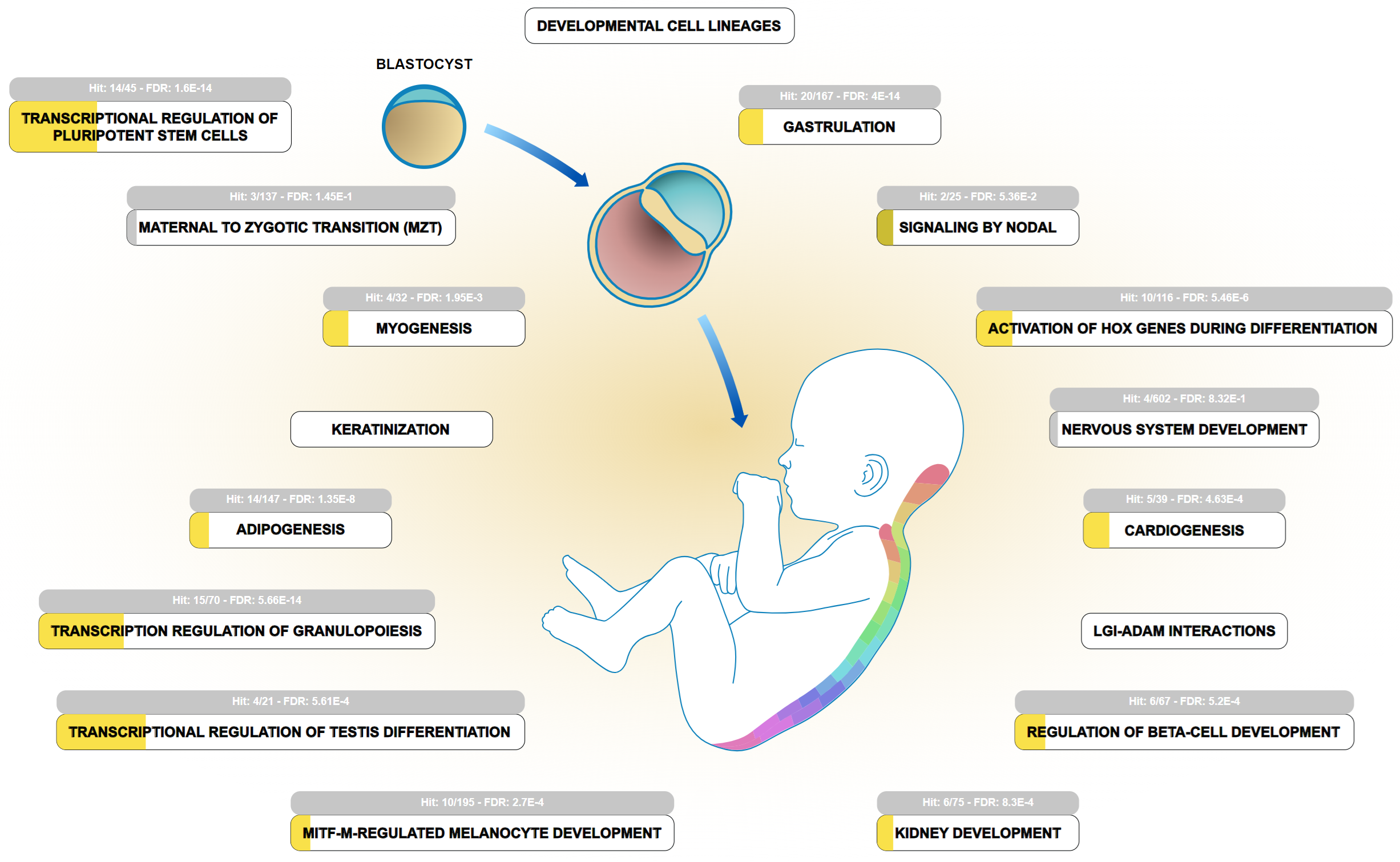 Fig. 1.
Fig. 1.
The developmental biology pathway is one of the most significant pathways related to TFs participating in PRSS23 gene regulation. The TFs were extracted from databases hosting datasets from chromatin immunoprecipitation assays and TF silencing assays. PRSS23 was added to the combined dataset uploaded to Reactome v. 90 [16, 17]. Some pathways, e.g., transcriptional regulation of pluripotent SCs, gastrulation, transcriptional regulation of granulopoiesis, and adipogenesis, are listed in Table 1 and serve as subpathways under the developmental biology pathway. The width of yellow color code bars in the subpathway icons represents the proportion of overrepresentation. Light gray color indicates insignificant overrepresentation. LGI-ADAM, leucine-rich glioma-inactivated-a disintegrin and metalloproteinase; MITF-M, microphthalmia-associated transcription factor-M.
| Pathways | Number TFs1 | FDR p-value2 |
| Transcriptional regulation of pluripotent stem cells | 8 | 1.60 × 10–14 |
| RNA polymerase II transcription | 43 | 1.60 × 10–14 |
| Generic transcription pathway | 42 | 1.60 × 10–14 |
| Gene expression (transcription) | 48 | 1.60 × 10–14 |
| Developmental biology | 47 | 1.60 × 10–14 |
| Gastrulation | 14 | 4.00 × 10–14 |
| Transcriptional regulation of granulopoiesis | 10 | 5.66 × 10–14 |
| Germ layer formation at gastrulation | 6 | 6.22 × 10–12 |
| Nuclear receptor transcription pathway | 6 | 1.63 × 10–11 |
| Interleukin-4 and interleukin-13 signaling | 9 | 2.28 × 10–9 |
| Regulation of PTEN gene transcription | 10 | 5.78 × 10–9 |
| Adipogenesis | 8 | 1.35 × 10–8 |
| Diseases of signal transduction by growth factor receptors and second messengers | 18 | 1.94 × 10–8 |
| Transcriptional regulation by RUNX1 | 13 | 1.28 × 10–7 |
| Specification of primordial germ cells | 4 | 1.28 × 10–7 |
| Estrogen-dependent gene expression | 12 | 1.76 × 10–7 |
| ESR-mediated signaling | 13 | 2.01 × 10–7 |
| Transcriptional regulation of white adipocyte differentiation | 7 | 3.79 × 10–7 |
| Transcriptional regulation by RUNX3 | 8 | 3.86 × 10–7 |
| PIP3 activates AKT signaling | 13 | 3.88 × 10–7 |
| Signaling by interleukins | 15 | 4.95 × 10–7 |
| Regulation of NFE2L2 gene expression | 4 | 1.11 × 10–6 |
| PTEN regulation | 10 | 1.44 × 10–6 |
| POU5F1 (OCT4), SOX2, NANOG activate genes related to Proliferation | 5 | 2.12 × 10–6 |
| Intracellular signaling by second messengers | 13 | 2.24 × 10–6 |
1Number TFs, number of uploaded transcription factors contained in a pathway; 2FDR p-value, false discovery rate p-value reporting results of the statistical overrepresentation test.
Further pathways include, e.g., interleukin-4 and interleukin-13 signaling, regulation of PTEN gene transcription, estrogen-dependent gene expression, and signaling by interleukins. It should be noted that these pathways are based on TFs that can be either transcriptional activators or repressors. For example, signal transducer and activator of transcription 1 (STAT1) is listed twice in the second dataset. Whereas STAT1 silencing in a skin cell line led to an increased PRSS23 expression, STAT1 silencing in an upper aerodigestive tract cell line led to a decreased PRSS23 expression. Furthermore, in the context of the second dataset, PRSS23 transcription may only be influenced indirectly through one or more other factors.
As of September 2024, the Biological General Repository for Interaction Datasets (BioGRID) database v. 4.4 curates 61 PRSS23-interacting proteins from human, mouse, and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (n = 52, 7, and 2 proteins, respectively) [19]. The two SARS-CoV-2 proteins are the accessory proteins open reading frame 8 (Orf8) and Orf9c, which are involved in immune evasion and antiviral response mechanisms [20, 21]. Given the fact that PRSS23 is moderately upregulated in endothelial cells under spike protein-stimulated conditions (section 2.8.1), it could be worth studying whether PRSS23 is implicated in response mechanisms provoked in the course of SARS-CoV-2 infection.
On the set of PRSS23 and its interacting proteins a Reactome v. 90 pathway
analysis was conducted using the Protein Analysis Through Evolutionary
Relationships (PANTHER) v. 19.0 knowledgebase [16, 22]. The resulting top
pathways, most significantly enriched in PRSS23 interacting proteins, are
implicated in various biological functions, e.g., membrane trafficking, viral
infections, diseases, and tumor suppressor TP53 regulation (Table 2). However,
immune- and infection-related pathways comprise nearly half of the listed
pathways. Although it has been reported that, e.g., the expression of
PRSS23 is enriched in gamma-delta (
| Pathways | Fold enrichment | FDR p-value1 | PRSS23 interacting proteins |
| Budding and maturation of HIV virion | 38.02 | 0.0248 | PPIA, CHMP4B, UBC |
| M Phase | 6.64 | 0.0274 | CEP152, UBE2, NCAPD3, PPP2R1A, CHMP4B, UBC, NIPBL |
| Modulation by Mtb of host immune system | 0.0284 | UBC, B2M | |
| Immune system | 2.71 | 0.0292 | PPIA, UBR1, UBE2I, CRP, PDPK1, APP, PPP2R1A, RACGAP1, CCL3, ACTG1, UBC, SOX2, B2M, CHI3L1, CRLF1, ACTB |
| Vesicle-mediated transport | 4.4 | 0.0295 | TGOLN2, LRP2, APP, RACGAP1, TRAPPC4, ACTG1, CHMP4B, UBC, ACTB |
| Regulation of TP53 expression and degradation | 29.57 | 0.031 | PDPK1, PPP2R1A, UBC |
| Membrane trafficking | 5.1 | 0.031 | TGOLN2, LRP2, APP, RACGAP1, TRAPPC4, ACTG1, CHMP4B, UBC, ACTB |
| Regulation of TP53 degradation | 30.41 | 0.031 | PDPK1, PPP2R1A, UBC |
| SARS-CoV-1-host interactions | 14.78 | 0.0313 | PPIA, UBE2I, PDPK1, UBC |
| Disease | 2.92 | 0.034 | FXR1, PPIA, UBE2I, SEC11C, PDPK1, APP, PPP2R1A, ACTG1, CHMP4B, UBC, PDGFRA, SLC25A6, B2M, SNW1, ACTB |
| Infectious disease | 3.76 | 0.034 | PPIA, UBE2I, SEC11C, PDPK1, APP, ACTG1, CHMP4B, UBC, SLC25A6, B2M, ACTB |
| Signaling by receptor tyrosine kinases | 6.19 | 0.0342 | COL5A1, BAX, PDPK1, PPP2R1A, ACTG1, TCF12, UBC, PDGFRA, ACTB |
| Clathrin-mediated endocytosis | 12.32 | 0.0348 | TGOLN2, LRP2, ACTG1, UBC, ACTB |
| SARS-CoV-1 infection | 12.58 | 0.042 | PPIA, UBE2I, PDPK1, CHMP4B, UBC |
| Cytokine signaling in immune system | 4.16 | 0.0426 | PPIA, UBE2I, APP, PPP2R1A, CCL3, UBC, SOX2, B2M, CRLF1 |
| Signaling by interleukins | 5.41 | 0.043 | PPIA, APP, PPP2R1A, CCL3, UBC, SOX2, CRLF1 |
| Diseases of signal transduction by growth factor receptors and second messengers | 6.39 | 0.043 | FXR1, PDPK1, PPP2R1A, ACTG1, UBC, PDGFRA, SNW1, ACTB |
1FDR, false discovery rate p-value.
Although several human microRNAs (miRNAs) have been predicted to bind to PRSS23 sequences, evidence to bind to PRSS23 and affect its expression has only been revealed so far for a few miRNAs [24]. For example, two miRNAs, i.e., miR-192-5p and miR-215-5p are microarray-validated PRSS23 mRNA interactors from the miRNA targets dataset of the MiRTarBase 8.0, whereas miR-532-5p and miR-1246 have been shown in two herein reviewed studies to directly interact with PRSS23 mRNA sequences in luciferase reporter assays [24, 25, 26].
A single cell transcriptomics dataset analysis, which was conducted by the Human
Protein Atlas project, detected enhanced PRSS23 expression particularly
in lymphatic endothelial cells, ionocytes, basal respiratory cells, granulosa
cells, endothelial cells, and club cells [27, 28]. An RNA-sequencing (RNA-seq)
profiling study performed by the Human Protein Atlas project in different types
of immune cells found an enriched expression of PRSS23 in
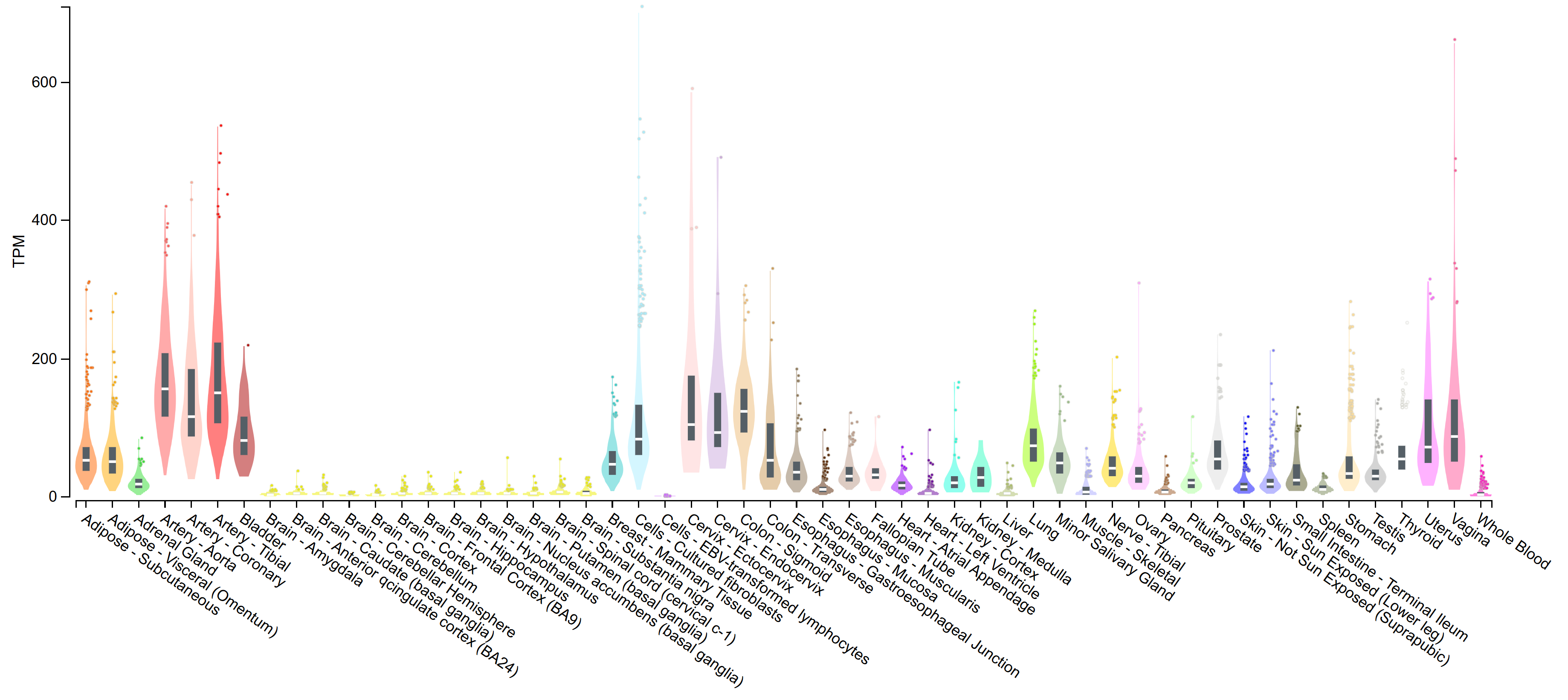
The Genotype-Tissue Expression (GTEx) portal, curating tissue expression data from approximately 1000 adult individuals, was utilized to provide a detailed overview of the tissue expression of PRSS23 [28, 32, 33]. The serine protease exhibits a wide range of tissue expression in humans with the highest levels examined in some major arteries suggesting the involvement of PRSS23 in vascular biology (Fig. 2, Ref. [32]).
 Fig. 2.
Fig. 2.
Expression of PRSS23 mRNA in 54 tissue compartments and cultured cells. Highest PRSS23 expression is determined in the aorta, tibial artery, coronary artery, sigmoid colon, endo- and ectocervix, bladder, cultured skin fibroblasts, vagina, uterus, and lung. High expression of PRSS23 in some tissue compartments might suggest further biological roots and functional implications. TPM, transcripts per million. The GTEx portal refers to analysis release V8, dbGaP, accession phs000424.v2.p1 [32].
To identify the abundance of protein classes for genes that are most similar to
PRSS23 in terms of co-expression, the top 100 co-expressed genes were
downloaded from the ARCHS4 gene expression collection [34]. Using the
overrepresentation test for the protein class annotation dataset of the PANTHER
v. 19.0 knowledgebase, the following protein classes were found to be
significantly (p
Research performed by Chen et al. [3] in zebrafish detected prss23 expression during cardiac valve formation in the ventricle, atrium, and atrioventricular canal. Morpholino knockdown of Prss23 rendered the EndMT at the atrioventricular canal, and further experiments indicated that snail family transcriptional repressor 1 (SNAI1, alias SNAIL) is functionally located downstream of PRSS23 during EndMT. Subsequently, applying human PRSS23 and SNAI1 in zebrafish embryos revealed that both molecules were functionally competent indicating the evolutionarily conserved role of PRSS23 in valvulogenesis.
In situ hybridization in mouse ovary localized Prss23 mRNA expression primarily in granulosa cells of the secondary/early antral follicles [1]. In contrast, Prss23 expression was not detectable in primary follicles, and expression was low in antral follicles. This led to the suggestion that Prss23 is possibly involved in transition processes of pre-antral to antral follicles. A complementary study in mouse ovary employed a terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay to assess apoptosis and detected upregulated Prss23 expression specifically in atretic follicles [35].
In situ hybridization in uterine compartments at preimplantation gestation day 3.5 (D3.5) of wild-type (wt) mice detected Prss23 expression only in the uterine luminal epithelium [36]. A knockout mouse model that has a delayed implantation period was used as reference. Prss23 was downregulated in the luminal epithelium upon embryo implantation, which normally initiates around D4.0 in wt mice.
Employing scRNA-seq to investigate PRSS23 expression in single endothelial cells, PRSS23 has been described as a new marker for venule endothelial cells in adult human brain, and, based on an expression gradient along the arteriovenous axis, PRSS23 has been reported as a pulmonary endothelial vein marker in mice [37, 38].
Aiming to identify new bone metabolism-related genes, an scRNA-seq study was conducted on primary femoral head tissue cells derived from patients who had undergone hip replacement surgery [39]. Based on the differentially expressed gene (DEG) set from osteoblastic lineage cells and a protein-protein interaction network analysis, the researchers found that expression levels of two new bone metabolism-related genes, namely PRSS23 and matrix remodeling associated protein 8 (MXRA8), were significantly upregulated during in vitro osteogenic differentiation. Particularly, PRSS23 was highly expressed in osteoblast precursors and in early osteoblasts.
An experimental study, aiming to improve development of in vitro-produced bovine embryos, found that conditioned media containing progesterone-stimulated endometrial cells or histotrophic molecules present in the conditioned media that included bovine PRSS23 may be suitable for improving in vitro-produced embryo development [40].
Forde et al. [41] studied in a same research field the effect of low progesterone levels on conceptus elongation by comparing the endometrial transcriptome of beef heifers with low progesterone levels to those of a control group with normal progesterone levels. Subsequently, quantitative real time-polymerase chain reaction (qRT-PCR) and in situ hybridization revealed that bovine PRSS23 expression was confined to the luminal epithelium, and expression increased from day 7 to day 13 of the estrous cycle, especially in the control group. The researchers noted that PRSS23 could be involved in breaking down proteins into amino acids that can serve as nutrients for the conceptus during elongation.
A wide range of recently conducted molecular biology studies has identified PRSS23 expression pattern and functions in various non-cancerous diseases and conditions, expanding the spectrum of PRSS23 implications beyond previously known indications. Section 2.8 includes studies on some pandemic or common diseases and conditions such as viral infections, cardiac heart failure, diabetes, and Alzheimer’s disease. Drug-related and toxicological studies in cardiac tissue, oocytes, and liver tissue are listed in sections 2.10.1, 2.10.6, and 2.10.7.
A number of molecular expression assays were employed in a research study to assess whether the spike protein of SARS-CoV-2 induces endothelial inflammation via binding to angiotensin converting enzyme 2 (ACE2) [42]. Besides activation of pro-inflammatory molecules and signaling, proteome analysis revealed that PRSS23 is one of the moderately upregulated proteins in endothelial cells under spike protein-stimulated conditions. In this context, it might be worth studying the function of PRSS23 during immune responses in view that the protease is capable of interacting with the accessory proteins Orf8 and Orf9c of SARS-CoV-2, which are known to be involved in immune evasion and antiviral response mechanisms (section 2.3).
An RNA-seq study investigated the host responses to influenza and vaccinia viruses in different types of immune cells, e.g., peripheral blood mononuclear cells (PBMCs), monocytes, B cells, and cluster of differentiation 8+ (CD8+) T cells [43]. For this purpose, the different types of immune cells were separated in vitro from peripheral blood obtained from healthy donors who previously had received both seasonal influenza and smallpox vaccine. Bioinformatics analysis then identified PRSS23 as one of the top downregulated genes in influenza-stimulated CD8+ T cells compared to unstimulated controls.
A research team studied the mechanisms underlying the unsafety of Newcastle disease virus (NDV) vaccine for in ovo vaccination [44]. Among five trypsin-like proteases that can activate NDV in chicken embryos, complement factor D (Cfd) and Prss23 were found to be accountable for the broad tissue tropism and high pathogenicity of the NDV in chicken embryos.
The functional significance of exosomal miR-1246 and PRSS23 in the context of cardiac heart failure (CHF) was studied by Wang et al. [26] using cell culture experiments and a CHF rat model. For these experiments, exosomes were utilized from human umbilical cord mesenchymal stem cells (hucMSCs) following oxygen and glucose deprivation. Besides equivalent in vitro findings, the researchers demonstrated that hucMSC exosomes could limit myocardial injury and promote angiogenesis in CHF rats. In this rat model, Prss23 expression in myocardial tissue was significantly increased after left anterior descending artery surgery; however, decreased upon hucMSC exosome application. Furthermore, a dual-luciferase reporter assay illustrated that mir-1246 binds to PRSS23 mRNA sequences. In addition, the presence of miR-1246 in hucMSC-derived exosomes could be demonstrated, while application of an miR-1246 inhibitor could reverse the protective function of hucMSC exosomes. In this regard, application of MSC-derived exosomes is widely assessed as a therapeutic option for repairing cardiac injuries [45].
A meta-analysis of scRNA-seq data on pancreatic islets from T2DM donors and
metabolically healthy donors identified sets of DEGs and upstream regulators in
To gain more insight into the complex pathobiological features of Alzheimer’s disease, a transcriptome profiling study was conducted in the hippocampus and retrosplenial cortex of an Alzheimer’s disease mouse model [47]. Both brain regions are involved in memory processes. Male and female mice were investigated separately and compared to each other, and all categories were compared to the respective categories in control mice. A distinct gender- and tissue-specific gene signature was revealed and notably, PRSS23 was only significantly upregulated in the hippocampus of female Alzheimer’s disease mice.
A number of scRNA-seq studies investigated the composition of specific cell clusters and cell populations in localized scleroderma and systemic sclerosis to identify proteins and processes underlying the disease.
In an scRNA-seq study, conducted in localized scleroderma and healthy skin control samples, the researchers identified different fibroblast subclusters [48]. One of the identified subclusters was a myofibroblast-like subcluster that was characterized by the profoundly expressed markers secreted frizzled related protein 4 (SFRP4) and PRSS23 and by containing features shared with SSc. This subcluster was more abundant in localized scleroderma than in control samples.
An scRNA-seq profiling study in SSc and healthy skin identified a fibroblast cluster that was mostly confined to SSc fibroblasts and defined by upregulation of PRSS23 and secreted frizzled related protein 2 (SFRP2) expression [49]. Gene ontologies related to this fibroblast cluster included, e.g., ECM and structure organization and collagen fibril organization. In SSc, these fibroblasts represent progenitor cells of myofibroblasts, and the transition to myofibroblasts most likely occurs via transforming growth factor beta (TGFB) pathway signaling. In a similar context, high level of PRSS23 expression in myofibroblasts was observed in a more recent study on SSc [50]. The myofibroblasts originated from a fibroblast subtype, characterized by high expression of collagen genes, and Hippo signaling pathway factors were likely promoters of the transition process as well as EndMT.
It should be noted that markedly increased expression of PRSS23 in specific subpopulations of localized scleroderma and SSc was identified in studies using scRNA-seq. In contrast, a larger microarray study, which used skin biopsies and was analyzed by employing the GEO2R tool, detected only a moderate 1.7-fold overexpression of PRSS23 in SSc compared to control samples (Gene Expression Omnibus accession no. GSE58095) [51].
In a mouse model for kidney fibrosis, LeBleu et al. [52] found that the whey acidic protein (WAP) four-disulfide core domain 2 (Wfdc2, alias HE4) protease inhibitor is highly upregulated in fibrosis-associated myofibroblasts compared to the normal control. WAP domain-containing proteins are known to inhibit a wide range of serine and cysteine proteases. Among other upregulated molecules in fibrosis-associated fibroblasts and fibrotic kidney were some ECM proteins as well as Prss23 and Prss35. In a hydroxyproline release assay, recombinant human WFDC2 was capable of partially inhibiting Prss23 and Prss35 from degrading type I collagen. The inhibitory activity could be reversed by an anti-WFDC2 antibody. WFDC2 expression is present in the epididymis and in a number of other tissues, e.g., in regions of the respiratory tract and in some lung cancer cell lines [53].
A research team investigated the molecular pathogenesis of IgA nephropathy by performing scRNA-seq on PBMCs and kidney cells from three pediatric patients [54]. IgA nephropathy is characterized by IgA deposits in the glomerular mesangium. The results revealed an increased expression of PRSS23, C-C motif chemokine ligand 2 (CCL2), and EMT-related genes in podocytes from the patients compared to control samples.
To assess new therapeutic approaches for focal segmental glomerulosclerosis (FSGS), a study team performed RNA-seq analysis in podocytes, mesangial cells, and glomerular endothelial cells of mutant mouse models for FSGS [55]. The results showed that Prss23 was strongly upregulated in podocytes of mutant mice, which had a homozygous knockout of the Cd2 associated protein (Cd2ap), and Prss23 was also strongly upregulated in mesangial cells of double mutant mice, which had a heterozygous knockout of Cd2ap and a homozygous knockout of the Fyn proto-oncogene (Fyn). The researchers suggested that upregulated PRSS23 might bear important functions in FSGS.
With the objective to gain insight into the molecular pathogenesis of pterygium, which is a triangular growth of fleshy tissue on the conjunctiva, a research team interrogated publicly deposited expression datasets [56]. Subsequent qRT-PCR analysis in pterygium from patients confirmed the presence of PRSS23 among six significantly upregulated genes compared to control samples.
A study, applying proteomic profiling of brain parenchymal micro vessels in cerebral amyloid angiopathy type 1, detected, among other molecules and compared to controls, accumulation of the high-temperature requirement protein A1 (HTRA1), serum amyloid P component (encoded by the APCS gene), and PRSS23 [57]. HTRA1 is a serine protease showing in an assay proteolytic activity against APCS and PRSS23.
Research in breast and gastric cancer is currently one of the main topics for studying PRSS23 implications in malignancies. Drug-related research in breast and gastric cancer, lung adenocarcinoma, esophageal squamous carcinoma, and Ewing sarcoma is discussed in section 2.10.
An immunohistochemical study conducted by Chan et al. [2] in breast cancer specimens detected PRSS23 expression in the nucleoplasm, and this expression was on average profoundly higher in ESR1-positive breast cancer compared to ESR1-negative breast cancer. In subsequent research, different PRSS23 promoter constructs were used to detect the critical estrogen-responsive region of the PRSS23 promoter and upstream region. The most increased luciferase activity was obtained using a promoter construct containing approximately 2 kb upstream of the AUG start codon, which increased the activity by 40% in 17beta-estradiol-treated Michigan Cancer Foundation-7 (MCF-7) breast cancer cells compared to hormone-starved cells. A longer and two shorter promoter constructs showed less or no activity. ChIP assays then demonstrated that ESR1 directly interacts with a PRSS23 upstream promoter region. Furthermore, employing colony formation assays in MCF-7 breast cancer cells, PRSS23 silencing reduced tumor diameter on average by 30% compared to PRSS23-unsilenced controls. The authors noticed that the results of the study may provide a basis to evolve PRSS23 into a prognostic or therapeutic target for breast cancer.
In this regard, it is worth noting that PRSS23 was included in a pre-selected set of 36 genes, which showed prognostic significance in patients with Tamoxifen-treated, ESR1-positive breast cancers [58]. In contrast, this gene set had no prognostic significance in non-hormone-treated breast cancer patients.
A bioinformatics analysis of a publicly deposited microarray expression dataset assessed expression changes in MCF-7 breast cancer cells at 12, 24, and 48 h time points after 17beta-estradiol treatment [59]. PRSS23 was upregulated at all three time points and regarded as the central node of assembled gene networks that were related to signaling pathways including epidermal growth factor (EGF) receptor family (Erb), wingless-related integration site (Wnt), mitogen-activated protein kinase (MAPK), and inositol trisphosphate (IP3) pathways.
RNA binding proteins (RBPs)-focused CRISPR-Cas9 screening detected several RBPs, which have particular functions in promoting proto-oncogene MYC-driven cancer pathways [60]. A majority of the detected RBPs revealed an increased expression in MYC-amplified, basal-like triple negative breast cancer (TNBC) compared to other breast cancer types. Furthermore, prognosis of TNBC patients was more favorable when tumors with on average higher MYC levels had lower levels of YTH N6-methyladenosine RNA binding protein F2 (YTHDF2). YTHDF2 is an mRNA decay factor and N-6-methyladenosine (m6A) reader protein. Subsequent research revealed that depletion of YTHDF2 resulted in proteotoxic stress and in higher expression of PRSS23, which otherwise is subjected by YTHDF2 to mRNA decay, whereas co-depletion of YTHDF2 and PRSS23 regained proliferation rates of TNBC cells.
Using immunohistochemistry, Han et al. [61] found that low expression of PRSS23 in gastric cancer correlated with favorable prognosis. In a xenograft gastric cancer model, gastric cancer cells harboring a short hairpin (shRNA)-mediated knockdown of PRSS23 showed less tumor weight and volume compared to control mice. Subsequently, microarray expression data of gastric cancer cells, bearing the PRSS23 knockout, showed a number of significantly differentially expressed pathways compared to control including downregulation of the following processes and signaling pathways: estrogen-mediated S-phase entry, eukaryotic initiation factor 2 (EIF2), bone morphogenetic protein (BMP), CD40, and ultraviolet A (UVA)-induced MAPK. Corroborative with these findings, PRSS23 knockdown inhibited cell viability and this was reversed by overexpressing eukaryotic translation initiation factor 4E (EIF4E). The authors stated that PRSS23 has prospects of serving as a target for gastric cancer treatment.
An analysis of The Cancer Genome Atlas (TCGA) datasets from gastric cancer by Qin et al. [62] indicated that PRSS23 overexpression is significantly associated with shorter overall and disease-free survival. Furthermore, scratch wound healing and transwell invasion assays demonstrated that small interfering RNA (siRNA)-mediated knockdown of PRSS23 significantly impaired migration and invasion of gastric cancer cells. In addition, PRSS23 knockdown in gastric cancer cells decreased expression of secreted fibroblast growth factor 2 (FGF2). Then, co-culture experiments of gastric cancer cells with tumor-associated macrophage (TAM)-like cells, indicated that PRSS23 promotes TAM/M2 macrophage infiltration by positively regulating FGF2 expression and secretion in a yet to be defined manner.
An enriched pathway analysis was conducted on DEGs derived from a publicly deposited microarray dataset of stage I and II renal cell carcinoma (RCC) [63]. This analysis found that six pathways, i.e., phosphoinositide 3-kinase (PI3K)/Akt, fork head box O (FOXO), endocytic, MAPK, tight junction, and cytokine-cytokine receptor interaction pathways were among the most common signaling cascades altered in stage I and II RCC. PRSS23 and several other genes were then accounted for the observed pathway alterations.
Transfection of miR-30c-5p and miR-30c-2-3p, known to be downregulated in pancreatic ductal adenocarcinoma, into pancreatic cancer cell lines inhibited cell proliferation, migration, and invasion [64]. PRSS23 and DNA topoisomerase II alpha (TOP2A) were among 18 genes that were regarded as putative targets of miR-30c-2-3p regulation; however, a luciferase reporter assay was conducted only for TOP2A, indicating a direct binding of miR-30c-2-3p to 3′-UTR sequences of TOP2A. Furthermore, PRSS23 and TOP2A were included in a set of 10 genes, whose high expression was found to be an independent prognostic factor for short-term survival of patients.
An scRNA-seq study identified in non-small cell lung cancer (NSCLC) a CAF subcluster, which produces the ECM protein periostea, encoded by the periostin (POSTN) gene. This subcluster promotes various aspects of NSCLC progression by establishing an immunosuppressive tumor microenvironment in connection with a specific subset of TAMs [65]. PRSS23 was one of the top 10 upregulated genes in POSTN-positive CAFs, which were associated with lower T cell infiltration in NSCLC and more unfavorable prognosis of patients.
Using datasets from the TCGA database and other publicly available resources, 15 overexpressed and prognostically relevant genes in pleural mesothelioma were identified through a selection process that included univariate and multivariate Cox regression analyses [66]. Among the selected genes, a subset of genes, including PRSS23, was found to be abundantly expressed in malignant pleural mesothelioma cell lines, and therefore it was noticed that the clinical relevance of these genes should be further assessed.
The molecular mechanisms underlying the malignant transformation of oral submucous fibrosis to oral squamous cell carcinoma were investigated in a transcriptomic and metabolomic profiling study [67]. This study revealed a landscape of intratumoral heterogeneity. Significant expression of some mesenchymal markers, including PRSS23, was observed in those oral squamous cells that underwent a partial EMT.
Analysis of microarray expression datasets and weighted gene co-expression network analysis (WGCNA) identified six upregulated and three downregulated key genes associated with papillary thyroid cancer (PTC) [68]. PRSS23 was one of the upregulated key genes, and, based on the gene expression signature, the estimated abundance of immune infiltration indicated that expression of four key genes, including PRSS23, positively correlated with the expression of B cells, CD4+ T cells, neutrophils, and dendritic cells. Furthermore, PRSS23 was also included in predicted relationships between key genes and regulatory miRNAs.
Based on a microarray expression study and employing iterative algorithms, PRSS23, named SPUVE in the study, was included in a classifier set of 19 genes that was competent to most accurately discriminate PTC from normal thyroid samples [69].
Bioinformatics analysis of publicly accessible microarray expression and scRNAseq datasets from anaplastic thyroid carcinoma (ATC) showed that expression of sialic acid binding Ig like lectin 15 (SIGLEC15) was highest among SIGLEC family members [70]. ATC cells with high SIGLEC15 expression were characterized by high expression of PRSS23 and cancer stem cell (CSC) marker CD44 as well as comparable higher interaction with cells from the tumor microenvironment, including endothelial cells and T cells. An anti-SIGLEC15 antibody, administered to zebrafish xenograft model and ATC-bearing mice, was capable of significantly reducing tumor growth.
Transcriptome profiling of vestibular schwannomas revealed enrichment of overexpressed metal- and serine proteases, including PRSS23, in the nonmyelinating, aggressive form of vestibular schwannomas [71]. This led to the suggestion that overexpressed proteases are in this context molecular drivers of ECM remodeling.
Point mutations in hotspot regions of the isocitrate dehydrogenase 1 (IDH1) and IDH2 are major oncogenic drivers in low-grade gliomas and to a lesser degree in some other malignancies [72]. A meta-analysis on publicly deposited datasets that compared expression profiles between IDH-mutant and IDH-wt conditions in six isogenic disease models found significantly higher expression of PRSS23 in four of the six model systems. The researchers noted that this observation may gain relevance for further research in IDH-mutant tumors.
To identify new molecular mechanisms responsible for ultraviolet B (UVB) induced-skin carcinogenesis, cultures of keratinocytes and melanocytes were irradiated with UVB [73]. RNA-seq detected PRSS23 as one of the downregulated genes in both keratinocytes and melanocytes from different individuals, whereas, e.g., the tumor suppressor cyclin dependent kinase inhibitor 1A (CDKN1A) was upregulated under the same conditions.
Aiming to identify biomarkers in osteosarcoma cells related to processes of matrix mineralization, a research team analyzed a publicly accessible microarray expression dataset by comparing DEGs between osteosarcoma cells attached to the demineralized osseous surfaces and those attached to the mineralized osseous surfaces [74]. PRSS23 was identified as one of 12 hub genes upregulated in the demineralized component; however, only the hub gene transforming growth factor beta receptor 1 (TGFBR1) possessed prognostic significance.
To detect gene signatures of CSCs in comparison to those found in hiPSCs, a study induced CSCs from hiPSCs using either conditioned media from cancer cell lines or primary cell cultures of cancer tissues that were incubated with reprogramming factors [75]. Gene expression analysis and data filtering identified PRSS23 as one of several genes upregulated in CSCs compared to hiPSCs.
The response mechanism of PRSS23 and miR-532-5p to the application of the
Using publicly accessible gene expression datasets and bioinformatics analysis, Zeng et al. [76] established a time course of gene expression changes in MCF-7 breast cancer cells that develop upon tamoxifen treatment. PRSS23 was strongly downregulated in approximately the first four weeks of tamoxifen treatment and thereafter expression levels recovered nearly to those in control samples. Notably, 46 genes, affected by tamoxifen treatment, are listed in the BioGRID database as PRSS23 interacting proteins.
A meta-analysis was conducted on microarray expression datasets derived from breast cancer cell lines to identify genes that are likely involved in acquired lapatinib resistance [77]. Lapatinib is an anti-cancer drug that inhibits signaling of erb-b2 receptor tyrosine kinase 2 (ERBB2) and epidermal growth factor receptor (EGFR). A co-expression and interaction network survey could condense the DEG set to six deregulated key genes. Four of the six genes were upregulated, i.e., PRSS23, galectin 1 (LGALS1), caveolae associated protein 1 (CAVIN1, alias PTRF), and four and a half LIM domains 2 (FHL2), whereas two genes were downregulated, i.e., transducer of ERBB2, 1 (TOB1), and suppressor of cytokine signaling 2 (SOCS2).
In search for a new gastric cancer biomarker, Xiong et al. [78] identified high expression of PRSS23 in gastric CSCs compared to differentiated cells derived from serum-induced gastric CSCs. High expression of PRSS23 was then found to significantly correlate with poor prognosis of gastric cancer patients. Further experiments, employing different human immunodeficiency virus (HIV) protease inhibitors as well as the chemotherapeutic agents 5-fluorouracil and cisplatin, indicated that the HIV drug tipranavir significantly and effectively inhibited the viability of the gastric CSCs at low concentrations. A gastric CSC xenograft model substantially sustained the in vitro findings. Of notice, cell culture experiments indicated that tipranavir exhibits also an inhibitory viability effect on lung, esophageal, colorectal, breast, liver, and paclitaxel-resistant prostate cancer cells. Otherwise, normal gastric epithelial cells were insensitive to tipranavir treatment. Further experiments in gastric cancer cells and gastric CSCs indicated that tipranavir treatment, by decreasing the expression of PRSS23, induces the activation of the interleukin-24-dependent mitochondrial apoptotic pathway.
Tipranavir is a sulfonamide-containing dihydropyrrole co-administered for inhibiting the processing of viral Gag and Gag-Pol polyproteins in HIV-1 infected cells [79]. Specifically, tipranavir blocks the cleavage of the viral polyprotein precursors into active HIV-1 proteins by binding to the catalytic site of the HIV-1 protease [80]. One of the advantages of tipranavir is its capacity to generate a structure of especially strong hydrogen bonds with invariable regions of the HIV-1 protease, and this capacity is preserved in HIV-1 mutants [81]. Tipranavir was approved for the first time in June 2005 for the treatment of HIV-1 infections. Perinatal HIV clinical guidelines issued for tipranavir include the recommendation that the drug should not be used during pregnancy [82].
To identify patients who may benefit from cisplatin-containing neoadjuvant chemotherapy in lung adenocarcinoma and esophageal squamous carcinoma, the researchers established a neoadjuvant chemotherapy score where higher values indicated a higher sensitivity to chemotherapy [83]. This score was established on 12 genes, including PRSS23, which originated through a selection process from a larger set of genes that were differentially expressed between pre- and post-chemotherapy.
A research team investigated the resistance mechanisms to the small molecule inhibitor YK‑4‑279 that is observed in those Ewing sarcomas carrying the fusion gene EWS‑FLI1 [84]. An Ewing sarcoma cell line could be established showing resistance to the inhibitor. Subsequently, RNA-seq detected PRSS23 as one of the three most downregulated genes in the resistant cell line.
A research team analyzed by liquid chromatography-mass spectrometry the proteome of culture media containing mouse oocytes within the primordial/primary follicles to characterize mechanisms that could protect oocytes from damages induced by cancer chemotherapy [85]. Prss23 was one of the compounds that became less abundant under cisplatin-induced apoptotic conditions, while either luteinizing hormone (LH) or LH + cisplatin-induced conditions prevented Prss23 expression from decreasing.
In a proteomic study, a model system consisting of normal hepatocytes and MYC-silenced hepatocytes was employed to identify biomarkers for carcinogenic effects caused by fine particulate matter in liver tissue [86]. PRSS23, which was downregulated in normal hepatocytes and upregulated in MYC-silenced hepatocytes, and solute carrier family 39 member 10 (SLC39A10) were identified as putative biomarkers.
In human, PRSS35 is located on chromosome region 6q14.2 and is the only PRSS23 paralogue. Although PRSS35 exhibits some overlapping expression features with PRSS23, it acts under certain conditions as a tumor suppressor. In hepatocellular carcinoma, secreted PRSS35 mediated degradation of the chemokine C-X-C motif chemokine ligand 2 (CXCL2), which led to the impairment of pro-tumor functions of neutrophils [87].
In mouse CAFs, Prss35 was identified as one of the top upregulated genes [88]. Subsequent research in mouse models of skin carcinogenesis, harboring a Prss35 knockout gene, revealed aberrant collagen composition, i.e., increased thick collagen fiber deposits in the ECM and elevated tumor formation compared to control mice.
Despite wide expression of Prss35 in mouse ovaries, Prss35 knockout mice were fertile indicating that Prss35 is nonessential for fertility, and the unaffected litter size of Prss35 knockout mice was comparable to that of wt mice [36]. Wahlberg et al. [35] found a different expression regulation for Prss23 and Prss35 during ovulation in mice. Whereas Prss23 was downregulated by gonadotropins, Prss35 was upregulated.
The serine protease PRSS23 exerts diverse functions under healthy conditions and in various diseases. Specifically, its involvement in critical developmental and reproductive processes leverages the serine protease to a gene of interest. In this regard, the introduction of new molecular technologies, like the recently innovated scRNA-seq technique, is adding to identifying the abundance of PRSS23 expression in a narrow spatiotemporal and cell-specific context, which in turn assists in unraveling its functional implications.
In cancer, PRSS23 expression has been associated with unfavorable prognosis in a number of malignancies, including head and neck, renal, and pancreatic cancer [11]. However, PRSS23 was initially not detected as a specific cancer gene. For example, the Catalogue for Somatic Mutations in Cancer (COSMIC) database notices only infrequent and simple coding mutations for PRSS23 exhibiting no specific mutational signatures and no fusion events [89]. Likewise, copy number variations (CNVs) affecting the PRSS23 gene are not very common. The highest percentages of CNVs are less than 17% per entity and are observed in anaplastic melanoma, lung atypical carcinoid tumor, and lactotroph pituitary neuroendocrine tumor [90]. However, its capacity to participate in key cancer pathways, e.g., in EMT, can be a critical component for promoting cancer. Furthermore, PRSS23 has been repeatedly implicated in EndMT, and in recent years it has become increasingly evident that EndMT is resistant to various forms of cancer therapies and therefore represents a likely target for therapeutic intervention [91]. In addition, functional involvement of PRSS23 in essential developmental processes could be prone to regain functional implications in cancer development or progression; however, therapeutic agents targeting PRSS23 may interfere under certain conditions with such developmental processes during pregnancy.
Further research is required to characterize PRSS23 in more detail, e.g., to determine its substrate specificity and activity. This research is critical for assessing inhibitory proteins and compounds to establish a spectrum of prospective therapeutic applications, e.g., to survey PRSS23 inhibitors for anti-fibrotic therapies. In addition, the function of PRSS23 could be specifically studied using PRSS23 knockout mice, to investigate its dispensable or indispensable significance in developmental processes, or diseases more comprehensively.
ATC, anaplastic thyroid carcinoma; BioGRID, Biological General Repository for Interaction Datasets; CAF, cancer-associated fibroblast; CD, cluster of differentiation; ChEA3, ChIP-X enrichment analysis 3; CHF, cardiac heart failure; ChIP, chromatin immunoprecipitation; CNV, copy number variation; COSMIC, Catalogue for Somatic Mutations in Cancer; CSC, cancer stem cell; DEG, differentially expressed gene; ECM, extracellular matrix; EMT, epithelial-to-mesenchymal transition; EndMT, endothelial-to-mesenchymal transition; FDR, false discovery rate; FSGS, focal segmental glomerulosclerosis;
All data presented are derived from referenced, publicly available resources.
HJS concepted and designed the review study, collected and analyzed data, wrote the manuscript, and approved the final manuscript. The author agreed to be accountable for all aspects of the work.
Not applicable.
I gratefully acknowledge the support provided to conduct the review.
This research received no external funding.
The author declares no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.