1 College of Grassland Science, Xinjiang Agricultural University, 830052 Urumqi, Xinjiang, China
2 Institute of Hybrid Wheat, Beijing Academy of Agriculture and Forestry Sciences, 100097 Beijing, China
3 Institute of Plant Nutrition, Resources and Environment, Beijing Academy of Agriculture and Forestry Sciences, 100097 Beijing, China
4 Laboratory of Plant Breeding and Genetics, Department of Agricultural and Environmental Biology, The University of Tokyo, 113-8657 Tokyo, Japan
†These authors contributed equally.
Abstract
Nitrate transporter NRT1/PTR family (NPF) proteins are crucial for plant nitrogen uptake and utilization. As an important hexaploid crop for grain and forage, oat (Avena sativa L.) requires substantial levels of nitrogen. However, the oat nitrate transporter 1 (NRT1) family remains uncharacterized.
In this study, the oat NRT1 subfamily members were identified through the Hmm and Pfam databases. Bioinformatics analysis was performed using the MEGA 11 and TBtools software to elucidate the physicochemical properties, evolutionary relationships, chromosomal localization, and gene structures. Furthermore, quantitative real-time polymerase chain reaction (qRT-PCR) analysis and the green fluorescent protein (GFP) fusion expression vector were utilized to investigate the candidate oat NRT1s.
Phylogenetic classification categorized oat NRT1s into eight subfamilies, with the most abundant being the NPF5 subfamily. Physicochemical property analysis revealed that the number of amino acids in the proteins encoded by these genes ranged from 235 to 673, with their molecular weights (MWs) ranging from 26 kDa to 74 kDa. Chromosomal localization revealed that these genes were unevenly distributed across all 12 oat chromosomes. Promoter analysis revealed that light-responsive elements appeared most frequently in the promoters of these genes (39.3%), followed by abscisic acid (ABA)-responsive elements (13.5%) and methyl jasmonate (MeJA)-responsive elements (9.4%). qRT-PCR analysis revealed that most of the genes exhibited tissue-specific expression patterns. Among them, AsNPF2.6 was highly expressed in the leaves at 1 h post-low nitrogen (LN) treatment, while AsNPF4.5 was highly expressed in the leaves at 12 h. Both these genes exhibited low expression levels in the roots. However, AsNPF7.16 and AsNPF7.19 were both highly expressed in the roots at 9 h post-LN treatment but exhibited low expression in the leaves. Subcellular localization revealed that all five proteins (AsNPF2.6, AsNPF4.5, AsNPF7.16, AsNPF6.8, and AsNPF7.19) were localized to the cytoplasm and cell membrane.
Our results demostrate the involvement of AsNRT1 family members in nitrogen transport in oat, providing theoretical support for further investigation into the functions and molecular mechanisms of action of oat NRT1s in nitrogen transport.
Keywords
- Avena
- nitrate transporters
- nitrogen
- phylogeny
- genomics
- cluster analysis
Nitrogen is the most demanded nutrient element by plants and is a key limiting factor for plant growth [1, 2]. However, currently, only developed countries have a nitrogen fertilizer utilization rate of 50–60% [3], while developing countries, such as India [4] and Brazil [5], have an average nitrogen fertilizer utilization rate of only 30–40%. The low utilization rate of nitrogen fertilizer not only leads to a waste of resources and economic losses but also causes a series of environmental problems, including acidification of soil crusts, eutrophication of water bodies, nitrate contamination of groundwater, and intensification of greenhouse gas emissions [6, 7]. Therefore, fully exploring the potential of plants to enhance their nitrogen utilization efficiency is a key issue that must be addressed in contemporary agricultural production.
The primary form of nitrogen that plants obtain from the soil is nitrate, and nitrate transport proteins primarily facilitate the uptake and transportation of this essential nutrient [8, 9]. Nitrate is a key component of plant-synthesized proteins, chlorophyll, and various other compounds. Additionally, nitrate stored in vesicles plays an important role in ion homeostasis and osmoregulation. The nitrate transporter 1 family (NRT1/PTR family, abbreviated as NPF family) is the largest nitrate transporter protein group in plants, mainly responsible for the transport of nitrate and plant hormones [10, 11].
In the dicotyledonous plant Arabidopsis thaliana, the NPF family comprises 53 NRT1 members [12]. Among these, 11 proteins have been reported as essential for nitrogen uptake and transport in Arabidopsis thaliana [13, 14]. For instance, AtNRT1.1 (AtNPF6.3) represents a seminal discovery within the NRT1 protein family, playing a crucial role in nitrate uptake by plants [15]. AtNRT1.1 (AtNPF6.3) is an integral part of the signaling cascade response and is actively involved in coordinating the adaptive response of plants to fluctuating nitrate concentrations in the environment, highlighting its significance for sustainable plant growth and nutrition [16]. Furthermore, AtNRT1.4 is involved in regulating nitrate homeostasis in the leaves and affecting the leaf development processes [17], while AtNRT1.5 is responsible for mediating nitrate transport from roots to aboveground parts [18]. In addition, AtNRT1.6 is involved in regulating early embryonic development processes in plants [19]. AtNRT1.7 promotes the transport of nitrate between old and young leaves and participates in its reuse process, while AtNRT1.9 is responsible for the transport of nitrate to the base through the phloem [20]. Notably, the expression levels of these genes are induced by nitrate ions (NO3–) and are strongly regulated by phytohormones. However, few studies have examined the relationship between NRT1 and NO3– in crops other than wheat and rice, primarily due to the research limitations associated with non-model plants in terms of mutant materials and gene-cloning technology. Previously, response characteristics of NRT1.1 and NRT1.2 under different nitrogen supply conditions have been investigated in wheat [21], and the biological function of OsNRT1.1 has been elucidated in rice [22].
Low nitrogen utilization efficiency is a significant constraint on the sustainable development of oat (Avena sativa L.). Screening and characterizing key nitrate transporter genes is crucial for breeding nitrogen-efficient oat varieties. While oat quality is largely determined by its genetics, it is also influenced by the external environment. Oat is often grown in arid and infertile soils and is commonly fertilized with NH4+. This practice can lead to stunting during the early reproductive stages of the plant due to the slow biotransformation of NH4+ caused by low soil temperatures in early spring. Incorporating a certain percentage of NO3– into the nitrogen fertilizer might promote early oat development [23, 24]. Nitrogen fertilizers are critical in dry matter accumulation, morphology establishment, and fresh grass yield quality of oat [25, 26]. At present, there is an imbalance between forage supply and demand, and with the continuous development of animal husbandry, the demand for forage is increasing. Excessive application of nitrogen fertilizers to increase forage production results in the production of large amounts of ammonium nitrogen, which wastes resources and increases the pressure on the environment. Therefore, it is imperative to explore the cloning of nitrogen transporter genes for various molecular improvements and yield increases.
The whole genome sequencing of oat has been successfully completed [27]; however, the function of the NRT1 family in oat growth and development is still unclear. At present, there are few research reports on the response patterns of the members of this family under different nitrogen treatment conditions and their mechanisms of action in maintaining nitrogen homeostasis in oat. Therefore, in the present study, we identified the oat NRT1 family members and performed bioinformatics analysis and gene cloning of several candidate genes involved in nitrate uptake and transport. Our findings provide excellent genetic resources for oat nitrogen-efficient molecular breeding and are important for the high and stable yield of oat.
The sequences of oat proteins were downloaded from the Ensembl Plants database (https://plants.ensembl.org/index.html) [28] and was used to build a local protein database. Subsequently, a hidden Markov model (PF00854) of the NRT1 family was obtained from the Pfam database, and the local database was searched using the HMMER v3.0 (Sean Eddy, Seattle, WA, USA) (E value
Using the sequences of 53 NRT1s involved in nitrate uptake and transport in Arabidopsis thaliana, Oryza sativa L., Zea mays L., and Setaria italica miltiorrhiza as probes, homologous sequence searches were conducted with the oat whole genome database using BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Combined with phylogenetic tree analysis, 197 oat NRT1s were ultimately identified. Chromosomal localization (including the start and end sites of the 197 oat NRT1s on the chromosome), encoded protein length, and other basic information of oat were obtained from the Ensembl Plants database (https://plants.ensembl.org/). The ExPASy platform (https://www.expasy.org/) was used to predict and analyze the molecular weight (MW), isoelectric point (pI), mean hydrophobicity value (grand average of hydropathicity value, abbreviated as GRAVY value), instability index, and lipid index of oat NRT1s [32]. The sequences of the proteins were submitted to the WoLF PSORT website (https://wolfpsort.hgc.jp/) [33] and Cell-PLoc 2.0 website (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) [34] to predict their subcellular localization.
To elucidate the relationships of NRT1s between wheat and other species, a total of 197, 53, 3, 3, 3, and 8 NRT1 family members of oat, Arabidopsis thaliana, rice, maize, wheat, and S. italica, respectively, were obtained from the Ensembl Plants database and imported into MEGA 11 (MEGA Software, Tempe, AZ, USA) [35]. Phylogenetic analysis of NRT1 family members was performed using the Clustal W multiple sequence alignment method combined with the neighbor-joining (NJ) method. When building a phylogenetic tree, the bootstrap value was set to 1000, and the default configuration was used for all other parameters. The Newick (NWK) format file of the phylogenetic tree generated by the MEGA 11 software has been uploaded to the Evolview online platform (https://evolgenius.info//evolview-v2/#login) to optimize the visualization effect [36].
Using the “GTF/GFF Gene Localization Visualization” plugin in TBtools (CJ Chen, Guangzhou, Guangdong, China), the physical location information of oat NRT1 family members was input [37], and the gene localization map on chromosomes was generated and exported. The covariance relationships among oat, Arabidopsis thaliana, and rice genomes were analyzed using the MCscanX plugin in the TBtools software. A total of 197 AsNRT1s were highlighted, and covariance maps among genomes were plotted.
Exon-intron structure information was obtained from the oat genome annotation file using the “Visualize Gene Structure” tool from TBtools [37]. The AsNRT1 protein sequence was submitted to the MEME online tool (https://meme-suite.org/meme/tools/meme) to perform conservative motif prediction with the following parameter settings: The number of motifs was set to 15, and the length of motifs was set within the range of 6–100 amino acid residues. Subsequently, TBtools was used to visualize and analyze the predicted gene structure and conserved motifs [37, 38].
The regulatory sequence 2000 bp upstream of the transcription start site of AsNRT1 was extracted using the “GXF Sequence Extraction” tool in TBtools and submitted to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to analyze the cis-acting components [39]. The cis-acting elements in the AsNRT1s were analyzed, and the results were categorized according to the function of the elements and visualized using TBtools.
The coding sequences (CDSs) of all AsNRT1s in oat were extracted and compared with the diploid oat (A. longiglumis) CDS database, retaining the diploid oat genes with the highest matching degree to the oat AsNRT1s. Based on the diploid oat expression database (http://db.ncgr.ac.cn/oat/newel.php) by matching the gene names, the expression levels of NRT1s in different tissues of diploid oat (including root tips, roots, leaves, stems, spikes, and spikelets) were obtained (represented by the number of fragments per kilobase transcript read per million mapped reads, abbreviated as FPKM value), and a gene expression clustering heat map was drawn using the TBtools software.
The seeds of Bailyan VII oat material used in this experiment were provided by Prof. Zhao Guiqin of Gansu Agricultural University. It is suitable for cultivation in arid and semi-arid areas, alpine shady and humid areas, second-shade areas and similar ecological zones in central Gansu. After soaking the seeds in a 1% hydrogen peroxide solution for 3 min for surface disinfection, they were rinsed thrice with deionized water and then soaked in deionized water for 12 h. Post-soaking, the seeds were evenly arranged in petri dishes lined with moistened filter paper, oriented with ventral grooves facing downward. Seed germination was induced in the dark at 22 °C, with periodic moisture application by spraying deionized water. The germinated seeds were transferred to a light-controlled incubator supplemented with Hoagland nutrient solution for cultivation. The cultivation conditions were set as follows: relative humidity of 65%, light/dark period of 16 h/8 h, light period temperature of 22 °C, dark period temperature of 16 °C, and light intensity of 200–250 µmol
Total RNA was extracted from plant tissues using the RNAprep Pure Plant Total RNA Extraction Kit (B0226B, Tiangen Biochemical Technology, Beijing, China) for subsequent analysis of target gene expression. The Eco Real Time PCR system (Illumina, San Diego, CA, USA) was used for quantitative real-time polymerase chain reaction (qRT-PCR), with oat actin gene (AsActin) as the internal reference gene [40]. The primers used for qRT-PCR are shown in Supplementary Table 1.
Gene-specific primers were used to amplify the NRT1 CDS and clone it into the N-terminus of green fluorescent protein (GFP) in GFP expression vector (16318h-GFP-NOS). Protoplasts were isolated from wheat seedling leaves as described previously [41]. The recombinant plasmid 16318h-NRT1-GFP and the empty vector control 16318h-GFP were introduced into wheat protoplasts using the polyethylene glycol (PEG)-mediated transformation method [41]. The transfected protoplasts were cultured in the W5 solution at 23 °C for 18 h in the dark. Then, the GFP fluorescence signal was observed using a laser scanning confocal microscope (8100001684, Leica Microsystems GmbH, Wetzlar, Germany). The primer sequences are listed in Supplementary Table 2.
qRT-PCR data were normalized using actin, with vertical bars representing standard deviation. Significant differences in the variables were indicated by p
Through HMMER whole genome scanning combined with Conserved Domains Database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) conserved domain validation, a total of 197 oat NRT1 family members with complete PF00854 (PTR2) domain were identified. Subsequent screening via BLASTP alignment validated these 197 oat NRT1s. These genes encoded proteins ranging in size from 235 amino acids (AsNPF8.40) to 673 amino acids (AsNPF2.2) with MWs varying between 26 kDa (AsNPF8.40) and 74 kDa (AsNPF2.2) (Supplementary Table 3). Bioinformatic analysis revealed that the pIs of the proteins ranged from 4.89 (AsNPF8.28) to 11.06 (AsNPF7.4), indicating a broad range of acidic to basic properties within the NPF family.
Among the 197 identified NRT1s, 131 exhibited instability indices below 40, classifying them as stable proteins (Supplementary Table 3). Hydrophobic analysis showed that the average GRAVY value of the 195 proteins was positive, indicating that these proteins had hydrophobicity. Notably, only two proteins, AsNPF4.21 and AsNPF5.10, showed negative GRAVY values, indicating hydrophilic properties. Subcellular localization analysis using the Cell-PLoc 2.0 and the WoLF PSORT web tool predicted that nearly all AsNRT1s localize to vesicle membranes. In addition, 25 proteins were localized to the cell membrane, 7 to the endoplasmic reticulum, and 1 to the mitochondria. Thus, we hypothesized that the NRT1s in oat might play varying roles in nutrient transport.
A phylogenetic tree was constructed using the proteins from oat (A. sativa), rice (O. sativa), corn (Z. mays), common wheat (Tritium aestivum L.), S. italica and A. thaliana to analyze the evolutionary relationship of the oat NRT1 family (Fig. 1). Based on the subfamily classification system of Arabidopsis thaliana NRT1 family, phylogenetic analysis showed that the NPF5 subfamily of oat was the most abundant subfamily (with 56 members) in the oat NRT1 family, followed by the NPF8 subfamily (with 47 members). The NPF3 and NPF1 subfamilies were the least abundant, with only 11 and 1 family member, respectively. Phylogenetic analysis also showed that oat NRT1s in the same subfamily had closer genetic relationships but farther genetic relationships with the members in other subfamilies. This finding suggested that the oat NRT1 family might have undergone adaptive differentiation during its evolutionary process.
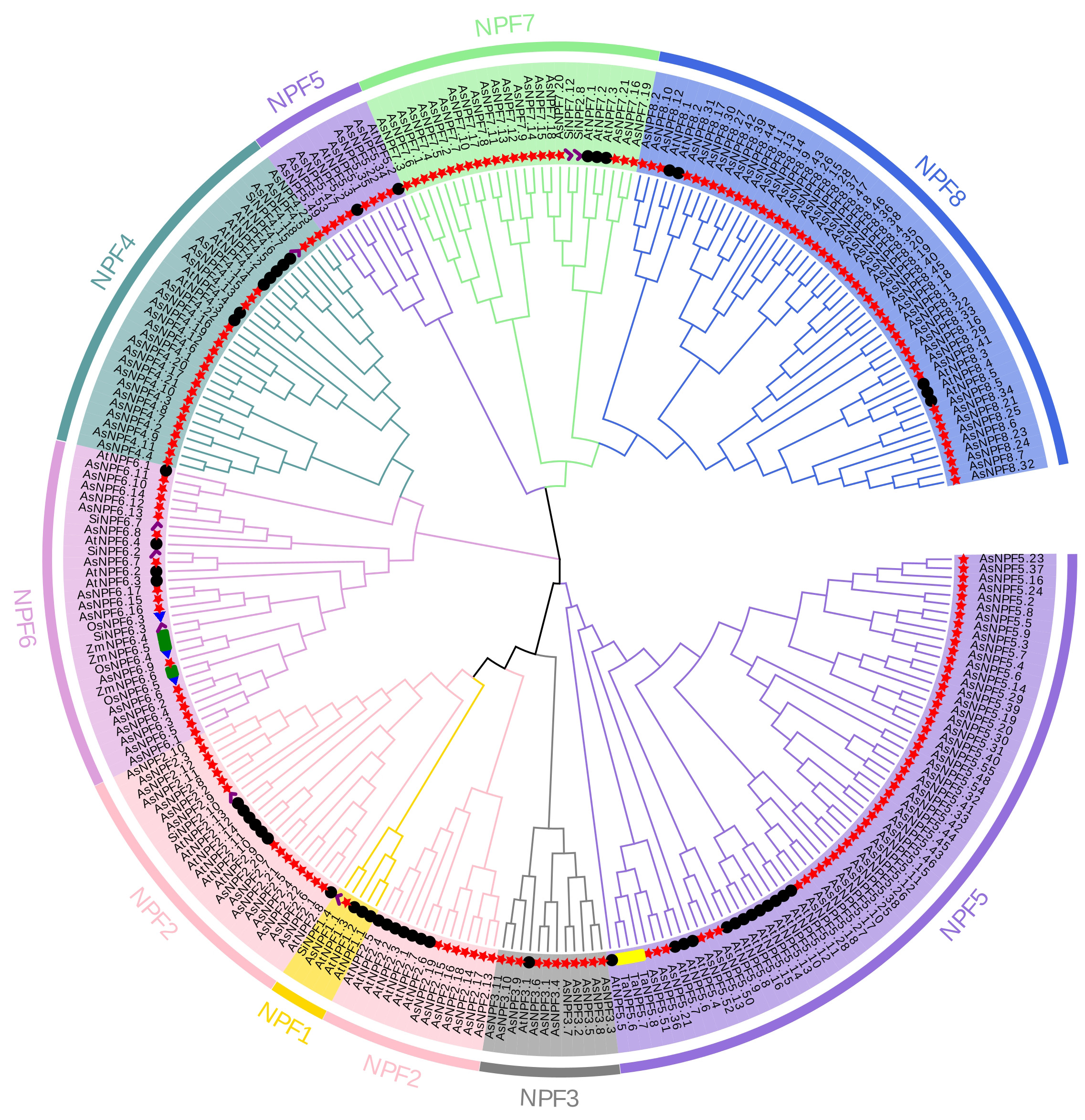 Fig. 1.
Fig. 1. Phylogenetic tree of NRT1s in monocots (Avena sativa L., Triticum aestivum L., Oryza sativa L., Zea mays L., and Setaria italica.) and a dicot (Arabidopsis thaliana). Oat NPF subfamilies are indicated by different colours: NPF1 (gold), NPF2 (pink), NPF3 (grey), NPF4 (cadet blue), NPF5 (medium purple), NPF6 (plum), NPF7 (light green), and NPF8 (royal blue). The red star, black circle, green rectangle, yellow strip, blue triangle, and purple check marks represent the AsNPFs, AtNPFs, ZmNPFs, TaNPFs, OsNPFs, and SiNPFs, respectively. NPF, nitrate transporter 1 family.
Next, we identified the genes in nitrate uptake and transport, including 11, 3, 3, and 8 genes in S. italica [42], Arabidopsis thaliana [15, 19, 43], O. sativa [44], and Z. mays [45], respectively. Phylogenetic analysis revealed distinct evolutionary relationships among oat NPFs. AsNPF2.1, AsNPF2.2, AsNPF2.4, AsNPF2.6, AsNPF2.7, AsNPF2.20, and AsNPF2.22 clustered closely with AtNPF2.11, exhibiting 89–95% sequence identity. AsNPF4.5, AsNPF4.18, and AsNPF4.23 were highly homologous to SiNPF4.6 (99%, 94%, and 99% identity, respectively). The AsNPF6.1–AsNPF6.3 subgroups demonstrated near-complete sequence conservation with OsNPF6.5 (99%, 92%, and 99% identity, respectively). AsNPF6.8, AsNPF6.13, and AsNPF6.14 clustered with SiNPF6.7 (93–97% identity). AsNPF6.9, AsNPF6.15, AsNPF6.16, and AsNPF6.17 aligned closely with OsNPF6.3 (93–95% identity). These proteins from the non-oat species were evolutionarily similar, with a high structural and functional similarity, and can be used as a reference for inferring the functions of oat NPF proteins.
Chromosomal localization analysis showed that 197 AsNRT1s were unevenly distributed across the oat chromosomes (Supplementary Fig. 1). Among them, 58, 38, 28, 27, 22, 14, and 9 genes were located on chromosomes 4, 1, 2, 7, 6, 3, and 5, respectively. We also detected one unassigned scaffold Un25. Notably, chromosome 4 exhibited the highest AsNRT1 density, with 23 AsNRT1s specifically clustered on its 4A subgenome.
Through collinearity analysis of the 197 oat NRT1s, their chromosomal distribution patterns were analyzed, homologous gene pairs were identified, and evolutionary relationships were inferred to identify potential gene duplication events. To investigate the evolutionary mechanisms of the oat NRT1 family, we generated a comparative phylogram within the genome of oat itself (Fig. 2a). To elucidate their evolutionary relationships with other species, we performed collinearity analysis of the 197 AsNRT1s across three species: The monocot O. sativa, diploid oat A. longiglumis, and the dicot A. thaliana (Fig. 2b). Our analysis revealed 114, 89, and 4 syntenic gene pairs with diploid oat, rice, and Arabidopsis thaliana, respectively. The differential number of conserved syntenic blocks reflects evolutionary divergence, with hexaploid oat A. sativa showing closer phylogenetic affinity to monocot rice than to the distantly related dicot Arabidopsis thaliana.
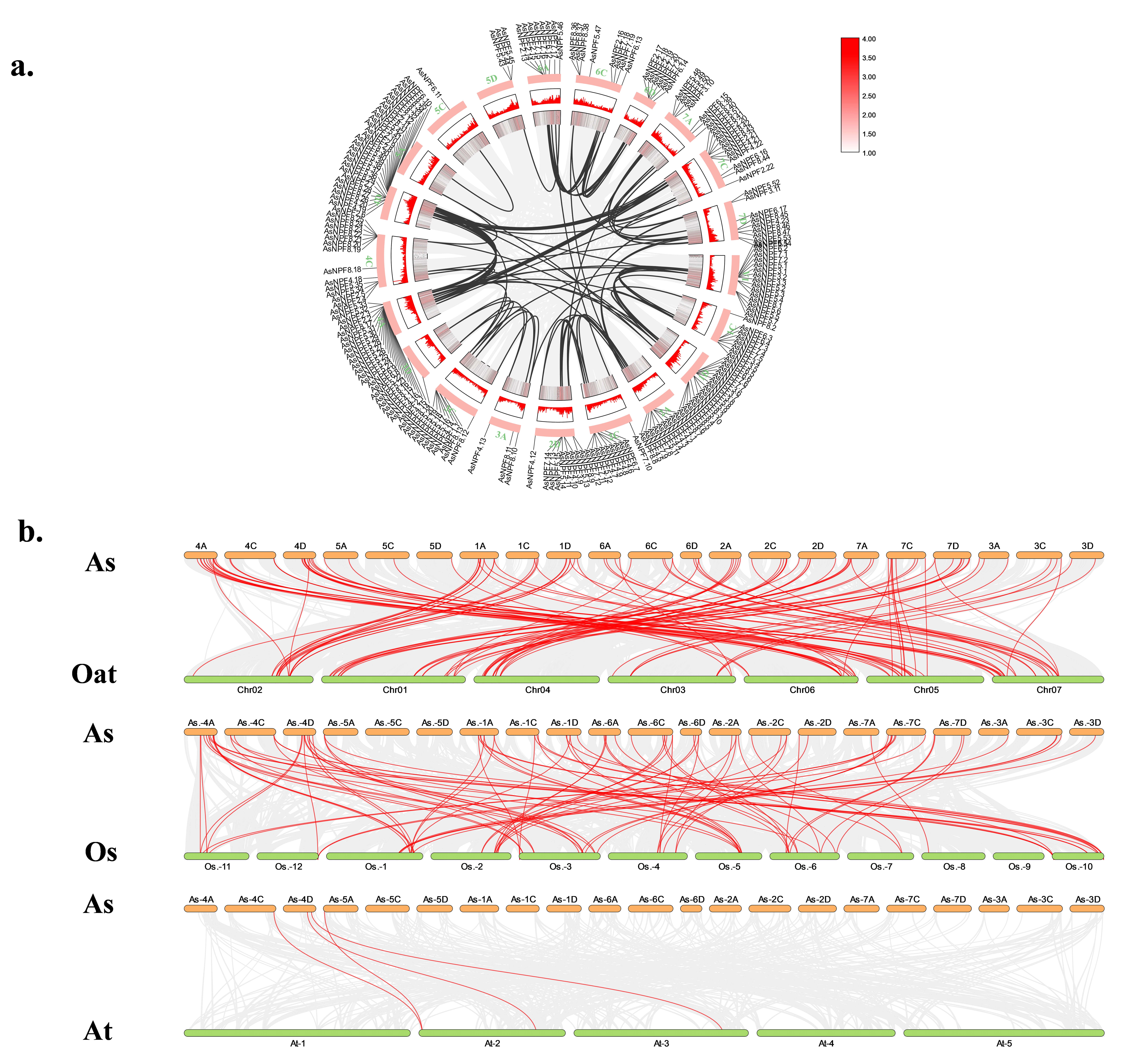 Fig. 2.
Fig. 2. Inter-species collinearity analysis of NRT1s in Avena sativa L., Oryza sativa, Arabidopsis thaliana, and A. longigluis. (a) Covariance analysis of AsNRT1s in the oat genome. The grey and black lines indicate all duplicated gene pairs in the oat genome and duplicate gene pairs among the 197 AsNRT1s, respectively. (b) Analysis of covariance between species of the AsNRT1s. The grey and red lines represent the region of co-collinearity between oat and other plants and the co-collinearity of AsNRT1s, respectively.
A 200-kb chromosomal region containing
To analyze the evolutionary selection pressure on the NRT1 family, we calculated the non-synonymous substitution rate (Ka), synonymous substitution rate (Ks), and Ka/Ks ratio of gene pairs (Supplementary Table 5). Among them, Ka/Ks of 1 indicated that the gene is in a neutral selection state, while Ka/Ks of
A phylogenetic tree based on the maximum likelihood (ML) method was constructed to analyze the structural domain characteristics of oat AsNRT1s. Firstly, ClustalW multiple sequence alignment was performed on the sequences of the 197 AsNRT1s, followed by phylogenetic analysis. The amino acid sequences of the AsNRT1s were submitted to the MEME database (Fig. 3). The evolutionary relationships among AsNRT1s were investigated based on their protein sequences, and the clustering results from the branches were consistent with those of Arabidopsis thaliana NRT1s. Using bioinformatic methods, a total of eight conserved motifs were identified in AsNRT1s and named motifs 1 to 8 (Supplementary Fig. 2).
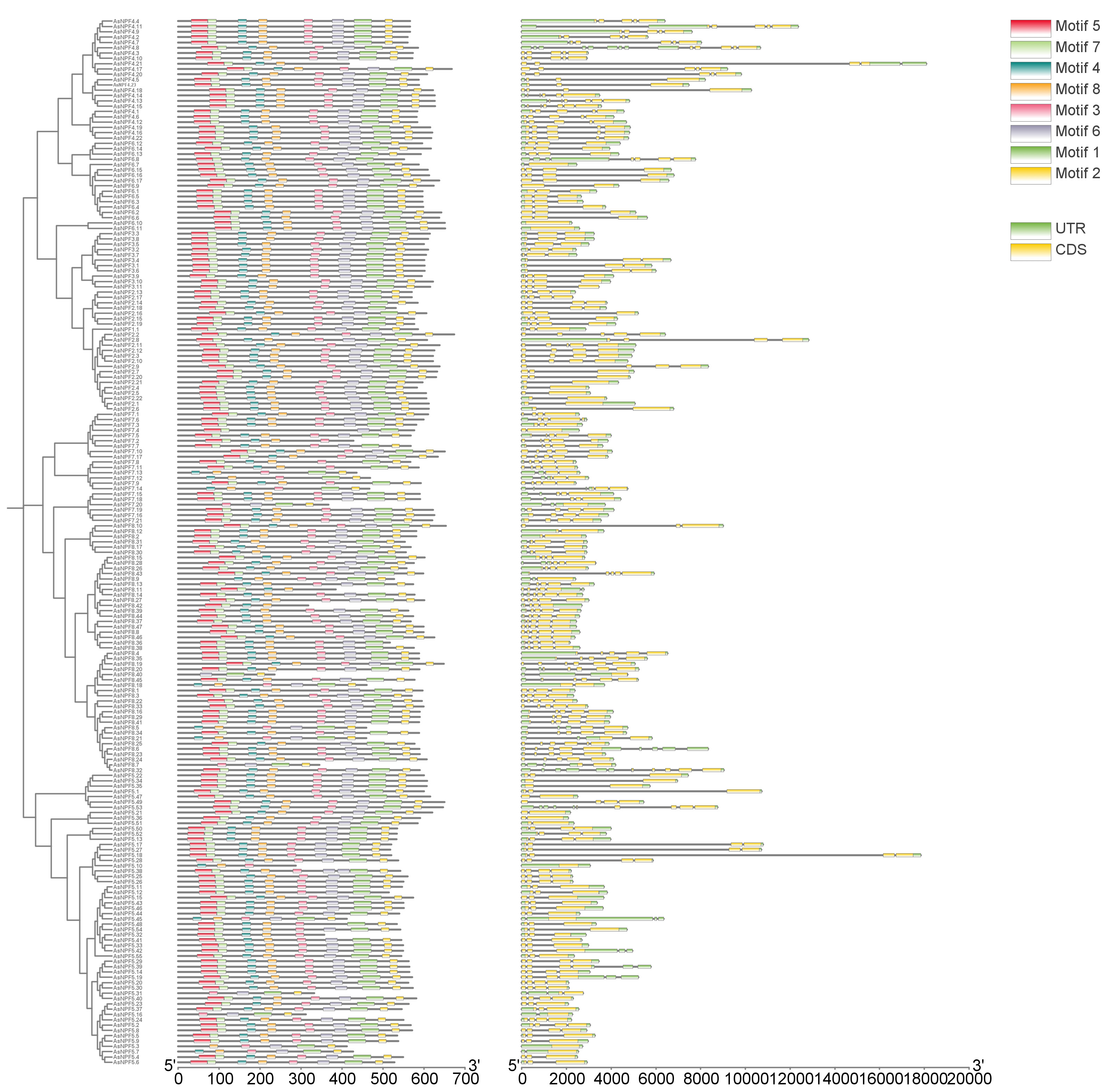 Fig. 3.
Fig. 3. Phylogenetic relationship and evolutionary trajectory analysis of AsNRT1s. Conservative motif analysis, annotated with different small boxes (each box representing an independent motif). In the genetic structure, introns are represented by black lines, while exons are distinguished by green and yellow boxes.
To further investigate the evolutionary characteristics of the oat NRT1 family, exon-intron structure analysis was performed on all identified AsNRT1s (Fig. 3). Our results showed that the number of exons in the genes of this family ranged from 1 to 6, with 10, 17, 55, 63, 52, and 3 genes containing 1, 2, 3, 4, 5, and 6 exons, respectively. By combining evolutionary trees and gene structures, we observed that the genes within the same group usually had similar structures.
The promoter sequences of the 197 AsNRT1s were submitted to the PlantCARE database to identify cis-acting elements to elucidate the transcriptional regulatory mechanisms of this gene family. As a key binding region of transcription factors, cis-acting elements directly participate in gene expression regulation (Supplementary Fig. 3).
We detected 23 elements associated with biological functions, and they were divided into four categories: Plant hormone-responsive, abiotic stress-responsive, plant growth and development-related, and light-responsive elements (Table 1). In addition, we detected ten elements associated with plant growth and development. These elements comprised GCN4_motif (related to endosperm expression), AACA_motif (specifically regulated by endosperm), GC-motif (related to hypoxia induction), HD-Zip 1 motif (related to palisade mesophyll cell differentiation), cis elements involved in anaerobic induction response, CAT-box (related to meristematic expression), NON-box (activated by meristematic tissue), root-specific expression motif I, RY-motif (regulated by seed specificity), and O2-site-a (regulated by zein metabolism). Among them, anaerobic induction-related elements were the most abundant functional components.
| Cis-regulatory element | Element | Element number | Classification |
| Abscisic acid responsiveness | ABRE | 644 | Hormone response related elements |
| Flavonoid biosynthetic genes regulation | MBSI | 14 | Hormone response related elements |
| Auxin-responsive element | AuxRE, TGA-box, TGA-element | 175 | Hormone response related elements |
| Gibberellin-responsiveness | GARE-motif, P-box, TATC-box | 175 | Hormone response related elements |
| Salicylic acid responsiveness | SARE, TCA-element | 95 | Hormone response related elements |
| The MeJA-responsiveness | CGTCA-motif, TGACG-motif | 450 | Hormone response related elements |
| Defense and stress responsiveness | TC-rich repeats | 54 | Abiotic stress response element |
| Drought-inducibility | MBS | 182 | Abiotic stress response element |
| Low-temperature responsiveness | LTR | 155 | Abiotic stress response element |
| Maximal elicitor-mediated activation (2 copies) | AT-rich sequence | 6 | Abiotic stress response element |
| MYBHv1 binding site | CCAAT-box | 136 | Abiotic stress response element |
| Wound-responsive element | WUN-motif | 4 | Abiotic stress response element |
| Endosperm expression | GCN4-motif | 40 | Plant growth and development elements |
| Endosperm-specific negative expression | AACA-motif | 5 | Plant growth and development elements |
| Anoxic specific inducibility | GC-motif | 106 | Plant growth and development elements |
| Differentiation of the palisade mesophyll cells | HD-Zip 1 | 16 | Plant growth and development elements |
| Essential for the anaerobic induction | ARE | 307 | Plant growth and development elements |
| Meristem expression | CAT-box | 175 | Plant growth and development elements |
| Meristem specific activation | NON-box | 2 | Plant growth and development elements |
| Root specific | motif I | 9 | Plant growth and development elements |
| Seed-specific regulation | RY | 32 | Plant growth and development elements |
| Zein metabolism regulation | O2-site-a | 122 | Plant growth and development elements |
| Light responsive element | I-box, Gap-box, Box II, L-box | 1883 | Elements associated with light response |
MeJA, methyl jasmonate; MBS, MYB binding sites; ABRE, abscisic acid responsive element; MBSI, MYB binding Site I; AuxRE, auxin response element; SARE, salicylic acid responsive element; LTR, low temperature response; ARE, antioxidant response element; WUN, wound-responsive element; GCN4, general control nonderepressible 4.
Furthermore, we detected six elements related to plant stress response, including TC-rich repeat sequences (related to defense and stress response), MYB binding sites (MBS)-elements (related to drought induction), LTR elements (related to low temperature response), MYBHv1 binding site (CCAAT-box), and WUN-motifs (related to wound response). These elements play key regulatory roles in plant development and environmental stress response. In addition, we detected six cis-sacting elements related to hormone response, including abscisic acid-responsive ABRE-element (related to abscisic acid), flavonoid biosynthesis-regulated MBSI-motif (related to MYB transcription factor-mediated regulation of flavonoid synthesis gene expression), auxin-responsive AuxRE-element (related to growth and development regulated by growth hormone) and TGA-box/TGA-motif (related to plant disease resistance response), gibberellin-responsive GARE-motif/P-box/TACT-box (related to gibberellin signaling pathway), salicylic acid-responsive SARE-element/TCA-motif (related to plant disease resistance response and abiotic stress response), and methyl jasmonate (MeJA)-responsive CGTCA-motif/TGACG-motif (related to plant defense responses and secondary metabolism). These components recognize specific stress signals within cells and mediate the regulation of related gene expression to adapt to unfavorable external environments. Further, we identified I-box, Gap-box, Box-II, and L-box components related to light response. Overall, the identified cis-acting elements were related to the response mechanisms of plants to internal hormone signals, external environmental stress, and the expression regulation patterns in specific growth stages and tissues, indicating that the AsNRT1 family might be involved in regulating multidimensional biological processes in oat.
By comparing the gene expression data of diploid A. longigluis and hexaploid A. sativa, the expression patterns of the AsNRT1 family in different tissues were analyzed, and their potential biological functions were explored. A total of 114 gene replication pairs were identified (Fig. 2). The expression data for these diploid oat genes were obtained from the oat database, and a tissue-specific expression heat map was drawn (Fig. 4). The expression levels of the AsNRT1s varied across different tissues. Most AsNRT1s exhibited tissue-specific expression, with distinct patterns observed for key members. AsNPF4.18 was highly expressed in the root tip, potentially mediating the initial uptake of nitrate by the root system. AsNPF7.16, AsNPF7.19, and AsNPF2.6 were highly expressed in the roots, potentially mediating nitrate uptake from the soil to the root system, intra-root transport, and xylem loading. AsNPF4.5, AsNPF2.7, and AsNPF6.8 were highly expressed in the leaves, potentially acting as unloading/loading hubs for vascular transport and, at the same time, participating in nitrogen signaling to coordinate metabolism and growth. AsNPF2.1 and AsNPF6.13 were highly expressed in the panicles and might mediate nitrate partitioning from the nutrient organs to inflorescences or act as signaling elements. AsNPF6.2 was highly expressed in the spikelets, potentially supplying nitrogen to the spikelets via the “source-deposit” transporter network to support floral organ development, pollen viability, and seed formation. Therefore, nitrogen uptake in oat might be initiated by AsNPF6.1 and AsNPF6.3, which take up nitrate from the soil. This process is followed by the activities of AsNPF7.16, AsNPF7.19, and AsNPF2.6 that mediate intra-root nitrate transport. Next, vascular nitrate transport is mediated by AsNPF4.5, AsNPF2.7, and AsNPF6.8, and AsNPF2.1 and AsNPF6.13 mediated nitrate partitioning to the florets. Finally, precise nitrogen transport to the spikelets was facilitated by AsNPF6.2 to mediate seed formation.
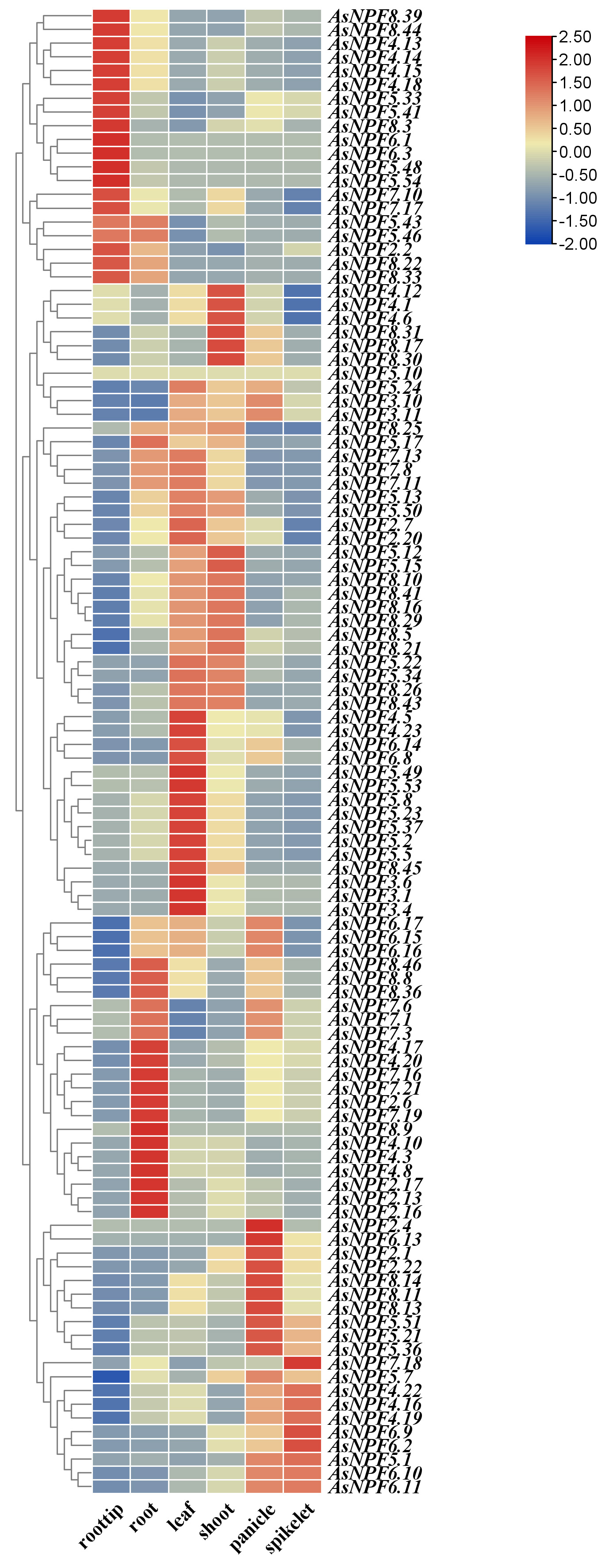 Fig. 4.
Fig. 4. Heat map of the expression profiles of AsNRT1s in different tissues. From left to right are the root tip, root, leaf, shoot, panicle, and spikelet. Orange and blue represent high and low expressions, respectively.
We selected 10 genes from the promoter region containing many adversity-related cis-acting elements to validate the bioinformatics data related to the expression analysis of oat NPFs. The genes showing large differences in their expressions across different tissues were found to be involved in nitrate uptake and transport in other plants. These genes were homologous with AsNPF6.2, AsNPF4.5, AsNPF2.1, AsNPF6.8, AsNPF4.18, AsNPF2.6, AsNPF2.7, AsNPF7.16, AsNPF7.19, and AsNPF6.13.
To verify the expression patterns of these ten genes under LN conditions in oats, we created a CK group (treated with 20 mM nitrogen) and an LN group (treated with 2 mM nitrogen) and analyzed the expressions of these genes in their root and leaf tissues. Our results showed that under LN conditions, the expressions of these genes were induced to varying degrees. In the roots of the LN group (Fig. 5), the levels of 8 of these 10 genes were significantly higher at 1 h post-treatment than those at 0 h (p
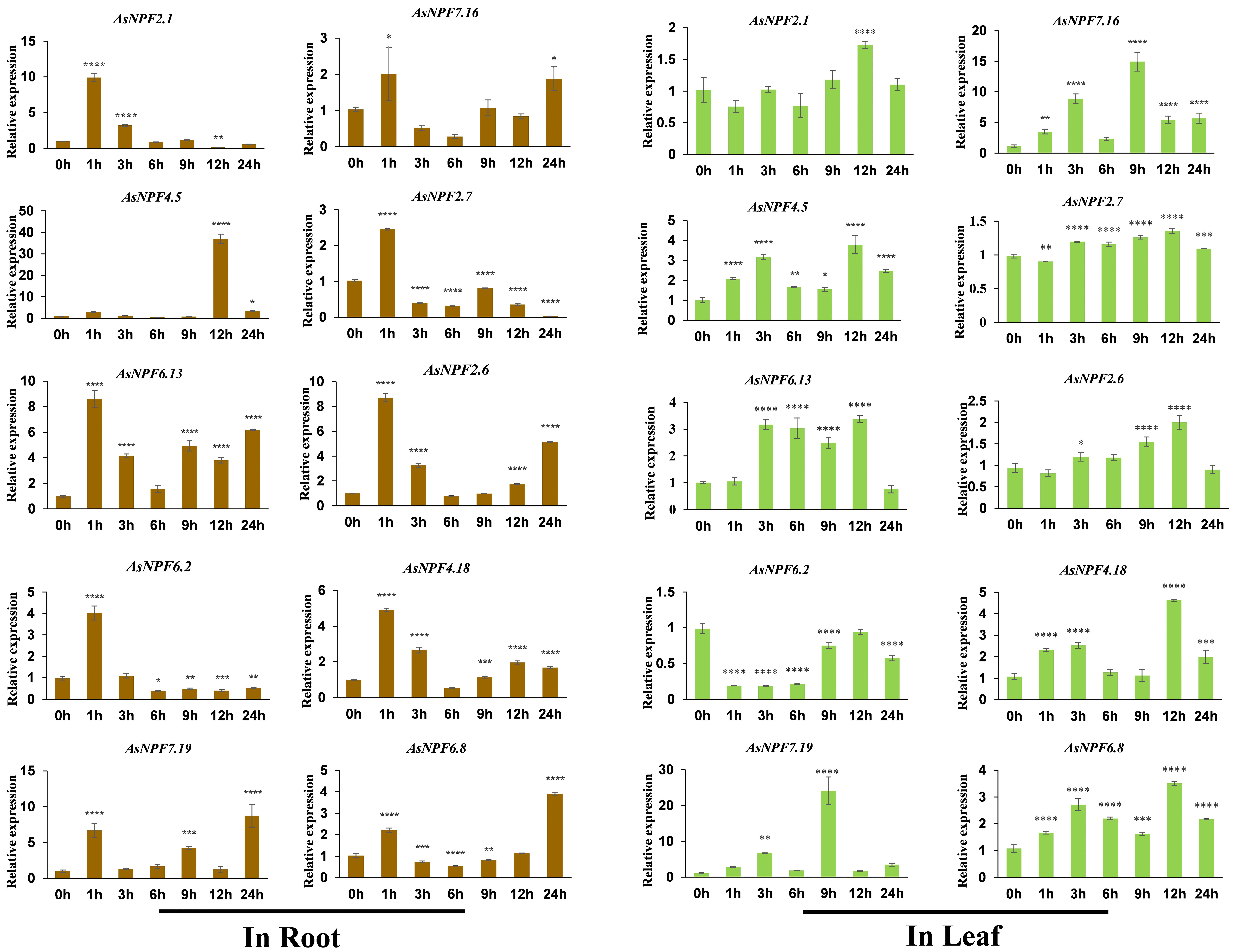 Fig. 5.
Fig. 5. The left and right figures show the expression profile of ten AsNRT1s in oat root and leaf tissue under low nitrogen (LN) treatment, respectively. The data is standardized using the actin gene as an internal reference, and the vertical error line represents the standard deviation (SD). Asterisks indicate significant differences in gene expression compared to the untreated control group (Student’s t-test: * p
Unlike the gene expression pattern in root tissue, the expression of AsNPF2.1 exhibited specific regulatory characteristics in leaf tissue under LN treatment (Fig. 5). At 12 h post-treatment, its levels were significantly higher than at 0 h (p
Next, we analyzed the subcellular localization of candidate proteins encoded by the AsNRT1s. Analysis using the wheat protoplast expression system showed that the GFP signal was distributed in the cytoplasm and cell membrane of wheat protoplasts (Fig. 6). Among them, weak GFP signals in the cytoplasm and cell membrane were observed only for AsNPF2.6 and AsNPF4.5. AsNPF6.8, AsNPF7.16, and AsNPF7.19 exhibited stronger GFP signals in the cytoplasm but weaker signals in the cell membrane. This finding was not in line with the biological prediction. These findings indicated that these 5 genes might primarily be involved in nitrogen transport and uptake in the oat cytoplasm and cell membrane.
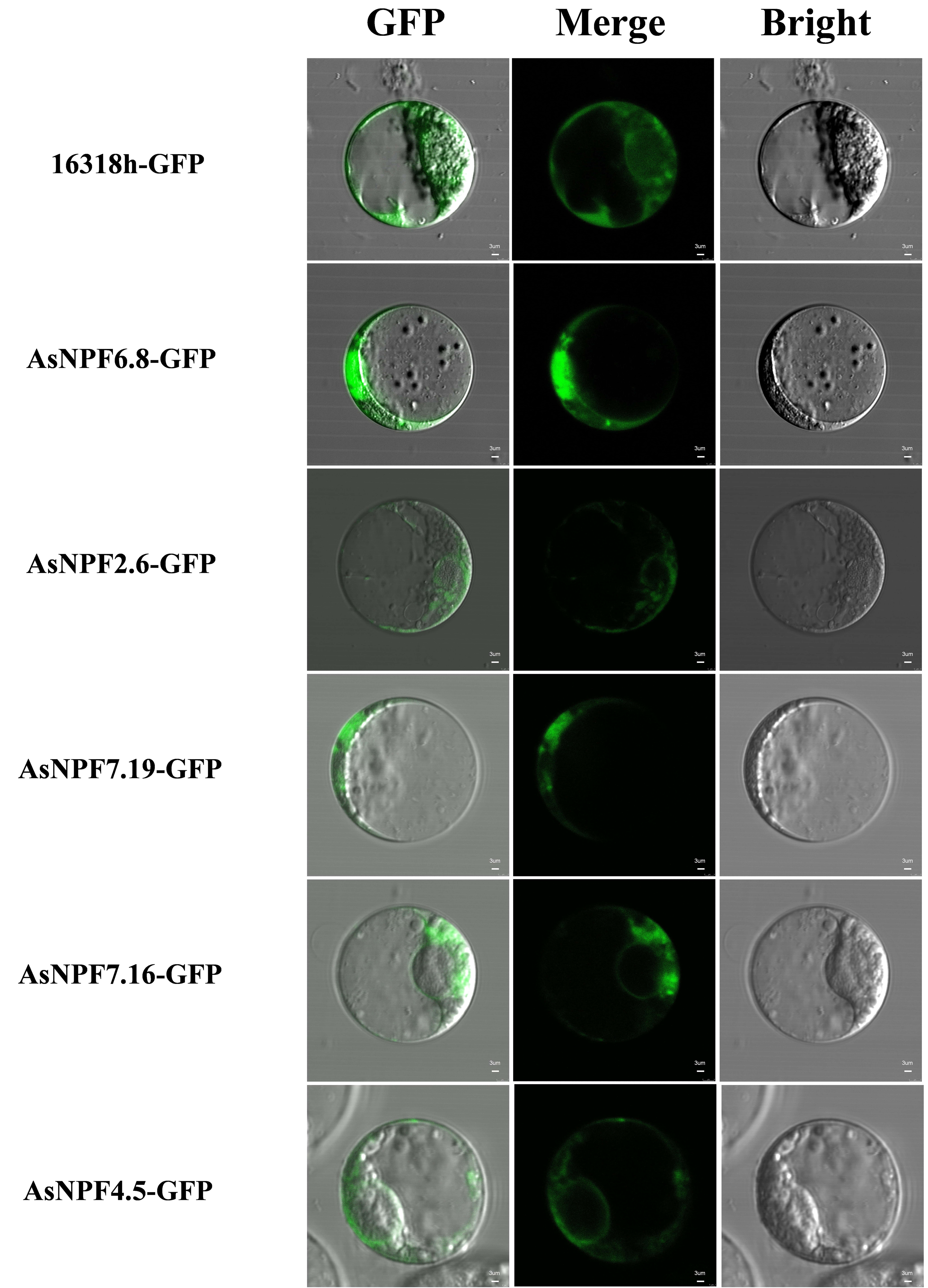 Fig. 6.
Fig. 6. Subcellular localization of AsNRT1s. Green fluorescent protein (GFP) stands for green fluorescence field, merge stands for superimposed field, and bright stands for bright field. GFP field: 488 nm. Scale bars = 3 µm.
The NRT1 family comprises critical genes involved in plant nitrogen uptake and utilization, and its members are engaged in nitrate transport and uptake. A study has shown that nitrate uptake and translocation are influenced by numerous factors, with light being a critical factor [23]. Bioinformatic analysis of the promoters of 197 AsNRT1s revealed that they contain abundant plant hormone and stress-responsive regulatory elements. The hormone-responsive elements were related to various hormone-related signaling pathways, such as gibberellin (GA)-, abscisic acid (ABA)-, auxin (IAA)-, MeJA-, and salicylic acid (SA)-related signaling pathways. Stress-responsive elements included drought-induced MBS, while environmental factors-responsive components comprised light-responsive regulatory components. Notably, among all the identified regulatory elements, the most abundant were the photoresponsive cis-acting elements (39.3%). This predominance might reflect photoperiod-driven metabolic coordination, where photosynthetic product accumulation creates elevated nitrogen demands for biomass synthesis. The abundance of these light-sensitive elements suggested their critical role in mediating plant–environment interactions via transcriptional regulation of nutrient acquisition pathways. This result was consistent with the genome-wide analysis of the NRT1 family promoters in grains [42]. In addition to photoresponsive elements, oat NRT1s were also regulated via multiple hormone signaling pathways. This finding was consistent with the results of previous studies. For instance, previous study [47] has shown that hormones significantly regulate the expression of cotton GhNPF family. In Arabidopsis thaliana, AtNRT1.1 transports growth hormone in lateral roots and responds to nitrogen starvation by inhibiting lateral root growth [48]. Cytokinins positively regulate the expression of AtNRT1.4 and AtNRT1.7, promoting nitrate distribution and transport [17, 20]. In the present study, analysis of hormone-responsive cis-elements revealed distinct regulatory signatures. ABA-responsive motifs constituted 13.5% of the identified elements, while MeJA-responsive elements constituted 9.4%. This finding indicated a strong functional linkage between NRT1 expression dynamics and phytohormone signaling pathways, particularly suggesting that ABA and MeJA mediated transcriptional regulation of nitrate transport processes. These results showed that the expression of AsNRT1s might be regulated by various plant hormones and environmental factors, thereby reflecting the functional diversity of this gene family.
The function of a gene is based on its expression, while the expression of a gene reflects its function [49]. We analyzed the tissue-specific expression patterns of 197 AsNRT1s. The seed- and spike-specific expressions of TaNPF1 in wheat have been shown to be related to its nitrate transport function during seed development [50]. The AsNRT1 family exhibited diverse tissue expression specificity. AsNPF6.1 and AsNPF6.3 exhibited preferentially high expression in the root tip, AsNPF8.9 was specifically upregulated in mature roots, and AsNPF2.4 displayed panicle-specific expression. These findings suggested that these genes might function predominantly in specific tissues to regulate nitrate transport and localized nitrogen metabolism. On the contrary, multiple AsNRT1 family members, including AsNPF2.20, AsNPF6.2, and AsNPF6.17, exhibited constitutive expression in various tissues, suggesting their potential involvement in the systemic nitrogen regulation during oat growth and development.
Under LN conditions, AtNRT1.1 (AtNPF6.3) exhibited opposite expression patterns in aboveground tissues and roots, suggesting varied regulatory mechanisms for AtNPF6.3 in these tissue types [2]. Previous studies on the maize homologous gene, ZmNPF6.6, reported similar results under LN conditions, showing opposite expression trends in the roots and leaves under LN treatment [45, 51]. The homologous gene in wheat, TaNPF6.3-6D, was significantly upregulated in the roots at 6 h and peaked at 24 h under LN treatment, but it exhibited an opposite expression pattern in the leaves [52, 53]. In the current study, under LN treatment, AsNPF6.2, AsNPF2.7, and AsNPF2.6 were first upregulated and then downregulated in the leaves, and first downregulated and then upregulated in the roots, indicating potential inhibitory effects of these genes on the nitrate uptake system [54]. These results indicated that AsNPF6.2, AsNPF2.7, and AsNPF2.6 exhibited opposite expression patterns in roots and leaves under LN treatment, which was in line with the findings in other species. This finding suggested that these oat genes might also play similar functions and roles as their corresponding genes in other species. Among them, the expressions of AsNPF7.16 and AsNPF7.19 were significantly higher in the roots than in other tissues, suggesting that they might play an important role in nitrate transport and uptake in the roots. In contrast, the expression of AsNPF4.5 was significantly higher in the leaves than in the roots.
The functional characteristics, metabolic activity, and interaction patterns of proteins are closely related to their subcellular localization. The functions of proteins vary based on their location within the cells. This implies that proteins must be present in specific subcellular structures to fulfill their biological functions. Thus, subcellular localization analysis of proteins is crucial to studying their functions. In Arabidopsis thaliana, AtNPF5.12 and AtNPF5.16 are located on the vesicle membrane and participate in the long-distance transport of nitrate from the root system to the aboveground tissues [55]. There are two splicing variants of OsNPF7.7 in rice, OsNPF7.7-1 and OsNPF7.7-2. Among them, OsNPF7.7-1 is located on the cell membrane, and its upregulation can promote nitrate absorption in the roots [56]. As a functionally homologous protein of Arabidopsis thaliana AtNPF6.3 (NRT1.1/CHL1), rice OsNPF6.5 is also located on the cell membrane, and its expression is significantly upregulated by nitrate induction [44]. There are subcellular localization differences between the tandem repeat genes ZmNRT1.1B and ZmNRT1.1C in maize. ZmNRT1.1B is located on the plasma membrane, while the fluorescence signals from ZmNRT1.1C-GFP fusion protein are distributed in the inner membrane-like structure, suggesting that these two protein subtypes might exhibit different functions [51]. In the present study, AsNPF4.5, AsNPF6.8, AsNPF2.6, AsNPF7.16, and AsNPF7.19 were transferred into the 16318h-GFP vector and transformed into wheat protoplasts using a seamless cloning method. Laser confocal microscopy revealed that these genes were predominantly present in the cytoplasm and cell membrane of the protoplasts. Among them, the AsNPF2.6 and AsNPF4.5 signals were weak in the cytoplasm. We observed strong AsNPF6.8, AsNPF7.16, and AsNPF7.19 signals in the cytoplasm but weak signals on the cell membrane. The localization paterns of these genes were different from those of NRT1s in other species. Therefore, we speculated that NRT1.1, which is partially distributed in the cytoplasm due to functional redundancy in leaf epidermal cells or in seedling-stage leaf cells, might be transiently retained in the cytoplasm due to the immature membrane transport system, while it becomes stably localized in the plasma membrane during the maturation stage [57]. We also speculated that deletions or missense mutations (for example,
Here, we performed a genome-wide characterization of the oat NRT1 family. A total of 197 AsNRT1 family members were identified, which were divided into 8 subfamilies based on their evolutionary relationships, with the members in the same subfamily exhibiting similar gene structures and conserved motifs. A comprehensive analysis of the chromosomal location, gene structure, and cis-acting elements of the 197 AsNRT1s showed that the oat NRT1s are involved in a coordinated response to adverse conditions and phytohormones. We further analyzed the expressions of ten candidate genes under LN treatment using qRT-PCR and found that five key oat NRT1s were induced by nitrogen treatment. Finally, the locations of the proteins encoded by these 5 NRT1s were identified as the cytoplasm and cell membrane by gene cloning and subcellular localization analyses. Our findings provided valuable information for further functional validation and laid a theoretical foundation for molecular breeding for optimal oat production.
CK, control; GRAVY, grand average of hydropathy; LN, low nitrogen; MW, molecular weight; NRT1, nitrate transporter 1; pI, isoelectric point.
All data generated or analyzed during this study are included in this published article and its supplementary information files.
RC and QX conceived the project. WZheng and SQG designed the experiments. RC and QX performed most of the experiments with the help of JG, RWS, YKD and WZhao. RC, SQG and JG analyzed the data and wrote the article. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Prof. Zhao Guiqin of Gansu Agricultural University provided and agreed to use Baiyan VII as experimental material for the experiment.
Not applicable.
This study was supported by the National Key Research and Development Program “Assessment of Carrying Capacity of Important Grassland Animal Husbandry and Potential for Ecological Animal Husbandry Base Construction in Xinjiang” (2022xjkk0404), “Integration of Highly Efficient Biological Breeding Technological Innovations and Application of Breakthrough Variety Breeding in Wheat” (KJCX20251004), Creation of new transgenic germplasm of important crops (KJCX20230203, KJCX20240305), Beijing Nova Program (20250484810) and National Natural Science Foundation of China’s “Research on Nitrogen Utilization Efficiency and Quality Improvement under the Mechanism of Root and Soil Action of Intercropping of Legume Forage and Kurrer’s Xiangshi Pear” (32060402).
The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/FBL39679.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.






