1 Xinjiang Key Laboratory of Lavender Conservation and Utilization, College of Biological Sciences and Technology, Yili Normal University, 835000 Yining, Xinjiang, China
2 School of Life Sciences, Xiamen University, 361102 Xiamen, Fujian, China
Abstract
Escherichia coli (E. coli) is a common opportunistic bacterial pathogen in both human and animal populations. Fatty acids serve as the central carbon and energy source, a process mediated by fatty acid-coenzyme A (CoA) ligases encoded by fad genes such as FadK. However, the function and the mechanism of FadK remain unclear.
The three-dimensional structure of FadK was modeled using AlphaFold2. After expression and purification, monomeric FadK was successfully isolated. The enzymatic activity was assayed, and real-time quantitative polymerase chain reaction (RT-qPCR) was performed to quantify FadK expression levels.
In enzymatic assays of fatty acid CoA ligase activity, caprylic acid was found to be the optimal substrate for FadK. We determined the optimal catalytic conditions for FadK, which include a pH of 7.4, ATP concentration of 0.6 mM, CoA concentration of 0.8 mM, and Mg2+ concentration of 0.8 mM at 37 °C. Notably, the activity of FadK showed a decrease with increasing concentrations of dodecyl-AMP, which was further confirmed by the RT-qPCR results.
Our findings will serve as a fundamental framework for the development of innovative therapeutics that target E. coli infections.
Keywords
- Escherichia coli
- fatty-acid CoA ligase FadK
- enzyme activity
- gene expression levels
- inhibitor dodecyl-AMP
Escherichia coli (E. coli) is the predominant opportunistic bacterial pathogen affecting both human and animal populations [1, 2]. This microorganism is responsible for a wide range of nosocomial and community-acquired diseases, particularly gastroenteritis, urinary tract infections, and bacteremia [3, 4]. Despite varying susceptibility profiles to conventional therapeutic antimicrobials, E. coli has the potential to acquire resistance determinants via horizontal gene transfer, both within and between species, accelerating the proliferation of drug-resistant bacterial strains [5, 6, 7]. Of great concern is the emergence of multidrug-resistant E. coli, which represents a major public health challenge due to the decreasing efficacy of existing antibiotic therapies [3, 4, 8, 9, 10]. Therefore, it is urgent to take immediate measures to control or even end the situations.
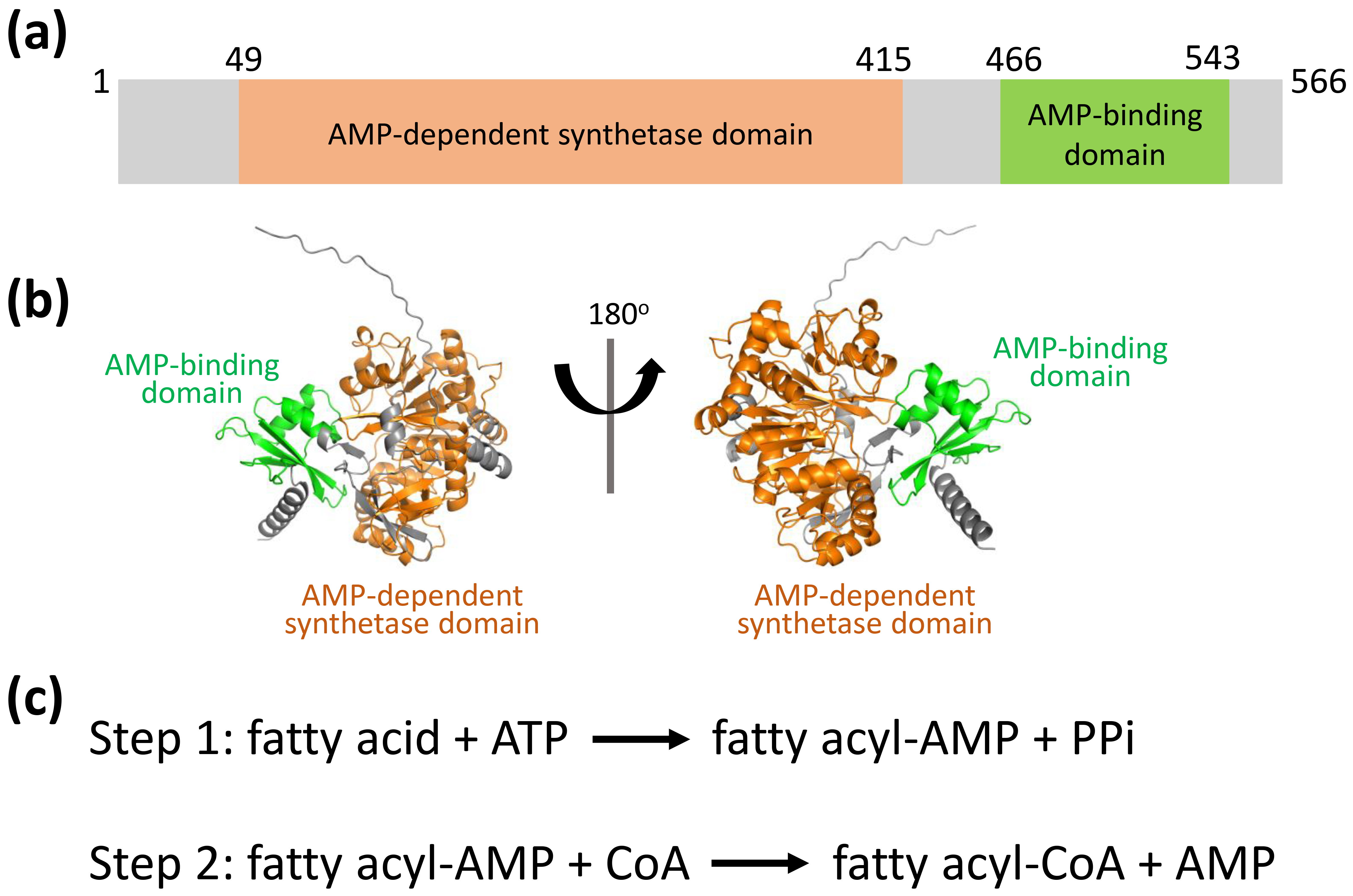
Bacterial species possess a diverse and distinct array of lipids, which play a critical role in their pathogenic mechanisms and disease development [11, 12, 13, 14, 15, 16, 17]. These microorganisms encounter numerous environmental stressors, including fluctuations in pH, reactive oxygen species, enzymatic breakdown, and shortages of vital nutrients or nitrogen compounds [16, 17, 18, 19, 20, 21]. Such hostile conditions frequently compromise the integrity of their lipid-dense cellular membranes, which are targeted by host defense systems [14, 20, 21, 22]. To ensure survival, bacteria have developed sophisticated mechanisms to adapt their lipid-dense cellular membranes. These structures are produced through the activity of key enzymes such as fatty acid-coenzyme A (CoA) ligases and acyl-CoA synthetases [12, 16, 17, 20, 21, 23, 24, 25, 26, 27, 28], which are important targets for developing new antibacterial drugs. In E. coli, the fatty acid-CoA ligase FadK exhibits substrate specificity for short-chain fatty acids (below C10) and contributes to their anaerobic metabolic processing [29]. E. coli FadK features two conserved domains: AMP-dependent synthetase domain (aa 49–415) and AMP-binding domain (aa 466–543) (Fig. 1a,b). The catalytic mechanism of FadK proceeds through a two-step reaction characterized by the formation of an acyl adenylate (acyl-AMP) intermediate (Fig. 1c). However, the function and the mechanism of E. coli FadK are still unknown.
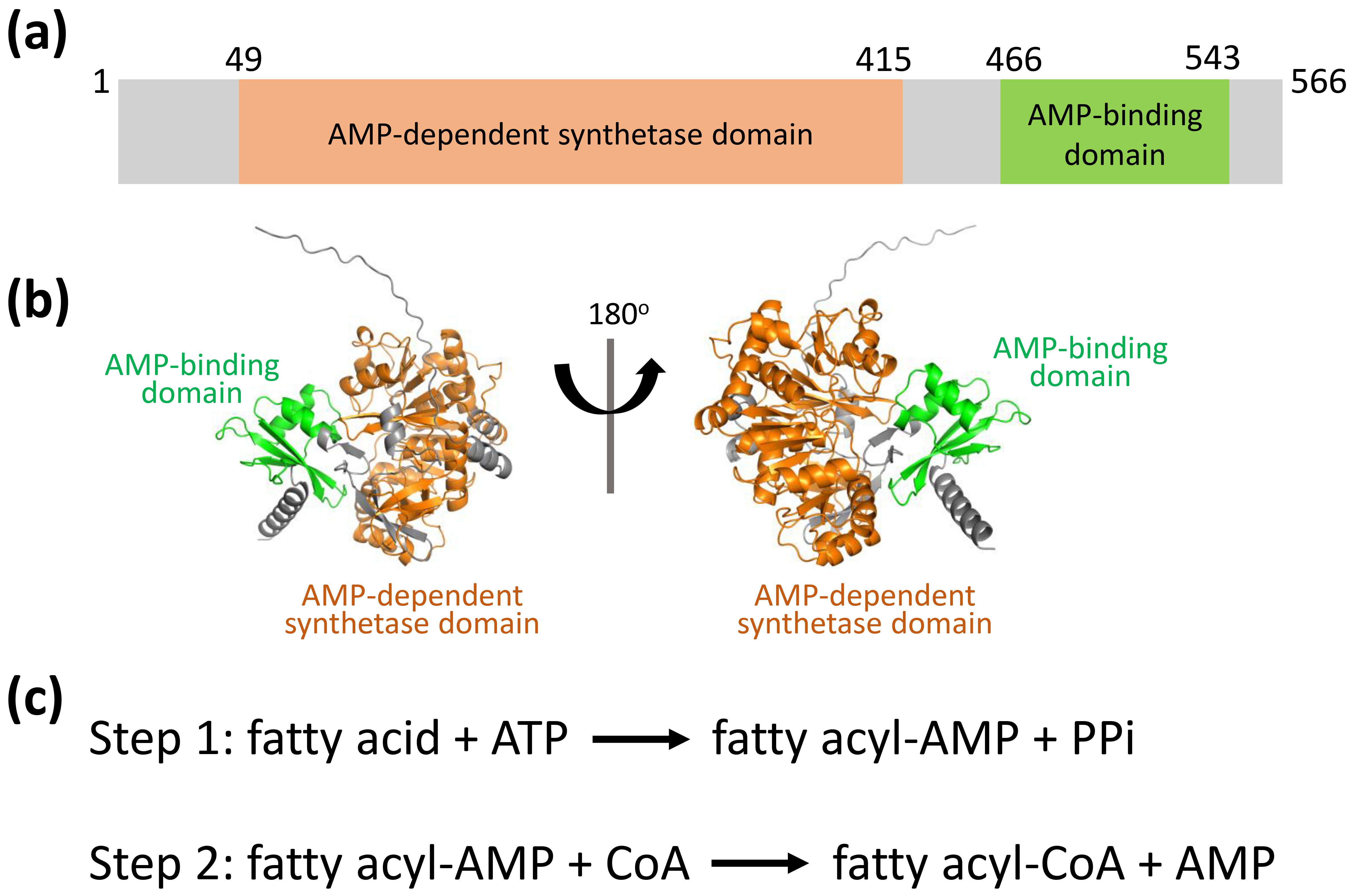 Fig. 1.
Fig. 1.
FadK structure predicted using AlphaFold2. (a) Schematic representation of the AMP-dependent synthetase domain (aa 49–415, in orange) and the AMP-binding domain (aa 466–543, in green). (b) FadK structure predicted by AlphaFold2, ribboned, in two orientations, with the two conserved domains colored as shown in Fig. 1a. (c) The enzyme mechanism is elucidated, involving a two-step reaction marked by the generation of a fatty acyl-AMP intermediate. CoA, coenzyme A; PPi, pyrophosphoric acid.
Herein, we successfully obtained monomeric FadK. We identified caprylic acid as the preferred substrate for FadK in fatty acid CoA ligase activity. The optimal conditions for the catalytic reaction were found to be pH 7.4, 0.6 mM ATP, 0.8 mM CoA, 0.8 mM Mg2+ at 37 °C. The enzymatic activity of FadK showed a decreasing trend with increasing concentrations of dodecyl-AMP. These findings offer substantial potential for designing innovative therapeutics targeting E. coli infections.
The primers specific to the recombinant FadK were custom-designed for PCR amplification and then synthesized by Shanghai Sangon Biotechnology (Shanghai, China). Subsequently, target genes were integrated into pET-28a(+) vector (Novopro, Shanghai, China) (Supplementary Table 1). PCR experiments were conducted utilizing Q5 polymerase (New England Biolabs, Ipswich, MA, USA). DNA sequence analysis was performed to confirm the correctness of the cloning process. The resulting plasmid constructs were then introduced into E. coli BL21(DE3) cells.
FadK expression on a large scale was conducted by inoculating bacterial cells
(20 mL) into LB medium supplemented (1 L) with kanamycin at 45 µg/mL,
followed by overnight incubation at 37 °C. Bacterial
cultures were incubated at 37 °C under constant agitation (220 rpm)
until achieving an optical density (OD) of 0.6–0.8 at 600 nm. Subsequently, the
culture was incubated at 25 °C with 1 mM Isopropyl
Nickel-Nitrilotriacetic acid (Ni-NTA) was employed to purify various protein expression constructs of FadK according to our previous reports [30]. Bacterial cells were suspended in a buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.4, 5 mM DTT, 5% glycerol) and lysed at 650 MPa for 3 min. After centrifugation, insoluble debris was removed and the soluble lysate was applied to a Ni-NTA affinity column pre-equilibrated with the same buffer. The column was washed with appropriate buffers to remove unbound proteins. FadK was eluted with 300 mM imidazole, concentrated and further purified by size exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare, Chicago, WI, USA).
FadK enzymatic activity was assessed through spectrophotometric measurement of coenzyme A (CoA) thiol group consumption, with 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) (Sigma-Aldrich, St. Louis, MO, USA) serving as the detection reagent. This approach, adapted from established protocols [31, 32, 33], was based on the fatty-acyl-CoA ligation reaction. The interaction between free thiols and DTNB produced a chromogenic response, which was measured by absorbance at 412 nm (OD412 nm).
The enzymatic activity assay was conducted using a 90 µL reaction solution
comprising 6 µg FadK, 10 mM ATP, varying fatty acid concentrations, 0.1 mM
Tris-HCl (pH 8.0), 10 mM K+ and 10 mM MgCl2. To enhance fatty acid
solubility, 0.001% (w/v) Plysurf A-210G was incorporated as a dispersant. For
fatty-acyl-CoA ligation activity determination, the inhibitor dodecyl-AMP was
included. After adding 10 µL of 5 mM CoA, the mixtures were incubated at 37
°C for 2 min, allowed to rest for 5 min, and heat-inactivated at 85
°C for 5 min. Post-cooling to ambient temperature, 70 µL of the
reaction sample was mixed with 600 µL of 400 µM DTNB in potassium
phosphate buffer (pH 7.0), and absorbance at 412 nm (OD412 nm) was recorded.
The DTNB extinction coefficient (13,600 M-1
Control reaction mixtures in which no protein was present were included for comparison. The results were evaluated at different concentrations of substrates, Adenosine triphosphate (ATP), CoA, and magnesium ion (Mg2+) concentrations, as well as different temperatures and pH values. Additionally, the inhibitor dodecyl-AMP was introduced to the bacterial suspension.
Bacterial cultures in the logarithmic growth phase were employed. Overnight
cultures were adjusted to inoculum densities of 1.73
Real-time quantitative polymerase chain reaction (RT-qPCR) was used to measure FadK gene expression in bacterial cultures harvested at late logarithmic growth. Cultures were exposed to dodecyl-AMP at concentrations of 2.5, 5 or 10 µM for 1 or 6 h, with the inhibitor administered as a single or split dose. Total RNA was isolated with the TransZol RNA extraction kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Next, cDNA was generated from the purified RNA using the PrimeScript 1st Strand cDNA Synthesis Kit (Takara, Kyoto, Japan). RT-qPCR was performed with PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) and primers detailed in Supplementary Table 3. Amplification and quantification were carried out on an Applied Biosystems QuantStudio 5 system (Thermo Fisher Scientific, Waltham, MA, USA). Gene expression levels were analyzed via 2-ΔΔCT method [30, 34, 35, 36, 37, 38]. with results displayed as log2-transformed values in histograms. Ratios exceeding zero denoted up-regulated expression, whereas values below zero indicated down-regulation. The housekeeping gene gyrA [39, 40, 41, 42] served as the reference for normalization, and its inclusion as a positive control confirmed assay validity.
All experiments were performed at least in triplicate, with results presented as
means
We designed and constructed prokaryotic expression plasmids to facilitate the expression of recombinant FadK protein, employing designated primers and restriction endonucleases (Supplementary Table 1). The induction of FadK protein expression was orchestrated within E. coli host cells. Subsequently, the supernatant was subjected to purification via Ni-NTA agarose affinity chromatography following cell lysis and centrifugation. To obtain highly pure FadK, purification parameters were systematically optimized. A buffer containing 150 mM NaCl, 5 mM DTT, 5% glycerol and 20 mM Tris-HCl (pH 7.4) was found to be critical to minimize non-specific hydrophobic and ionic interactions between FadK and co-purifying contaminants during chromatography. Additionally, 300 mM imidazole was added for FadK elution.
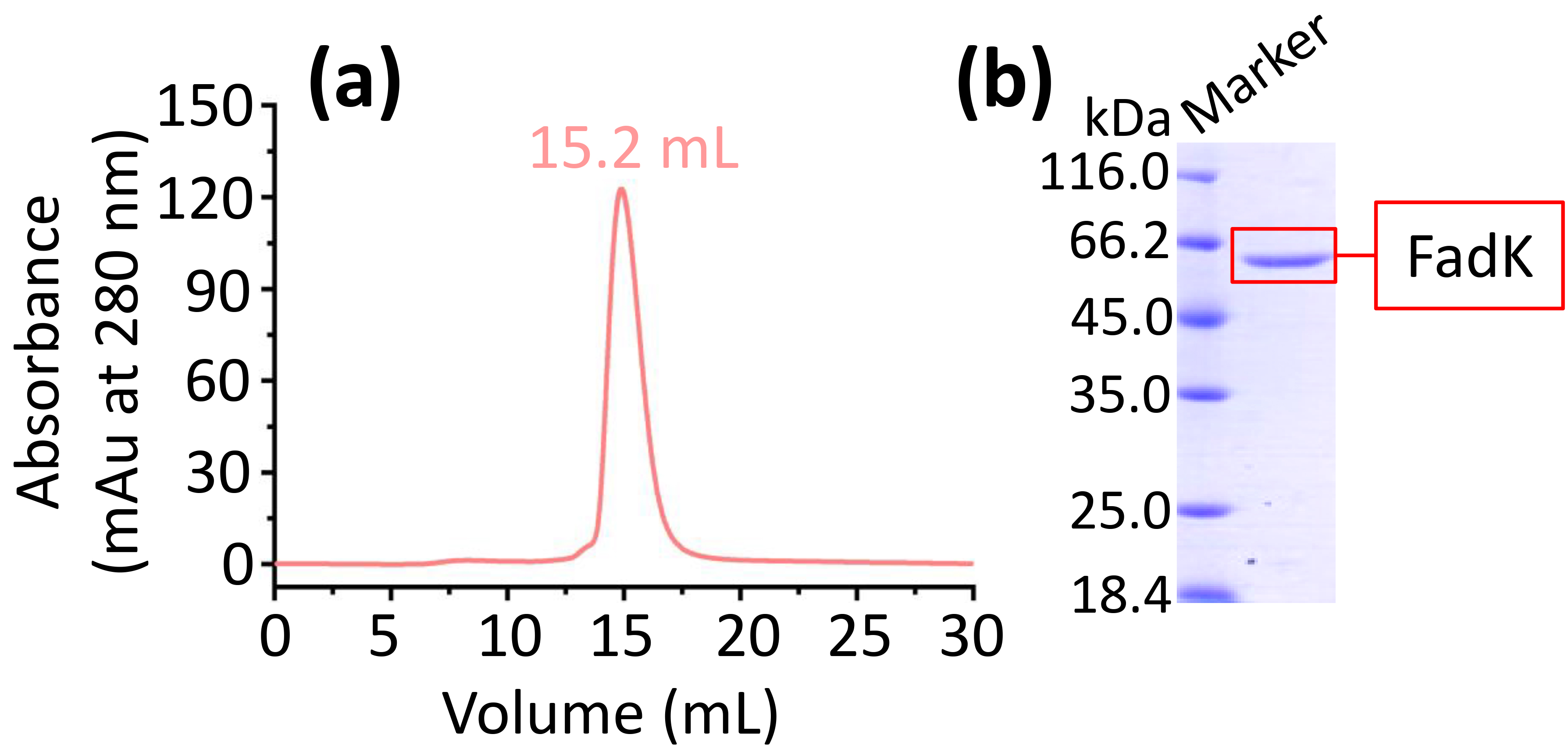
Furthermore, FadK purification was refined using the ÄKTA purification system. We found that the retention volume was 15.2 mL (Fig. 2a), implying the presence of monomeric forms of FadK protein.
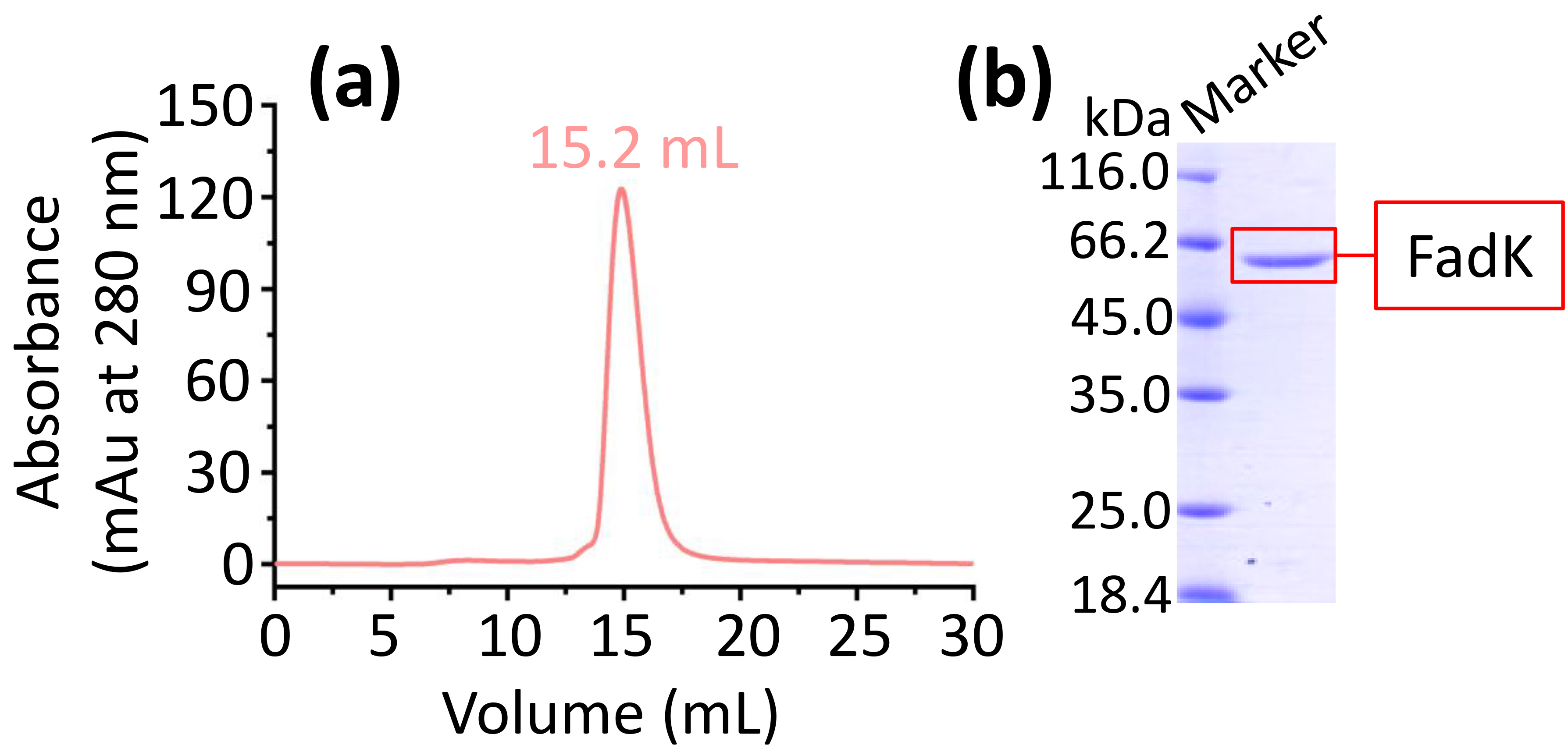 Fig. 2.
Fig. 2.
The expression and purification of FadK. (a) FadK was purified by size-exclusion chromatography on a Superdex 200 10/300. A single peak at 15.2 mL confirmed the monomeric state of FadK. (b) The peak fraction isolated from the ÄKTA purifier-based gel filtration was further analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) via a 12% gel.
Protein was collected based on the single peak observed in gel filtration chromatography. Subsequently, the protein was analyzed using 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), showing a band (Fig. 2b). Therefore, the high purity FadK was obtained and then used in the following experiments.
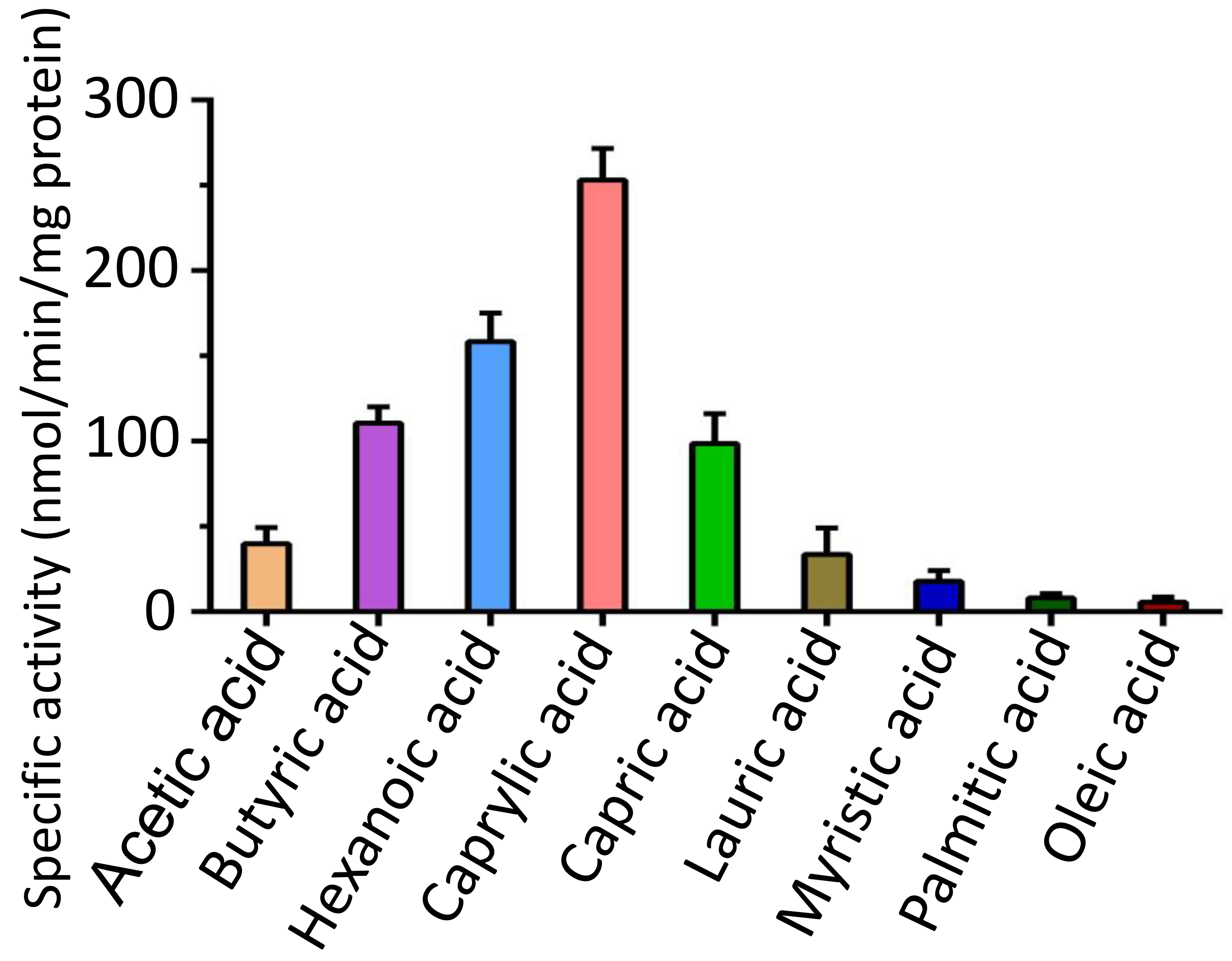
To assess the catalytic activity of FadK, we monitored the depletion of free thiol groups in CoA during FadK-catalyzed fatty acid-CoA ligation reactions [32, 33, 43, 44, 45]. We observed that specific activities of FadK at 253.1, 158.3, 110.6, and 98.7 nmol/min/mg protein, respectively, when utilizing caprylic acid, hexanoic acid, butyric acid, and capric acid as substrates (Fig. 3, Supplementary Fig. 1). In contrast, specific activities ranged from 6.4 to 39.8 nmol/min/mg protein when employing other fatty acids as substrates (Fig. 3). These showed the significantly enhanced catalytic efficiency of FadK with caprylic acid, hexanoic acid, butyric acid, and capric acid substrates compared to alternative fatty acids. Notably, FadK exhibited its utmost catalytic proficiency with caprylic acid, indicating that caprylic acid is the most stable of these fatty acid substrates (Fig. 3).
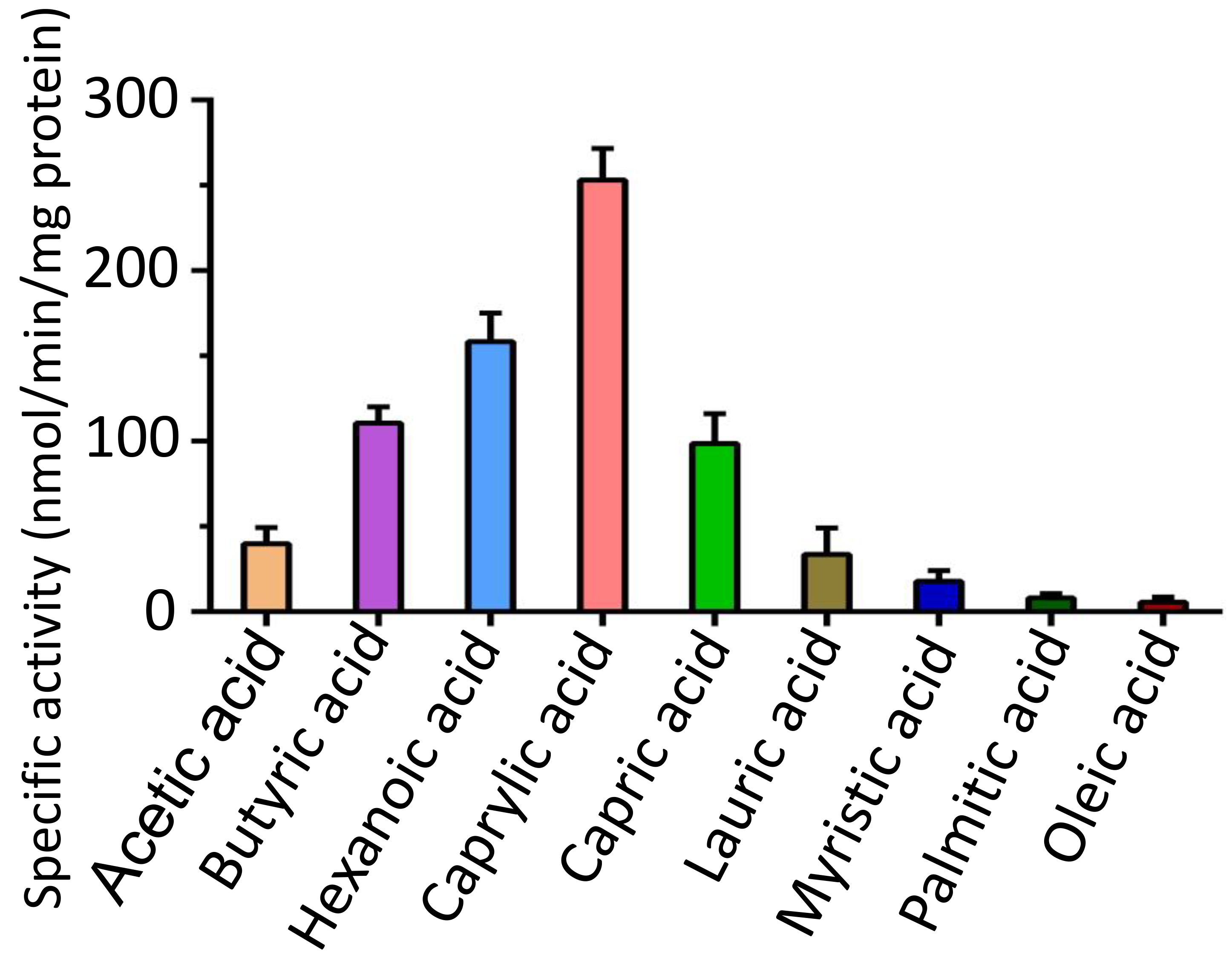 Fig. 3.
Fig. 3.
FadK different substrate specificity. Enhanced enzymatic activities were observed with caprylic acid, hexanoic acid, butyric acid, and capric acid substrates in comparison to other fatty-acid substrates. Caprylic acid was identified as the preferred substrate for FadK among these substrates.
The enhanced catalytic efficiency of FadK observed with caprylic acid as substrate is likely due to several factors, such as the participation of ATP and/or CoA in the catalytic process and the three-dimensional structural arrangement of the enzyme.
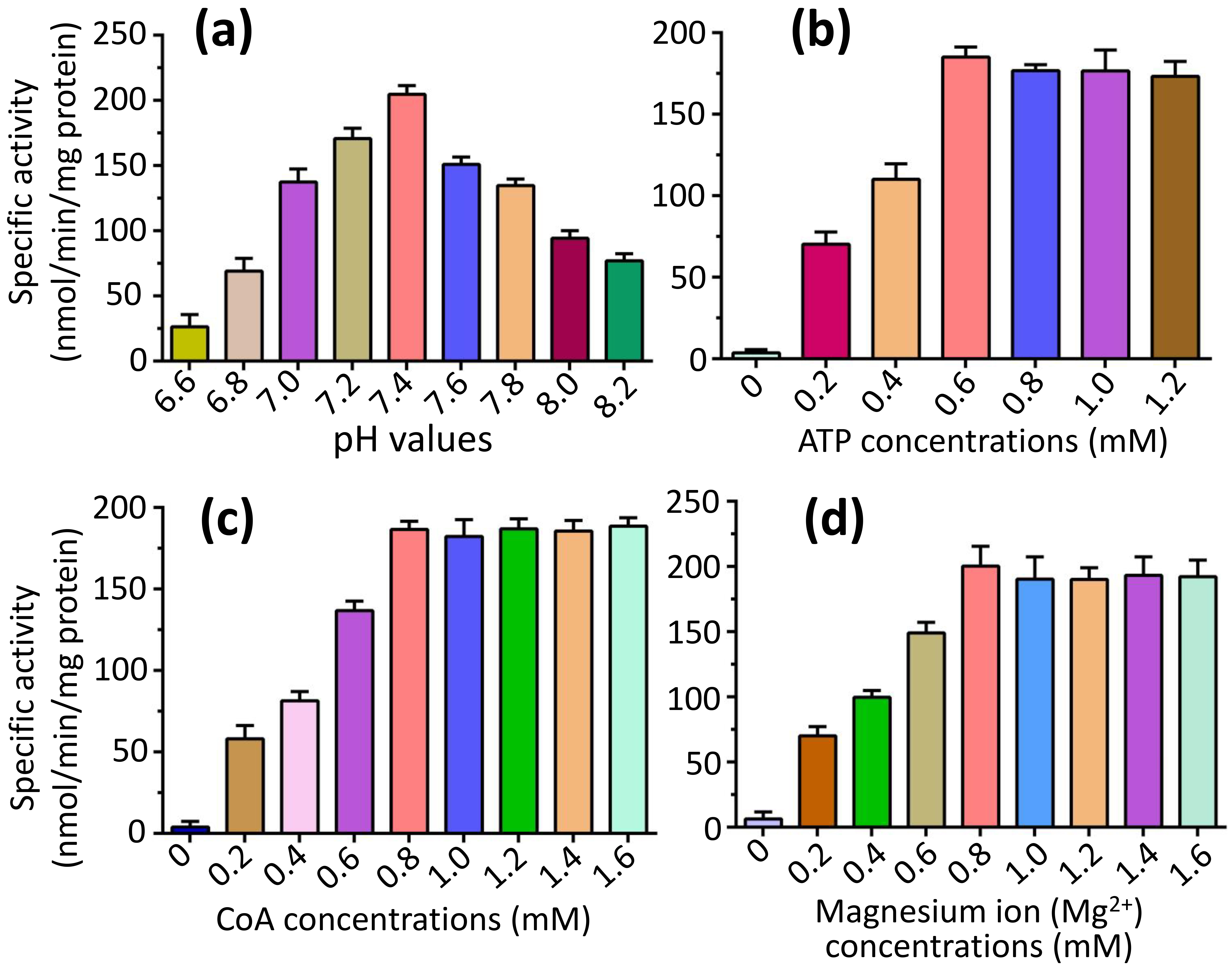
To explore the optimal parameters of the fatty-acid CoA ligase activity of FadK, we focused our investigation on the catalytic properties of the substrate caprylic acid, since it’s the optimal substrate for FadK (Fig. 3). We found that the fatty-acid CoA ligase activity of FadK exhibited an incremental trend with increasing pH, peaking at pH 7.4 before declining (Fig. 4a). Additionally, we observed a proportional increase in activity with increasing concentrations of ATP, reaching a peak at 0.6 mM ATP, beyond which no further increase was observed (Fig. 4b). Similarly, elevations in CoA concentration resulted in augmented activity, culminating at 0.8 mM CoA, with subsequent concentrations eliciting no additional effect (Fig. 4c). Furthermore, the activity of FadK surged with increasing Mg2+ concentrations, attaining maximal activity at 0.8 mM Mg2+, beyond which no further variations were noted (Fig. 4d). Consequently, we adopted these optimal conditions (pH 7.4, CoA at 0.8 mM, ATP at 0.6 mM and Mg2+ at 0.8 mM) for subsequent assessments of the activity.
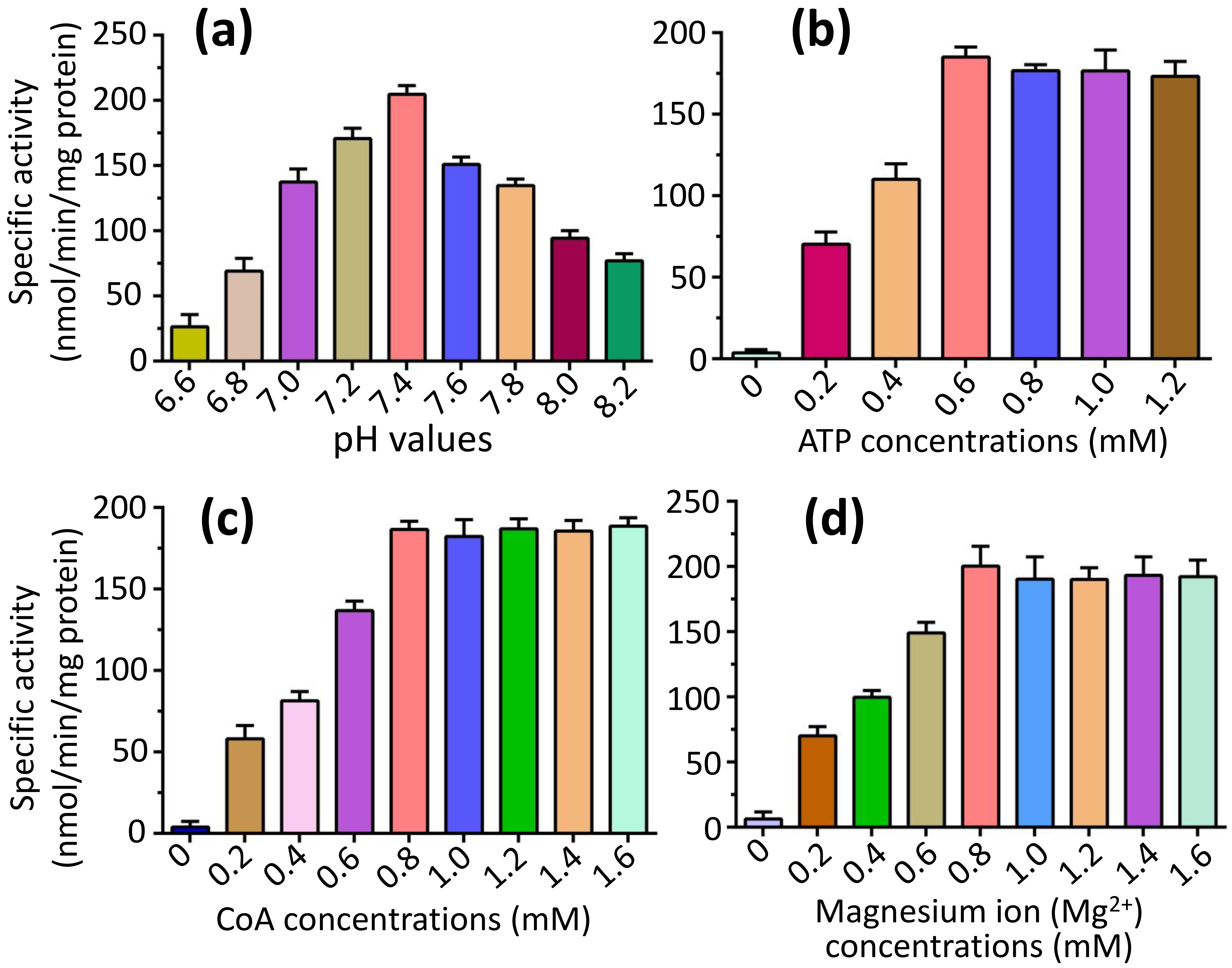 Fig. 4.
Fig. 4.
Comparison of the fatty-acid CoA ligase activity of FadK under varying conditions, encompassing different pH values, and concentrations of ATP, CoA, and Mg2+. (a) The activity of FadK exhibited an initial increase followed by a decrease with rising pH values, attaining maximal activity at pH 7.4. (b) Increasing concentrations of ATP led to a corresponding increase in activity, peaking at 0.6 mM ATP, beyond which no further change was observed. (c) Similarly, elevating CoA concentrations resulted in heightened activity, reaching a maximum at 0.8 mM CoA, with no subsequent alterations. (d) With increasing Mg2+ concentration, the activity of FadK escalated, achieving maximum activity at 0.8 mM Mg2+, followed by stability in activity levels.
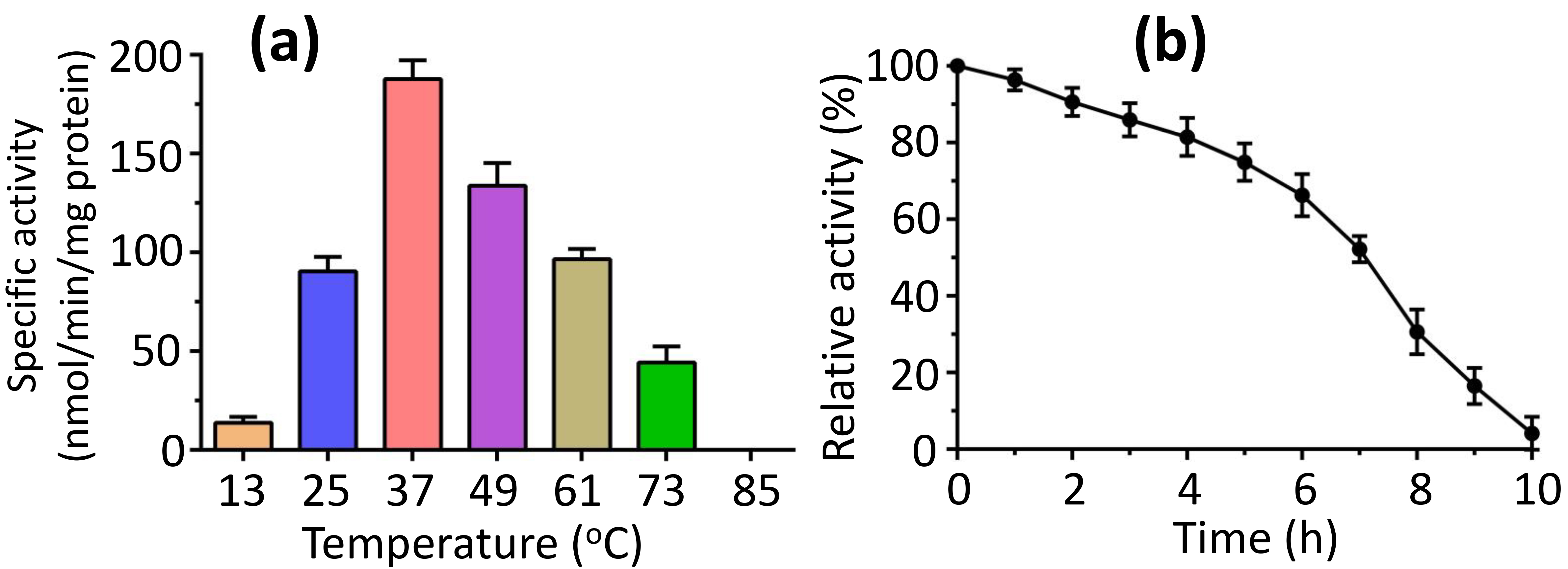
To explore how temperature affects activity, we measured activity over a temperature range of 13 to 85 °C. We found that the peak activity at 37 °C, with complete loss of the activity noted at 85 °C (Fig. 5a). Conversely, we evaluated the thermostability of FadK activity at 37 °C, finding that over 90% of the maximum activity persisted following a 2-h incubation at this temperature, and more than 50% of the peak activity of FadK remained intact after 7 h of incubation at 37 °C (Fig. 5b).
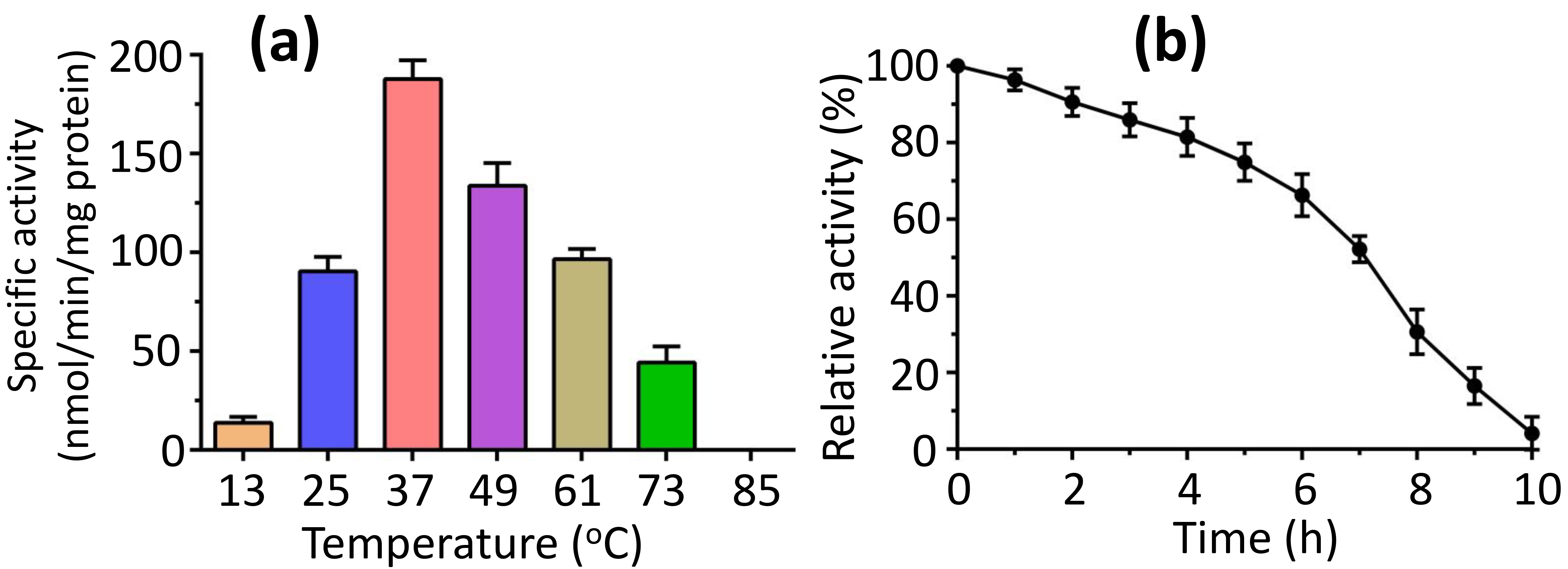 Fig. 5.
Fig. 5.
Temperature stability of the fatty-acid CoA ligase activity of FadK. (a) The impact of temperature on FadK activity revealed optimal activity at 37 °C, while FadK demonstrated complete inactivity at 85 °C. (b) Analysis of temperature stability indicated a decline in FadK activity over time.
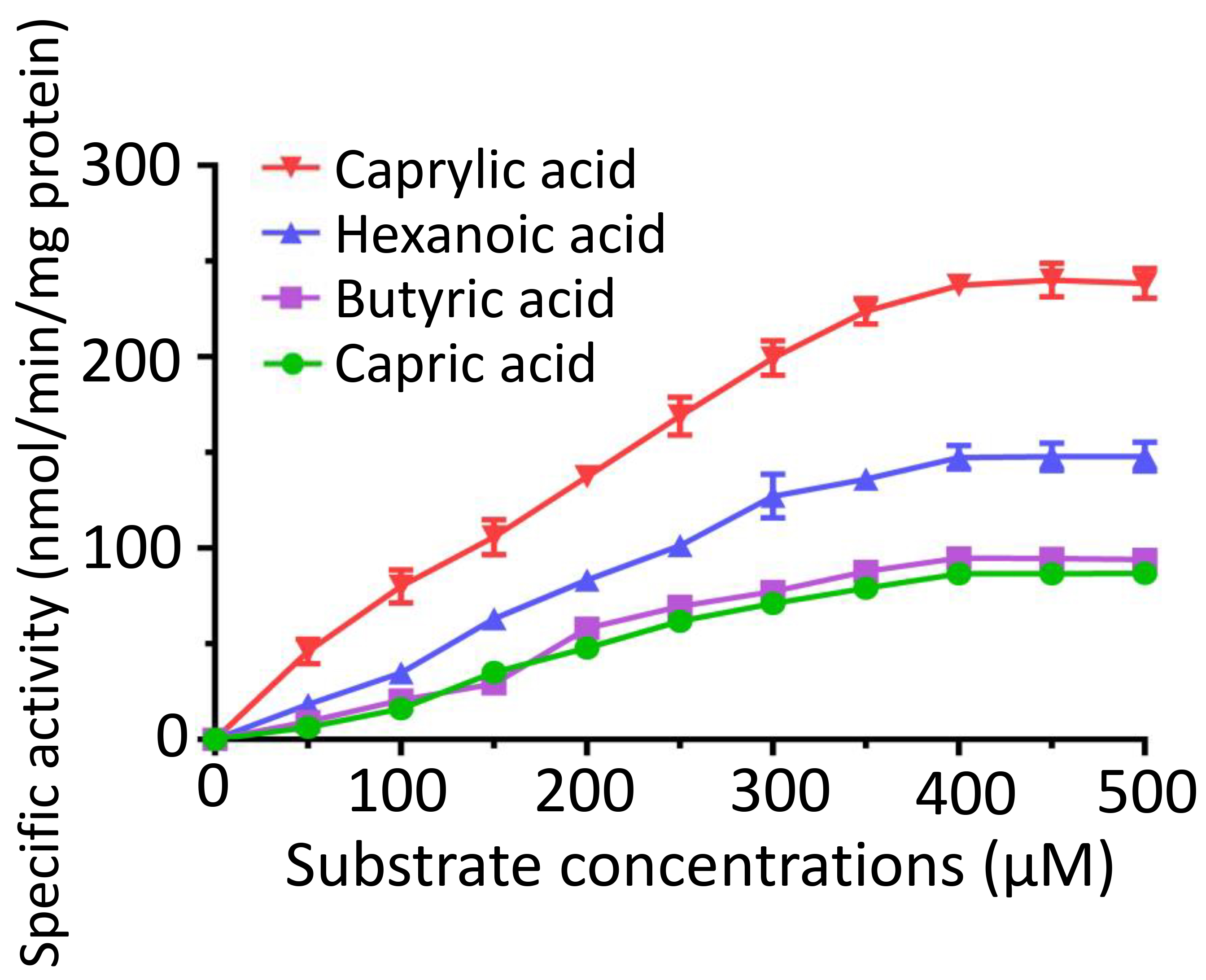
We conducted a kinetic analysis of FadK across various concentrations of caprylic acid, hexanoic acid, butyric acid, and capric acid. We observed an increase in the specific activities of FadK with an increase in the substrate concentration, plateauing at 400 µM (Fig. 6). The Michaelis constant (K𝑚) for caprylic acid was 330.2 µM, notably lower than those of other tested substrates, including butyric acid (946.4 µM), hexanoic acid (857.2 µM), and capric acid (749.5 µM) (Supplementary Table 2). This suggested that of the four substrates, FadK showed the highest level of activity on caprylic acid. Among these substrates, caprylic acid was found to be the optimal substrate for FadK (Fig. 6), in agreement with our previous results (Fig. 3).
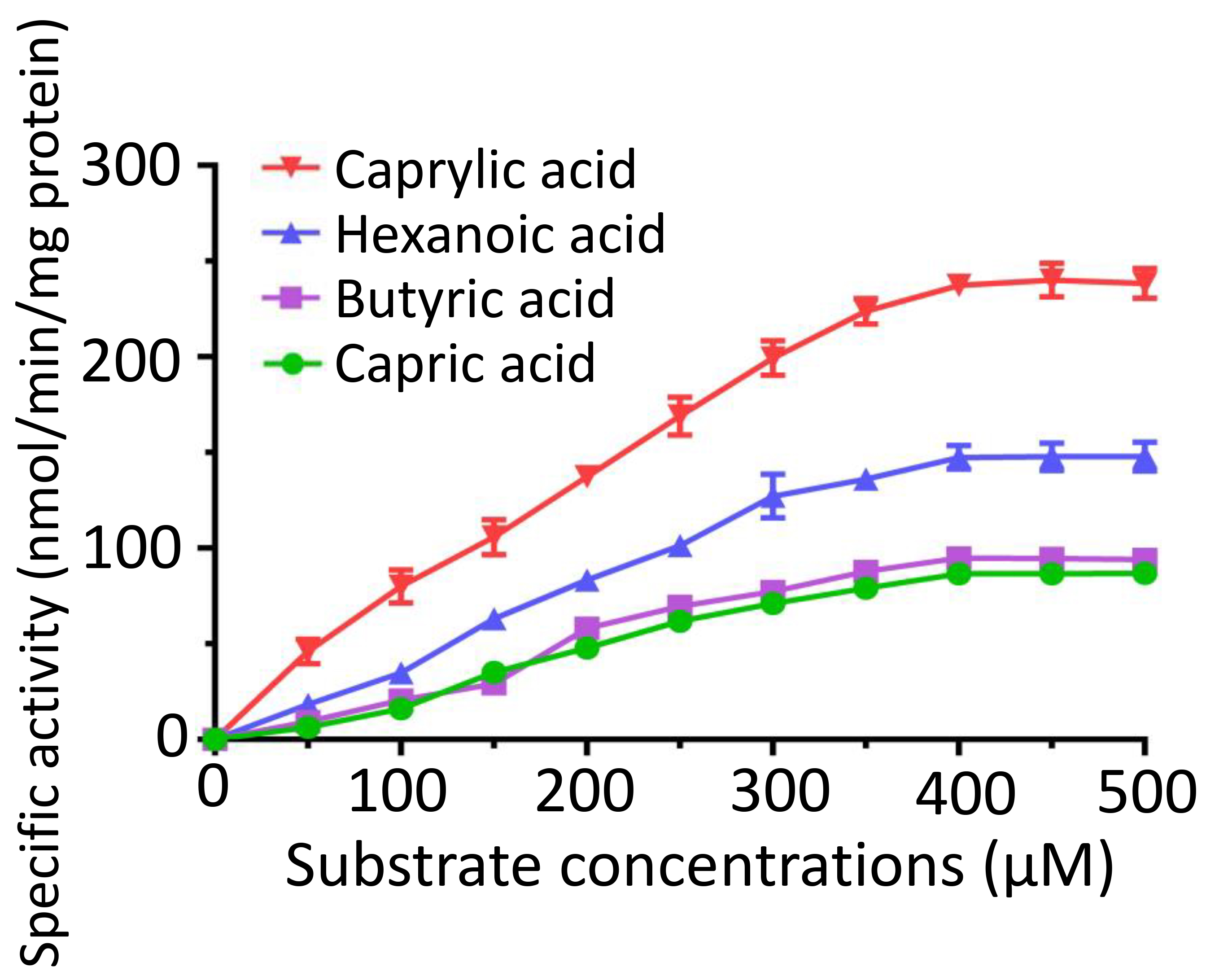 Fig. 6.
Fig. 6.
Enzymatic characterization of FadK across different substrate concentrations. Kinetic profiles were constructed by assessing the activity of FadK using multiple substrates (caprylate, hexanoic acid, butyrate, and capric acid).
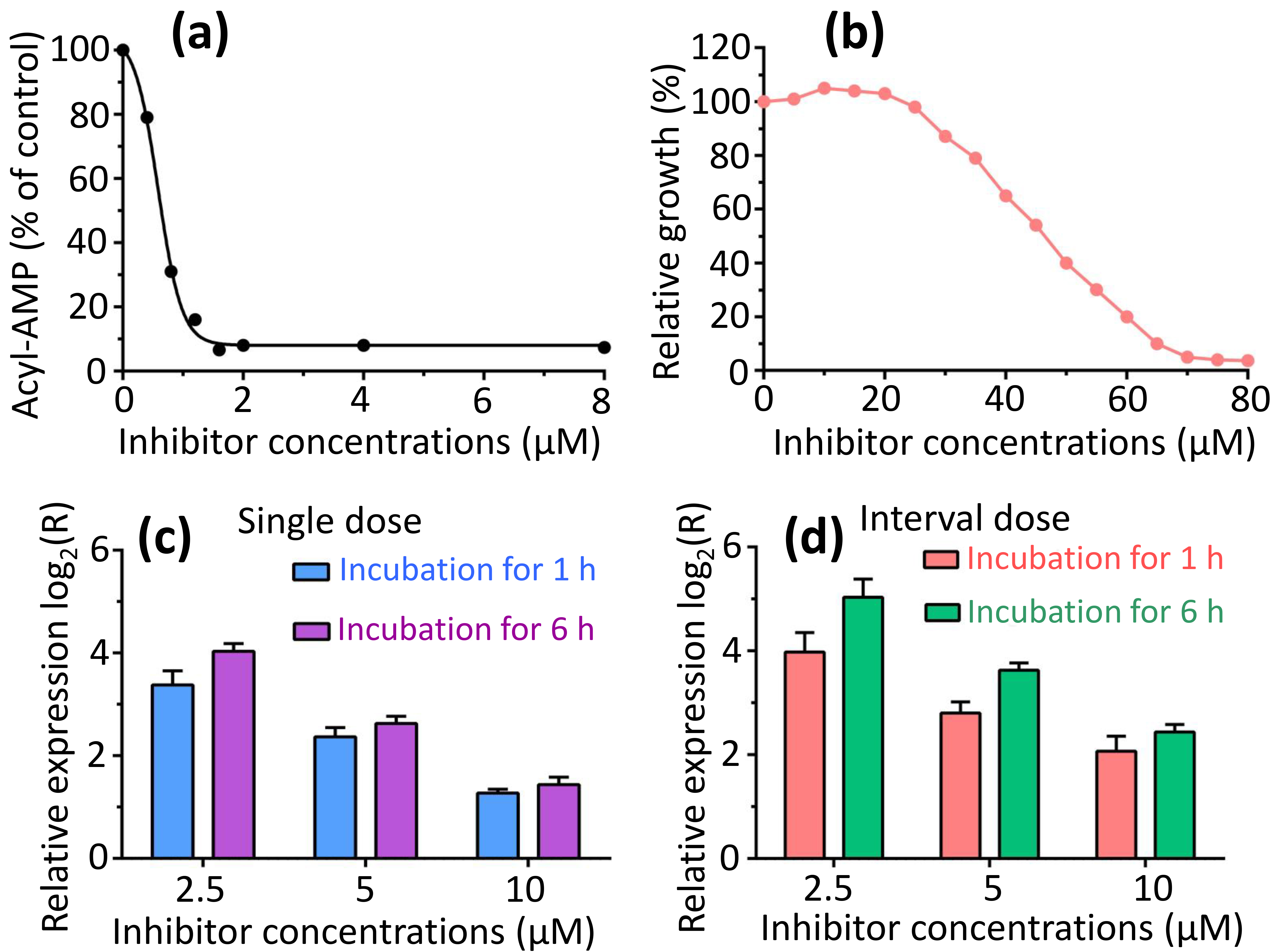
The primary aim in identifying and characterizing specific proteins lies in their potential as novel targets for the development of therapeutics. FadK emerges as one such newly discovered candidate. Recent investigations have utilized various nonhydrolyzable acyl-AMP analogs, including dodecyl-AMP, as mechanism-based inhibitors, which have demonstrated potent inhibitory effects on related adenylate-forming enzymes [46]. We examined the inhibitory impact of dodecyl-AMP on FadK activity, and observed a concentration-dependent inhibition, with over 90% reduction in activity at 2 µM (Fig. 7a). The half-maximal effective concentration (EC50) of the inhibitor dodecyl-AMP was calculated to be 0.56 µM, notably less than 10 times the concentration of FadK employed in the experiments. Furthermore, we investigated whether the inhibitor influenced bacterial growth. We found a dose-dependent inhibition of growth when the inhibitor was added to bacterial suspensions, with almost 90% loss of viability at 65 µM (Fig. 7b). These results underscore the impact of dodecyl-AMP inhibitor on the fatty acid CoA metabolism of bacterial strains, thereby influencing bacterial growth.
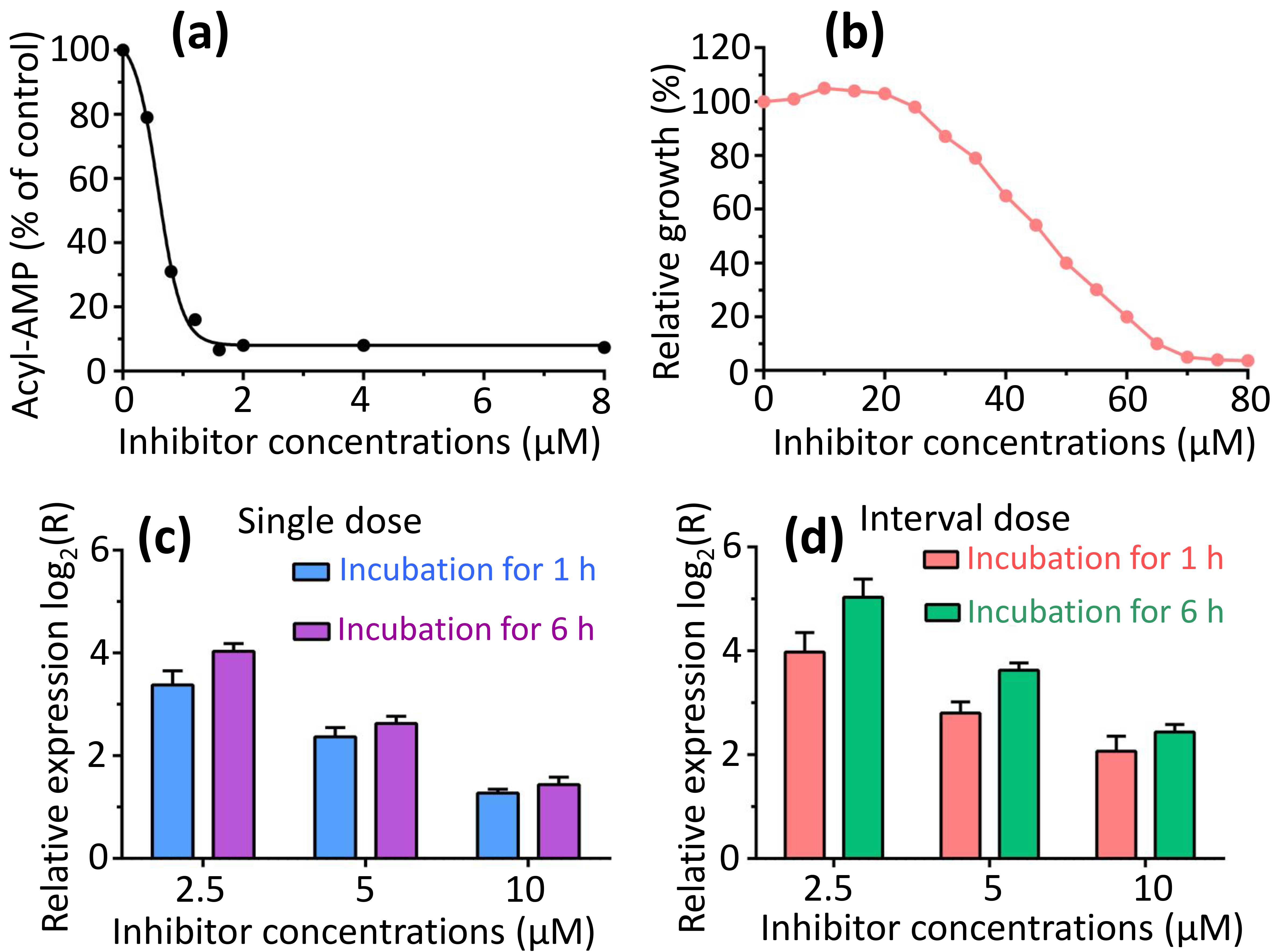 Fig. 7.
Fig. 7.
Dodecyl-AMP inhibited both FadK enzyme activity and bacterial proliferation. (a) The fatty-acid CoA ligase activity of FadK was assessed with the increase of dodecyl-AMP concentrations, revealing a decline in activity with increasing inhibitor concentrations. (b) Bacterial growth exhibited progressively stronger inhibition with rising inhibitor concentrations. (c,d) Gene FadK levels were measured via real-time quantitative polymerase chain reaction (RT-qPCR). Gene FadK expression levels were evaluated under single dose (c) and interval dose (d) conditions.
The central goal of the identification and functional characterization of essential, distinct proteins in E. coli is their potential as therapeutic targets for novel antibacterial agents [47, 48]. Among these candidates, FadK has attracted attention as a viable drug target. Non-hydrolyzable acyl-AMP mimetics screened as mechanism-driven inhibitors show potent inhibition against adenylate forming enzymes such as FadK, highlighting their therapeutic relevance [47, 48, 49]. We hypothesized that dodecyl-AMP would serve as a potent inhibitor of the fatty-acid CoA ligase activity of FadK.
To delve into the antibacterial mechanism of the dodecyl-AMP inhibitor at a molecular level, we quantified the expression of the FadK gene via RT-qPCR across a range of inhibitor concentrations. To ensure the viability of E. coli during qPCR signal measurement, we employed two approaches: (1) using the inhibitor at low concentrations (2.5, 5 and 10 µM), and (2) involving interval addition of the inhibitor.
We found a dose-dependent up-regulation of the FadK gene with increasing concentrations of the inhibitor (Fig. 7c,d). At singular doses, the FadK gene exhibited up-regulation by 10.2-, 5.3-, and 2.4-fold at 2.5, 5 and 10 µM inhibitor after incubation for 1 h, respectively. Similarly, following a 6-h incubation, up-regulation was noted at 16.2-, 6.2-, and 2.7-fold for the same concentrations (Fig. 7c). With interval dosing, the FadK gene displayed up-regulation by 15.7-, 7.0-, and 4.2-fold at 2.5, 5 and 10 µM inhibitor after incubation for 1 h, while after 6 h, up-regulation was observed at 32.7-, 12.4-, and 5.4-fold for the respective concentrations (Fig. 7d). These differential expression profiles of the FadK gene underscore its physiological importance in the fatty acid CoA metabolism for the survival of the bacteria.
In this study, we obtained FadK in monomeric form and identified caprylic acid as its optimal substrate, exhibiting maximum activity at 400 µM concentration. The optimal conditions for the fatty-acid CoA ligase activity of FadK were determined as pH 7.4 at 37 °C, with 0.8 mM CoA, 0.6 mM ATP and 0.8 mM Mg2+, respectively. We observed a decrease in FadK activity with increasing concentrations of dodecyl-AMP. Our work elucidates the catalytic behavior of FadK and sheds light on its preference for specific fatty acid substrates, providing critical insights for designing innovative therapies targeting E. coli infections.
A previous study has indicated that the expression of the FadK gene is suppressed during aerobic growth [29]. FadK gene expression reaches its peak under anaerobic conditions when fumarate, the terminal electron acceptor, is present. This enzyme preferentially catalyzes reactions with short-chain fatty acid substrates. In contrast, our study provides a more comprehensive analysis, identifying the optimal conditions for FadK catalytic activity as pH 7.4, 0.6 mM ATP, 0.8 mM CoA, 0.8 mM Mg2+, and 37 °C. Caprylic acid was found to be the most effective substrate for FadK activity. Moreover, dodecyl-AMP was shown to inhibit both FadK enzymatic function and bacterial growth. Additionally, we explored the thermostability and kinetic characteristics of FadK with various substrates.
Owing to the failure to generate protein crystals, structural predictions were derived using AlphaFold2. These persisting limitations prompted further exploration of FadK’s functional mechanisms. To identify structural homologs, SWISS-MODEL [50, 51, 52] was employed (Supplementary Table 4). Comparative analysis revealed that FadK exhibited amino acid sequence identities of 33.59% with sesquiterpene synthases from Mycolicibacterium smegmatis, 33.27% with those from Thermobifida fusca, 32.69% with Streptomyces gandocaensis, 32.35% with Escherichia coli, and 32.32% with Acinetobacter baumannii (Supplementary Table 4). Collectively, these findings offered critical clues regarding the structural and functional attributes of FadK in E. coli.
Overall, E. coli FadK, given its classification within the medium-chain fatty acid CoA ligase family, warrants further investigation to elucidate additional functional and mechanistic intricacies.
In this work, we reported mechanism and function of E. coli FadK: (1) FadK was obtained in monomeric form; (2) Caprylate represents the most suitable substrate for FadK regarding the fatty acid CoA activity; (3) Optimal conditions for FadK catalysis are pH 7.4, 0.6 mM ATP, 0.8 mM CoA and 0.8 mM Mg2+ at 37 °C; (4) The fatty-acid CoA ligase activity of FadK decreased with increasing dodecyl-AMP concentrations, which was further confirmed by the results of RT-qPCR. Our study elucidated the fatty acid-CoA ligase function of FadK and uncovered its distinct substrate specificity toward fatty acids. These insights enabled the design of novel therapeutics effective against severe, including terminal, E. coli infections.
The data presented in this study are available in this article (and Supplementary Materials), and further inquiries can be directed to the corresponding authors.
Conceptualization, DL; Data curation, DL; Formal analysis, DL; Investigation, DL, AA, LZ and FY; Methodology, DL; Resources, DL; Software, DL; Validation, DL; Writing — original draft, DL; Writing — review & editing, DL. All authors contributed to editorial changes in the manuscript. All authors have read and approved the final version of the manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Not applicable.
We thank the staff of BL02U1, BL18U1 and BL19U1 beamlines at Shanghai Synchrotron Radiation Facility, Shanghai, People’s Republic of China, for help with X-ray data collections.
This research received no external funding.
The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/FBL36701.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.