1 Department of Respiratory Medicine, Children’s Hospital, Zhejiang University School of Medicine, 310052 Hangzhou, Zhejiang, China
2 National Clinical Research Center for Child Health, 310052 Hangzhou, Zhejiang, China
3 Faculty of Dentistry, The University of Hong Kong, Hong Kong, S.A.R., China
4 Endodontic Department, Changzhou Stomatological Hospital, 213000 Changzhou, Jiangsu, China
Abstract
This review presents a comprehensive overview of single-cell RNA sequencing (scRNA-seq) analyses used to study tooth and periodontal tissues. The intricate cellular composition of both teeth and periodontium are revealed, leading to the identification of new cell types and tracing lineage profiles for each cell type. Herein, we summarize the progression of dental and periodontal tissue formation, tooth homeostasis, and regenerative mechanisms. scRNA-seq analyses have demonstrated that the cellular constituent ratio of dental and periodontal tissues transforms homeostasis or injury repair. Importantly, single-cell data in the diseased tissue demonstrated a change in both cell types and intercellular communication patterns compared to the normal state. These findings provide valuable insights into the underlying disease mechanisms at the cellular level in the context of single-cell vision, thereby facilitating the investigation of potential therapeutic interventions.
Keywords
- single-cell analysis
- tooth components
- stem cells
- regeneration
- cell differentiation
The sequencing technique can be traced back to 1975 when Frederick Sanger invented Sanger sequencing [1]. With technological advancements, bulk sequencing has become increasingly cost-effective and convenient, leading to its application in various medical disciplines. Bulk sequencing approaches have been used to discover novel biomarkers, disease prediction and diagnosis, and treatment optimization [2, 3, 4, 5]. Bulk sequencing represents average gene expression levels in specific tissues. However, the individuality and interaction of each cell are not captured. Additionally, bulk sequencing cannot discern the unique characteristics of individual cells. In 2009, Tang et al. [6] introduced an initial single-cell sequencing approach. Since then, single-cell sequencing has been extensively used for the systematic categorization of cells, identification of novel cell types [7, 8, 9], discovery of differentially expressed genes [10, 11], investigation of cell fate [12], and exploration of gene regulatory mechanisms [13]. This technique offers an accurate means of obtaining single cell genetic information, thereby impartially addressing previously unresolved issues [14].
Teeth and periodontium play crucial roles in mastication and digestion. Numerous studies have examined the histological and cellular aspects of teeth and periodontium morphogenesis and development [15, 16]. Nevertheless, there is a lack of understanding regarding the variations in gene expression among different cell types and their consequent impact on tissue development and repair.
Recently, single-cell RNA sequencing (scRNA-seq) has become the predominant strategy in most research endeavors focused on single-cell analysis [17, 18]. This review encompasses recent studies that used scRNA-seq to investigate homeostasis and repair processes in both the tooth and periodontium. Furthermore, the cellular heterogeneity within the diseased tissue was analyzed.
The classification of dental epithelial cells is based on four distinct types, namely, the outer enamel epithelium (OEE) and inner enamel epithelium (IEE), which converge at the turning corner of cervical loops, the stellate reticulum (SR) layer consisting of star-shaped cells, and the stratum intermedium (SI) layer that covers the ameloblasts. Subsequently, IEE cells differentiate into ameloblasts, which are responsible for enamel production. The classification of dental epithelial cells is primarily determined by their morphological characteristics [19]. However, the precise role of these cellular subtypes remains unclear.
The enamel formation process has four sequential stages: presecretory, secretory, transitional, and maturation. This classification considers the morphological and functional characteristics of the ameloblasts. During the presecretory stage, odontoblasts secrete pre-dentin, deposited at the dentinal enamel junction (DEJ). Subsequently, pre-ameloblasts form cytoplasmic projections that penetrate the basement membrane, substituting the mineralized pre-dentin. The pre-ameloblasts differentiate into ameloblasts, with Tomes’ process located at the apex closest to the most recently formed enamel. In the secretory stage, ameloblasts subsequently initiate the secretion of numerous enamel matrix proteins from the DEJ in a coronal direction. The enamel layer attains full thickness at the end of the secretory stage. During the transition and maturation stages, a significant portion of matrix proteins undergo hydrolysis, resulting in the mature mineralization of the enamel layer [20]. Continuous growth of the mouse incisor throughout its lifespan serves as a compensatory mechanism for enamel wear [21]. Consequently, the mouse incisor is an exemplary model for investigating the composition and regenerative properties of enamel (Fig. 1).
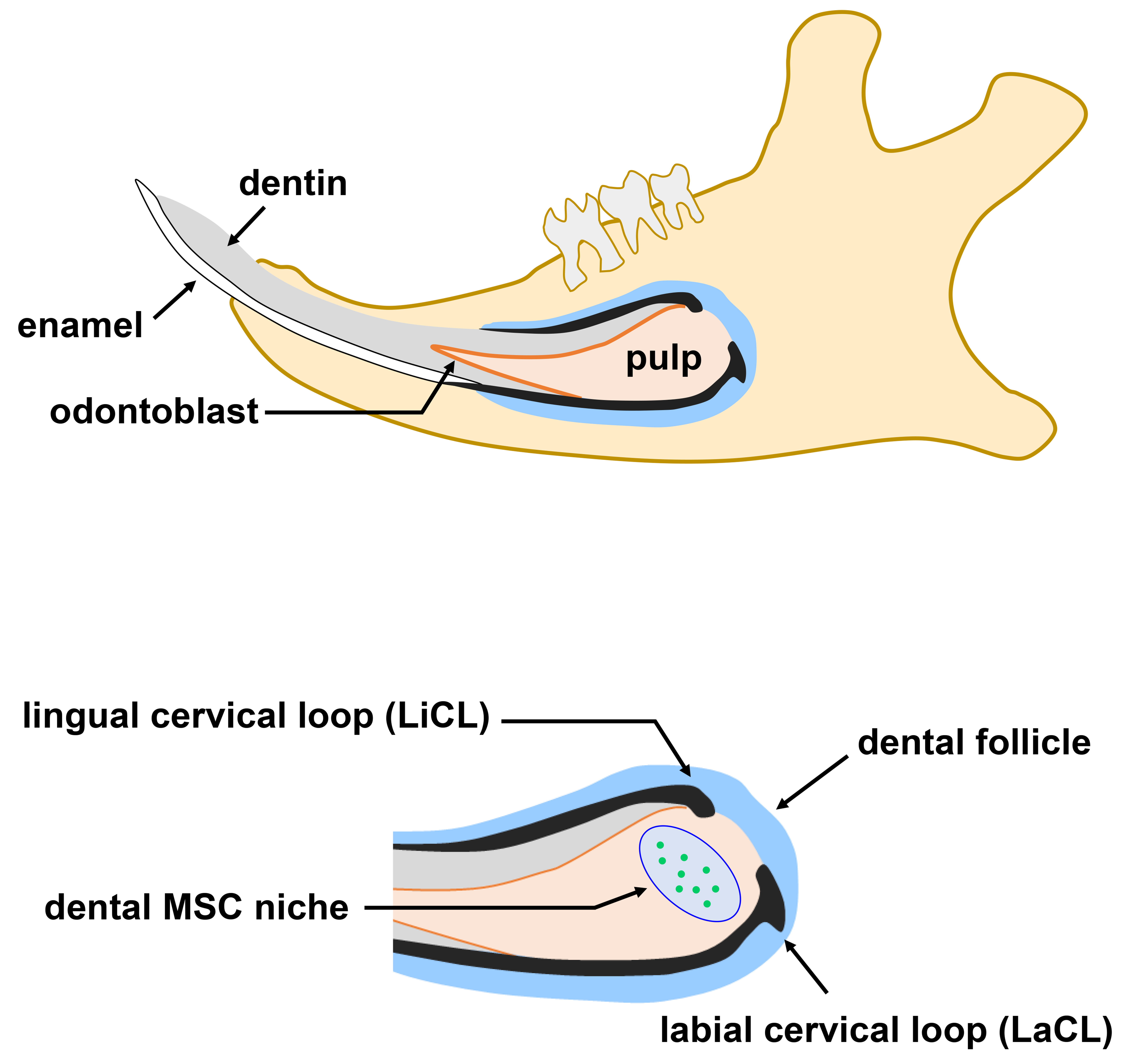 Fig. 1.
Fig. 1. Structure of mouse teeth. The morphology and structural features of mouse molars are similar to those of human molars. Mouse incisors undergo continuous growth throughout their lifespans. The regeneration process of the tooth is facilitated by the presence of cervical loop regions, primarily situated at the proximal end. Within these regions, dental epithelial stem cells predominantly present in the labial cervical loop (LaCL) differentiate into enamel-secreting ameloblasts. Conversely, dental mesenchymal stem cells, located between the LaCL and the lingual cervical loop (LiCL), produce dentin-secreting odontoblasts and contribute to the formation of the dental pulp. The figures are drawn by PowerPoint (Microsoft, office 2019, Redmond, WA, USA). MSC, mesenchymal stem cell.
Three recent studies investigated amelogenesis in mouse incisors using single-cell methods [22, 23, 24]. Chiba et al. [22] examined the mouse incisor epithelium using scRNA-seq and identified four epithelial clusters based on previously established markers. Two subpopulations were distinguished within the ameloblast population, with Dentin Sialophosphoprotein (DSPP) and Ameloblastin (AMBN) as the respective annotations. The IEE/OEE and SI/SR clusters were considered as the two epithelial cell types. The IEE/OEE cluster was characterized by Tbx1/frp5, whereas the SI/SR cluster was characterized by Notch1/Notch2. Krivanek et al. [23] identified 13 distinct subclusters of epithelial cells in mouse incisors that co-expressed Krt14 and Cdh1. Sharir et al. [24] used an unbiased approach to distribute cells, resulting in the identification of three clusters of dental epithelial cells: cycling cells, ameloblasts (including pre-ameloblasts), and non-ameloblast cells (Table 1, Ref. [22, 23, 24]). The spatial positioning of these populations was examined using single-molecule in situ hybridization and immunofluorescence staining (in all three studies).
| Sources of samples | Marker genes | Marked cell populations | Reference |
| Mouse aged seven days | Amel | Ameloblasts | Chiba et al. 2020 [22] |
| Dspp | Ameloblasts | ||
| Tbx1 | IEE/OEE cells | ||
| Sfrp5 | IEE/OEE cells | ||
| Pttg1 | IEE/OEE cells | ||
| Notch1 | SI/SR cells | ||
| Notch2 | SI/SR cells | ||
| Cldn10 | SI cells | ||
| Krt15 | OEE cells | ||
| Mouse aged 8–12 weeks | Ung | Cycling cells | Sharir et al. 2019 [24] |
| Top2a | Cycling cells | ||
| Cdc20 | Cycling cells | ||
| Ccnb2 | Cycling cells | ||
| Amelx | Ameloblast | ||
| Ambn | Ameloblast | ||
| Enam | Ameloblast | ||
| Mmp20 | Ameloblast | ||
| Cldn10 | IEE-OEE cells | ||
| Ighbpl1 | Upper IEE cells | ||
| Sparcl1 | VEE cells | ||
| Krt15 | VEE cells | ||
| Enpp2 | SI cells | ||
| Mouse ages 2–4 months | Shh | Pre-ameloblasts | Krivanek et al. 2020 [23] |
| Enam | Secretory ameloblasts | ||
| Klk4 | Maturation ameloblasts | ||
| Gm17660 | Postmaturation ameloblasts | ||
| RYR2 | Ameloblast | ||
| Thbd | Cuboidal layer of stratum intermedium | ||
| Slco4a1 | OEE cells | ||
| Vat1l | SR cells | ||
| Psmb10 | SI cells | ||
| Sox2 | Stem and progenitor cells | ||
| Acta2 | Stem and progenitor cells | ||
| Egr1 | Stem and progenitor cells | ||
| Fos | Stem and progenitor cells |
IEE, inner enamel epithelium; OEE, outer enamel epithelium; SI, stratum intermedium; SR, stellate reticulum; VEE, ventral enamel epithelium.
Conventional approaches for the identification and categorization of mouse dental stem cells use established stem cell markers found in human dental pulp, as well as bromodeoxyuridine (BrdU) or comparable long-term labels deposited within the stem cell niche to indicate the presence of slow-cycling populations [25, 26]. Mouse dental epithelial stem cells can be identified using pre-existing markers, namely Bmi1 and Sox2, which identify the slow cycling stem cells, whereas Lgr5 is an annotation marker for active stem cells [27].
In the classical paradigm, the cervical loop region serves as a niche for the stem cells. Epithelial stem cells reside in the SR or OEE close to labial cervical loop (LaCL) [28, 29]. These stem cells are believed to give rise to transit-amplifying cells (TACs) that migrated to the basal layer of the epithelium. These cells are then regulated by the microenvironment to differentiate into ameloblasts or root epithelium cells [30]. However, Sharir et al. [24] challenged the classical epithelial model by applying a single cell approach, where cycling cells within the enamel organ were identified as progenitors initiating the upregulation of differentiation-related genes are expressed in the IEE. Conversely, non-ameloblast cells were found to be distributed at the junction of IEE and OEE, as well as in the upper region of the IEE [24].
Chiba et al. [22] and Krivanek et al. [23] proposed innovative structures of the enamel organ. Novel marker genes have been identified to delineate dental epithelial cells. CLDN10 was observed in SI cells, whereas KRT15 expression was detected in OEE cells [22, 23, 24]. CLDN10 encodes the claudin-10 protein, which serves as the structural framework for tight junctions and facilitates the differentiation of SI cells [31]. Previous studies have identified KRT15 as a marker gene predominantly localized in the esophageal basal cell layer, suggesting its potential as a progenitor cell marker [32, 33].
Krivanek et al. [23] identified a cluster of RYR+ cells that expressed mechano-transduction-related genes, including Piezo2, Trmp2, and Trmp3. This observation revealed a strong association between these cells and the transduction of mechanical-chemical signals [34, 35]. Furthermore, Egr1/Fos+ cells were identified as the progenitors responsible for the formation of OEE. Moreover, a previously unknown cell type, specifically marked by Thbd, was identified in the cuboidal layer of the stratum intermedium. This cell type plays a crucial role in maintaining the interface between the ameloblasts and blood vessels, thereby improving our understanding of the epithelial profile.
A trajectory analysis performed by Chiba et al. [22] demonstrated that the IEE/OEE population serves as a precursor for the SI/SR populations and ameloblasts. Furthermore, this study revealed distinct functions of the two subtypes of ameloblasts. DSPP expression was observed in early secretory ameloblasts, but disappeared in fully differentiated ameloblasts. Conversely, AMBN expression gradually increased during differentiation from the early phase to mature ameloblasts. These findings provide a clearer understanding of the temporal characteristics of these two subpopulations [22].
Additionally, the expression of Wnt6 and Shh, signaling molecules known to promote differentiation, is typically observed in the enamel knots of molars [36, 37, 38]. Interestingly, these molecules were highly expressed in DSPP+ cells, suggesting their potential to differentiate into AMBN+ cells. Sharir et al. [24] used RNA velocity analysis to define the IEE cell source. These findings demonstrated that IEE progenitors serve as ancestors of both ameloblasts and OEE/SR cells. This conclusion was further supported by a quantitative analysis using EdU to track cell proliferating.
Krivanek et al. [23] categorized three primary mesenchymal populations in mouse incisors: the odontoblast population (one) and subpopulations within the pulp (two). The first subpopulation, located in the apical pulp between the cervical loops, was identified by Smoc2 and Sfrp2. Notably, genes associated with self-renewal (Thy1 and Gli1) were significantly expressed in this region, accompanied by a significant level of cellular division. The presence of Igfbp5 and Syt6 in the distal pulp indicates the existence of another subtype of pulp that exhibits a more advanced level of differentiation. Pseudo-time analysis revealed that odontoblasts undergo at least three stages of differentiation, progressing from pre-odontoblasts to early odontoblasts, and finally to late odontoblasts. This discovery holds potential for future investigations into dentinogenesis. Additionally, Foxd1 has been identified as a novel marker that characterizes progenitor cells confined to LaCL. A lineage tracing experiment determined the fate of Foxd1+ cells toward either odontoblast or dental pulp lineages. These findings suggest that the cells expressing Foxd1 predominantly differentiate into periodontal pulp cells and osteoblasts, which are responsible for dentin secretion [23]. Jing et al. [39] presented a comprehensive transcription atlas of mouse molars spanning embryonic 13.5 days to postnatal 7.5 days. During the progression of tooth bud development, the E14.5 mouse dental papilla cell population, identified by Crym, undergoes differentiation into the E16.5 mouse apical papilla population, distinguished by Lhx6, and the coronal papilla population, characterized by Lmo1. These distinct populations subsequently give rise to the odontoblast lineage and the pulp lineages [39].
Pagella et al. [40] successfully identified various cell types in human dental pulp tissue, including mesenchymal stem cells (MSCs), odontoblasts, fibroblasts, epithelial cells, endothelial cells (ECs), Schwann cells (ScCs), and immune cells. Notably, fibroblastic compartments, characterized by the expression of collagen-related genes, such as COL1A1, comprise most of the human dental pulp [40] (Table 2, Ref. [23, 40]). Besides, Ren et al. [41] performed sequencing on an immature human third molar with a root length of less than one-third that of mature molars to elucidate the developmental trajectories of young dental pulp cell populations. The analysis revealed a higher proportion of mononuclear phagocytes and lymphocytes in young pulp. Moreover, differential expression gene analysis indicated that young pulp exhibited a greater inclination for protein formation and immune system development. Pseudo-time analysis further elucidated that MSCs in the young pulp differentiate into two lineages, which were inferred to be the odontogenesis and myogenesis lineage based on pathway enrichment analysis [41].
| Sources of samples | Marker genes | Marked cell populations | Reference |
| Mouse aged 2–4 months | Dspp | Late odontoblasts | Krivanek et al. 2020 [23] |
| Dmp1 | Late odontoblasts | ||
| Wisp1 | Early odontoblasts | ||
| Col24a2 | Early odontoblasts | ||
| Sall1 | Pre-odontoblasts | ||
| Notum | Pre-odontoblasts | ||
| Smoc2 | Apical pulp | ||
| Sfrp2 | Apical pulp | ||
| Thy1 | Apical pulp | ||
| Gli1 | Apical pulp | ||
| Mki67 | Pulp near LaCL | ||
| Fgf3 | Pulp near LaCL | ||
| Foxd1 | Pulp near LaCL | ||
| Igfbp5 | Distal pulp | ||
| Syt6 | Distal pulp | ||
| Human dental pulp | Frzb | MSCs | Pagella et al. 2021 [40] |
| Notch3 | MSCs | ||
| Thy1 | MSCs | ||
| Myh11 | MSCs | ||
| Col1a1 | Fibroblasts | ||
| Mdk | Fibroblasts | ||
| Dspp | Odontoblasts | ||
| Dmp1 | Odontoblasts | ||
| Edn1/Cldn5 | Arterial ECs | ||
| Ackr1/CD234 | Postcapillary and collecting venules Ecs | ||
| Insr/Rgcc | Ecs | ||
| Ptprc | Immune cells | ||
| CD3e | Immune cells | ||
| Csf1r | Immune cells | ||
| Sox10 | ScCs | ||
| Krt14 | Epithelial cells | ||
| Krt5 | Epithelial cells | ||
| Hbb | Erythrocytes |
Cui et al. [42] used two distinct cell populations in their research, including freshly isolated and 10-day cultured human dental pulp cells. A significant variation in cellular composition was identified by comparing single-cell RNA sequencing results from these two groups. However, a subpopulation of stem cells characterized by MCAM, JAG1, and the absence of PDGFRA exhibited significant transcriptional characteristics in both freshly isolated and cultured pulp. The spatial distribution and expression levels of this specific subpopulation exhibited a notable degree of stability unaffected by senescence or inflammation. In addition, in vivo and in vitro experiments demonstrated that MCAM+JAG1+PDGFRA– cells possess an increased capacity for proliferation, osteogenesis, chondrogenesis, and adipogenesis compared to PDGFRA+ cells. These findings suggest that MCAM+JAG1+PDGFRA– cells retain their ability to proliferate and differentiate after in vitro transplantation, thereby providing potential insights for future regenerative therapies [42].
The periodontium, comprising the alveolar bone, gingiva, and periodontal ligaments, is important for attaching teeth to the bone and preventing oral diseases. Embryologically, periodontal tissue originates from the fibrous tissue surrounding the tooth bud, called the dental follicle (DF). Within the DF, progenitor cells generate periodontium during the developmental phase [43].
Lineage-tracing experiments were conducted to confirm the heterogeneity of DF cells. Subgroups of DF cells expressing parathyroid hormone-related protein (PTHrP), glioma-associated oncogene homolog 1 (Gli1), and osterix (Osx) have been identified [44, 45, 46]. These findings were supported by the scRNA-seq analysis conducted by Nagata et al. [47], who focused on seven clusters expressing mesenchymal markers in mouse periodontal tissue. These clusters included DF cells marked by Bmp3 and Spon1, periodontal ligament (PDL) cells marked by Scx and Postn, cementoblasts marked by Tubb3 and parathyroid hormone-like hormone (Pthlh), DP cells marked by Tac1, osteoblasts marked by Phex and Ifitm5, fibroblasts marked by S100a4, and marrow stromal cells marked by Ebf3. Nagata et al. [47] demonstrated the presence of PTHrP in DF cells, which exhibit characteristics of mesenchymal progenitor cells [46]. The localization of PTHrP+ DF cells was confirmed using fluorescent transgenic markers, demonstrating their expression in the vicinity of the developing tooth on postnatal day 6 (PN6) mice, as well as in the alveolar cryptal bone, acellular cementum, and PDL on PN25 mice. RNA velocity analysis was also performed using BMP3+ DF and Scx+ PDL cells as reference points. BMP3+ DF cells undergo osteoblast differentiation, whereas Scx+ PDL cells further differentiate into osteoblasts and marrow stromal cells [47].
Previous study has provided limited clarity regarding the distinctions between cementoblasts and osteoblasts, often called cells involved in forming hard tissues. It has been speculated that these cells represent the same cell type and occupy different anatomical locations [48]. Nagata et al. [47] identified specific genes as markers for cementoblasts (Wif1, Tubb3, and Pthlh) and osteoblasts (Phex, Nfib, and Pth1r). Specifically, PTHrP selectively labeled cementoblasts rather than osteoblasts. Consequently, these findings provide a unique opportunity to investigate the differentiation of periodontal stem cells in cementogenesis and osteogenesis [47].
Stem cells can generate progenitor cells that differentiate into various lineages [49]. The niche of mouse dental MSCs was situated within the epithelial cervical loop between the labial and lingual sides (Fig. 1) [50]. The co-expression of putative stem cell markers Thy1, Igfbp5, Gli1, and Lrig1 has been observed in this region. Differentiated cell populations can be generated from these MSC niches, including dental follicle cells, pulp cells, and odontoblasts [51]. Numerous studies have provided evidence of the heterogeneity of mouse dental MSCs. Chen et al. [52] and Zhao et al. [50] conducted studies on mouse incisors and identified that Gli1+ cells continuously produced of TACs. Gli1+ TACs proliferate to give rise to odontoblasts and dental pulp cells [50, 52]. An et al. [53] subsequently confirmed that the neurogenic CD90/Thy1 subtype originates from 30% of the differentiated cells during eruption and in injury cases. Seidel et al. [54] observed Lrig1+ stem cells exclusively in periodontium cells. Furthermore, Sharir et al. [24] proposed that commonly recognized stem cell markers, such as Sox2, Gli1, and Lrig1 lack specificity for any particular cell type.
Human dental-derived MSCs include MSCs isolated from various sources within the oral cavity, including dental follicles (DFSCs), apical papillae (SCAP), dental pulp (DPSCs), periodontal ligament (PDLSCs), and gingiva (GMSCs) [55]. However, current investigations at the single-cell level have been limited to the comparisons between DPSCs and PDLSCs. Notably, there is a lack of systematic research on SCAP, stem cells from human exfoliated deciduous teeth (SHED), and GMSC at the single-cell level. Human DPSCs and PDLSCs originate from the cranial neural crest during embryonic development. This process involves the epithelial-mesenchymal transition, during which the cranial neural crest cell population diverges from the ectodermal epithelium to form the mesenchyme. A small subset of this population differentiates into dental papilla and dental follicle, which subsequently give rise to the periodontium and dental pulp. Therefore, DPSCs and PDLSCs exhibited both similarities and heterogeneity.
Human dental-derived MSC niches were tracked in several studies (Fig. 2). In a comparative study using scRNA-seq, Lee et al. [56] observed that DPSCs exhibited elevated neurogenic ratings, whereas PDLSCs demonstrated higher ratings for osteogenesis and chondrogenesis and increased expression of endothelial growth factor. Additionally, DPSCs exhibited higher levels of CXCL14 and RARRES1. Moreover, Pagella et al. [40] demonstrated distinct migration and differentiation characteristics of DPSCs and PDLSCs. They proposed that this disparity may be attributed to the microenvironment rather than the inherent heterogeneity within the MSCs [56]. Dental pulp primarily comprises fibroblast-like cells, whereas the periodontal ligament predominantly comprises epithelial cells. Regulatory factors (FDCSP and WNT10A) were significantly expressed in fibroblast-like cells within the periodontal ligament, influencing MSC proliferation. The development of PDLSCs towards fibrogenesis and resistance to mineralization is primarily guided by epithelial cells and fibroblasts, whereas the osteogenesis of DPSCs is predominantly influenced by the pulp microenvironment [40].
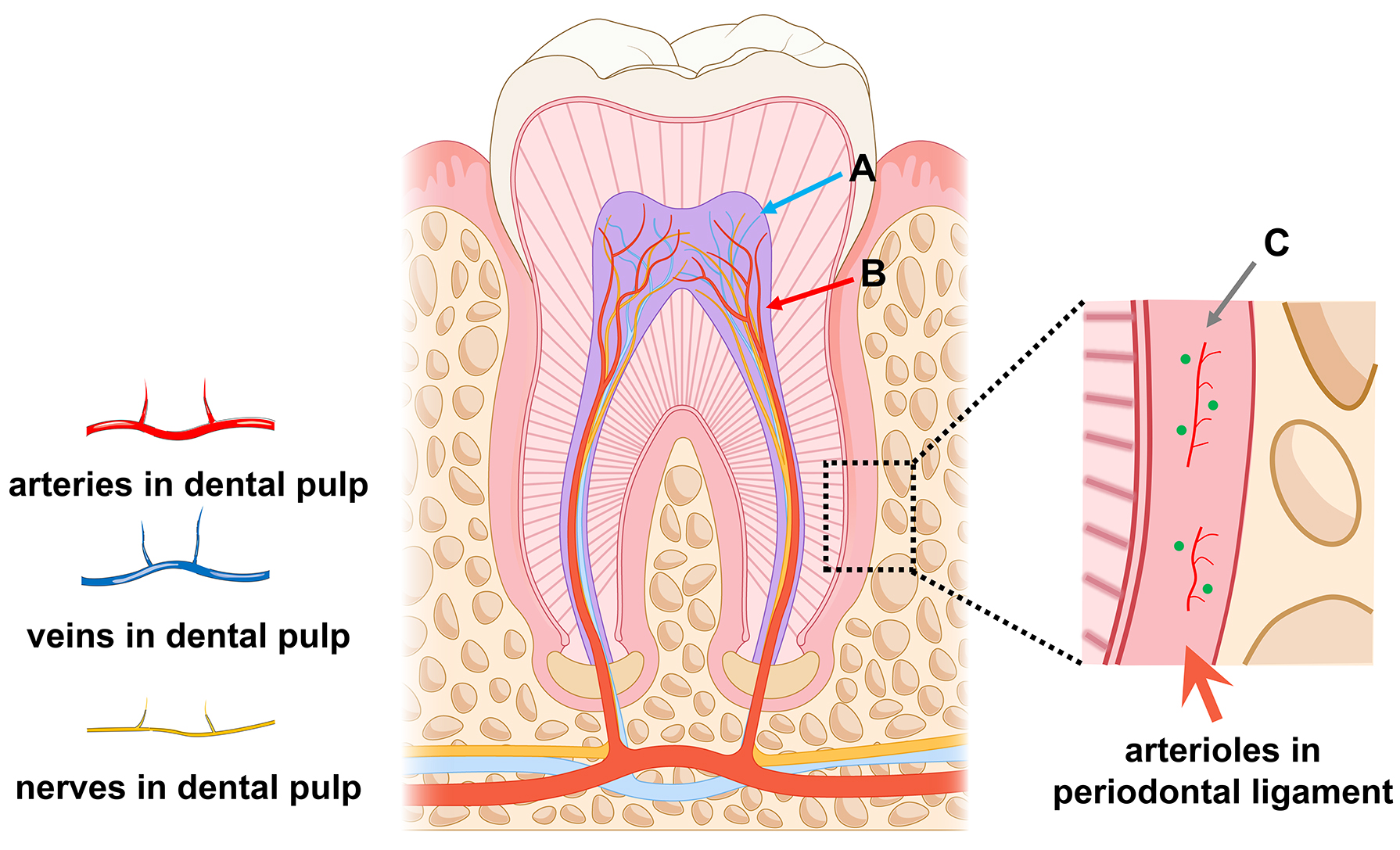 Fig. 2.
Fig. 2. Human dental-derived mesenchymal stem cell (MSC) niche. (A) Perineural MSC niche. (B) Perivascular MSC niche. (C) MSC niche surrounding arterioles in the periodontal ligament. The figures are drawn by PowerPoint (Microsoft, office 2019, Redmond, WA, USA).
Sharir et al. [24] introduced a novel model of epithelial populations in mouse incisors. Proliferation dynamics analysis of the incisor epithelium revealed that progenitor cells within the IEE exhibit active cycling dynamics. RNA velocity and genetic lineage tracing further confirmed that a more significant proportion of these progenitors produces functional ameloblasts, whereas a smaller subset generates non-ameloblast epithelial cells that are present in the SR and OEE. This differentiation process plays a crucial role in maintaining homeostasis in the enamel of the mouse incisors (Fig. 3, Ref. [24]). Sharir et al. [24] evaluated the regenerative process of incisor epithelial components in wild-type mice treated with 5-fluorouracil (5-FU) for four days [57]. In this model, 5-FU functioned as an antimetabolite impeding cellular proliferation. Conversely, the non-proliferative regions were not significantly affected. The 5-FU treatment effectively eliminated proliferative epithelial cells, particularly IEE cells, until the third day post-treatment, at which point they were observed to be regenerating. A scRNA-seq analysis was conducted at this stage, and no novel cell types were identified in the recovery group. An increase in the number of cycling cells was observed in the composition ratio of cell types, further supported by the expanded expression domains of Birc5 and Ccnb1. Conversely, the presence of pre-ameloblasts and ameloblasts was limited. The researchers hypothesized that ameloblast differentiation would be delayed based on the relocation of pre-ameloblast markers to the distal epithelium. Furthermore, the differentiation of Notch1+ SI cells into ameloblasts underscores the significant contribution of Notch1+ SI cells to injury recovery [24].
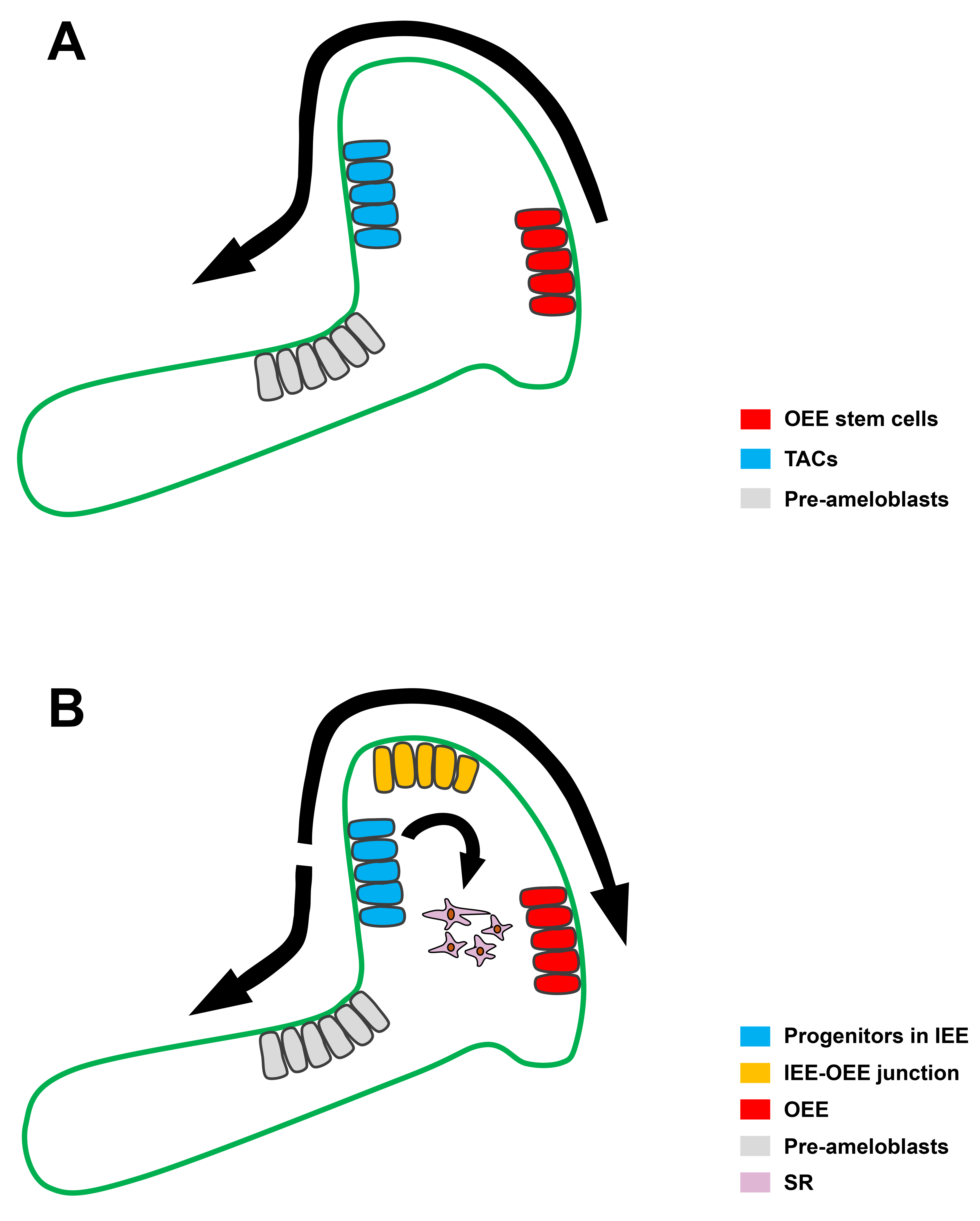 Fig. 3.
Fig. 3. Differentiation model of mouse incisor. (A) The classical model suggests that epithelial stem cells are located in the OEE near the LaCL, forming transit-amplifying cells in the IEE, which differentiate into ameloblasts. (B) Sharir et al. [24] suggested that using RNA velocity analysis, many progenitors in the IEE will further produce non-ameloblast epithelial cells in the SR and OEE. The figures are drawn by PowerPoint (Microsoft, office 2019, Redmond, WA, USA). OEE, outer enamel epithelium; SR, stellate reticulum.
Chen et al. [52] analyzed Runx2/Gli1+ cells within the niche of MSCs using mouse incisor pulp. ScRNA-seq revealed a significant upregulation of Runx2 in proximal Gli1+ cells. Although Runx2+ populations do not directly contribute to the formation of dental mesenchyme, they play a crucial role in maintaining mesenchymal homeostasis by regulating the proliferation and differentiation of TACs [52]. Pagella et al. [58] used scRNA-seq to confirm the enhanced interaction between NOTCH ligands and receptors in MSCs. Moreover, they identified NOTCH3 as a universal MSC marker implicated in maintaining tooth homeostasis and facilitating regeneration in vivo [58].
The periodontium endures continuous and dynamic remodeling owing to consistent mechanical stimulation associated with the masticatory function of the teeth. Cells originating from the periodontal ligament play a significant role in maintaining homeostasis of the periodontium and facilitating its repair following injury [59]. Zhao et al. [60] identified two distinct stem cell populations involved in cementogenesis at different stages in mouse molars. During normal homeostasis, cells expressing Wnt-responsive Axin2 gene in the periodontal ligament differentiate into cementoblasts. However, in a periodontitis model, CD90+ cells were responsible for forming cementoblasts in cases of mild periodontitis [60]. Takada et al. [61] focused on the role of the mohawk homeobox (Mkx) transcription factor, which is specific to tendons and ligaments, in maintaining the homeostasis of the periodontal ligament. This study revealed a decrease in the expression of fibrogenesis-related genes, including fibulin5 (Fbnl5), matrilin-4 (Matn4), and Tnn, in MSCs of Mkx-deficient mice. Conversely, the expression of osteogenic genes, such as Pthlh, bone sialoprotein (Ibsp), and insulin-like growth factor binding protein 5 (Igfbp5) was upregulated. Besides, Mkx-deficient mice exhibited increased expression of inflammatory genes in macrophages. These findings collectively indicate that Mkx regulates periodontal ligament homeostasis by inhibiting ossification and fiber formation [61].
Lin et al. [62] identified the molecular functions of nonimmune cells in chronic apical periodontitis (CAP). They identified four nonimmune clusters within the chronic apical periodontitis tissues: osteo-like cells (identified by the presence of OMD, COL1A1, and TNFRSF11B), basal/stromal cells (identified by BCAM, RGS5, and TAGLN), epithelial cells (identified by KRT19 and KRT14), and endothelial cells (identified by ACKR1, VWF, and CDH5). Osteo-like subclusters demonstrated the presence of osteoblastic and osteoclastic cell markers, implying the simultaneous occurrence of osteogenic and osteoclastic activities in CAP tissues. Basal/stromal cells express genes associated with basal fibroblasts and smooth muscle cell markers. Pseudo-time trajectories analysis on endothelial cells further demonstrated robust angiogenesis (evidenced by upregulated expression of KDR and VWF) and lymph angiogenesis (evidenced by upregulated expression of IGFBP5 and CLIC4) activity in CAP. Proliferation-related genes (IL2RG and NF
Chen et al. [63] used three distinct sources of periodontal tissue, namely patients with severe chronic periodontitis (PDs), patients with severe chronic periodontitis treated within one month (PDTs), and healthy controls (HCs) for single-cell procedure. They revealed a significant scarcity of MSCs, pre-OBs, and OBs in the PDT group, confirming impaired bone formation in periodontitis. Despite partial rescue, the PDT group exhibited elevated macrophage counts compared to the HC group. In the PD group, the number of venous and arterial ECs surpassed that of HCs. A notable decrease in T cell number was observed in the PD group compared to that in the HCs. This reduction was primarily observed in the decline of mucosal-associated invariant T cells (MAIT) and CD8+ terminally differentiated effector T cells (TEMRA). However, an increase in CD8+ tissue-resident memory (TRM) T-cells was observed in the PD group. Within the fibroblast subclusters of the PD samples, a significant presence of inflammation-related genes, such as CXCL1, CXCL13, and TNFRSF21, was identified. The PD group exhibited a higher abundance and distribution of CXCL12+ MSC-like pericytes than the HC group. These pericytes were also present in the PDT group, where MSCs were undetected. The PD group exhibited SPON2+ pericytes expressing numerous proliferation-related genes, providing evidence of a stressful state within this particular group.
In addition, upregulation of CSF3 was observed in venous endothelial cells in the PDT group compared to the PD group, and in the PD group compared to the HC group. Previous studies have demonstrated a significant association between CSF3 and bone mass reduction [64]. Additionally, gene set enrichment analysis revealed an increase in pathways associated with CD8+ T cells in individuals with periodontal disease compared to HCs, which were subsequently restored after the initial periodontal therapy. This study focused on the Ephrin-Eph signaling pathway between pre-osteoblasts and endothelial cells, indicating a strong communication between pre-osteoblasts and ECs during osteoblast generation [65, 66]. The PDT group exhibited a higher quantity of receptor-ligand pairs linked to Ephrin-Eph signaling than the other two groups. In conjunction with the Eph receptor, Ephrin ligands are believed to play a crucial role in fiber formation. Previous studies have established that pre-OBs can contribute to fibroblasts [67, 68]. This study suggests that the reduced bone regeneration observed in periodontitis may be attributed to either a decrease in the population of Pre-OBs or the fibroblastic inclination of Pre-OBs, influenced by the Ephrin-Eph signaling [63].
Qian et al. [69] conducted a study on human gingival tissue affected by periodontitis. They observed that the proportion of non-epithelial cells in the periodontitis tissues was higher than that in the gingival tissues of healthy donors. Additionally, diseased tissues exhibited high expression of MHC class Ⅱ genes in endothelial cells, which play a crucial role in antigen presentation [70]. Fibroblasts in periodontitis tissue were identified by their elevated expression of CXCL13, IL32, SFRP2 (indicating the Fibro1 subcluster), OGN, PRELP, and RUNX2 (indicating the Fibro2 subcluster). The pathway enrichment analysis demonstrated that Fibro1 subpopulations exhibited an augmented immune pathway, whereas Fibro2 subpopulations displayed an increased osteoblast generation pathway. Notably, in the healthy gingiva, the prevailing fibroblast subpopulation was OGN+ (Fibro2) fibroblasts, whereas in the diseased group, CXCL13+ (Fibro1) fibroblasts were predominant.
Besides, no additional myeloid subtypes were observed in the diseased group compared with the healthy group. Among the three distinct macrophage clusters, Macro_C1QA, Macro_PRDM1, and Macro_NLRP3, there was an observed increase in the proportion of CD11b+NLRP3+ macrophages in gingival tissue with periodontitis, and the Macro_NLRP3 subtype specifically contributed to angiogenesis. This finding suggested a robust association between CD11b+NLRP3+ macrophages and periodontitis. Additionally, patients with high expression of immediate-early genes (such as FOS and JUN) exhibited an increased presence of CD4+ T cells, whereas CD8+ T and NK cells from patients demonstrated heightened levels of CCL4, CCL4L2, and CCL3L3. In conclusion, these gene expression transformations are closely associated with the active reaction of the periodontal tissue against inflammation [69].
Previously, the identification of dental MSCs was limited to marker genes identified through animal experiments. However, the application of scRNA-seq enables the use of a broader range of MSC markers [71]. The potential for dental and periodontal regenerative medicine would be significant if it could be demonstrated that the cells identified by scRNA-seq possess stem cell properties in vitro and in vivo. Despite the increasing use of scRNA-seq in dental research, inherent complexities and obstacles remain. Dental structures comprise a significant amount of hard tissue and a restricted quantity of cellular constituents, necessitating a large number of samples to achieve a sufficient transcriptome count. The cells are intricately entangled with hard tissue in the dentin and periodontal ligament, rendering cell dissociation a formidable process. Notably, investigations pertaining to the tooth epithelium have predominantly focused on murine dentition. During enamel development in human teeth, epithelial components the gradually disappear. Although recent studies have effectively isolated and characterized cells from the human epithelial rest of Malassez (ERM) and Hertwig’s Epithelial Root Sheath (HERS), obtaining sufficient cells for single-cell analysis remains challenging [72, 73]. Moreover, regeneration using induced pluripotent stem cells (iPSCs) has emerged as a promising method in recent years [74]. The reprogramming of somatic cells derived from various tissues into iPSCs was achieved through artificial intervention, exhibiting developmental potential akin to embryonic cells. Notably, the successful generation of iPSCs has been reported using dental pulp and papilla cells [75, 76]. Through scRNA-seq, it is possible to gain insights into the genetic characteristics and differentiation trajectories of iPSCs at the single-cell level. Neavin et al. [77] used scRNA-seq to trace the expression of quantitative trait loci (eQTLs) and investigate the differential gene expression in iPSCs. Notably, the disappearance of eQTLs in the induced cell line and the emergence of novel eQTLs during the reprogramming of iPSCs were observed, suggesting a significant alteration in the gene expression patterns within iPSCs [77]. Hsiao et al. [78] developed a method for identifying the cell cycle of iPSCs using scRNA-seq. Furthermore, Schiebinger et al. [79] investigated novel cellular constituents and modified cell destinations during reprogramming. Consequently, further scRNA-seq exploration is imperative to elucidate the mechanisms underlying cell reprogramming.
The integration of inclusive atlases using single-cell sequencing of mouse and human teeth makes it possible to conduct interactive research on tooth development, disease, and regeneration [80]. This advancement provides opportunities for future exploration in multi-omics studies, wherein DNA sequencing and DNA methylation analysis can be integrated with scRNA-seq to obtain a more comprehensive understanding of hereditary information [81, 82]. In recent years, the introduction of targeted scRNA-seq has focused on identifying molecules with low expression levels that convey substantial biological signals. This enrichment of expression is achieved by labeling specific genes or genomic regions using oligonucleotide probes [83, 84]. Pokhilko et al. [85] used the scCapture-seq technique to construct a targeted single-cell sequencing library comprising probes targeting 972 transcription factors (TFs). This study demonstrated that scCapture-seq exhibited significant sensitivity towards TFs with low expression levels compared to scRNA-seq, enabling the depiction of gene regulation networks with enhanced resolution [85]. Furthermore, high-resolution spatial RNA-seq facilitates the simultaneous revelation of three-dimensional spatial information and transcriptomic characteristics [86, 87]. Moreover, single-cell proteomics promises to provide a more comprehensive depiction of protein expression [88, 89]. Using these inclusive techniques, a deeper comprehension of the tooth or periodontium’s development, repair, and disease condition can be attained, potentially facilitating the predictable regeneration of presently non-renewable tissues.
ScRNA-seq, single-cell RNA sequencing; OEE, outer enamel epithelium; IEE, inner enamel epithelium; SR, stellate reticulum; SI, stratum intermedium; DEJ, dentinal enamel junctions; BrdU, bromodeoxyuridine; MSCs, mesenchymal stem cells; ECs, endothelial cells; ScCs, Schwann cells; DF, dental follicle; PTHrP, parathyroid hormone-related protein; Gli1, glioma-associated oncogene homolog 1; Osx, osterix; Pthlh, parathyroid hormone-like hormone; TACs, transit-amplifying cells; HERS, Hertwig’s Epithelial Root Sheath; CAP, chronic apical periodontitis.
XXM, YFH and YLX designed the research study. XXM and KXG contributed to table preparation. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Not applicable.
Not applicable.
This research was funded by the Youth Science and Technology Project of Changzhou Health Commission, grant number QN202124.
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.



