1 Disease Control and Prevention, Zhangqiao Branch, Ningbo Ninth Hospital Medical Health Group, 315000 Ningbo, Zhejiang, China
2 Department of Doctor-patient Communication, The First Affiliated Hospital of Ningbo University, 315010 Ningbo, Zhejiang, China
3 Beijing STEPPIN Technology Co., LTD., 100195 Beijing, China
4 Infectious Disease Prevention and Control, Jiangbei Center for Disease Control and Prevention, 315000 Ningbo, Zhejiang, China
Abstract
The polarization of macrophages plays a critical role in the immune response to infectious diseases, with M2 polarization shown to be particularly important in various pathological processes. However, the specific mechanisms of M2 macrophage polarization in Mycobacterium tuberculosis (Mtb) infection remain unclear. In particular, the roles of Granulin (GRN) and tumor necrosis factor receptor 2 (TNFR2) in the M2 polarization process have not been thoroughly studied.
To investigate the effect of macrophage M2 polarization on Mtb infection and the mechanism of GRN and TNFR2 in M2 polarization.
Forty patients with pulmonary tuberculosis (PTB) and 40 healthy volunteers were enrolled in this study, and peripheral blood samples were taken to detect the levels of TNFR2 and GRN mRNA by Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR); monocytes were isolated and then assessed by Flow Cytometry (FC) for M1 and M2 macrophage levels. To further validate the function of TNFR2 in macrophage polarization, we used interleukin 4 (IL-4) to induce mouse monocyte macrophages RAW264.7 to M2 polarized state. The expression of TNFR2 was detected by Western Blot and RT-qPCR. Next, we constructed a GRN knockdown plasmid and transfected it into IL-4-induced mouse monocyte macrophage RAW264.7, and detected the expression of TNFR2, M1 macrophage-associated factors tumor necrosis factor-α (TNF-α), inducible nitric oxide synthase (iNOS), and interleukin 6 (IL-6), and the M2 macrophage-associated factors CD206, IL-10, and Arginase 1 (Arg1); Immunofluorescence staining was used to monitor the expression of CD86+ and CD206+, and FC was used to analyze the macrophage phenotype. Subsequently, immunoprecipitation was used to detect the binding role of GRN and TNFR2. Finally, the effects of GRN and TNFR2 in macrophage polarization were further explored by knocking down GRN and simultaneously overexpressing TNFR2 and observing the macrophage polarization status.
The results of the study showed elevated expression of TNFR2 and GRN and predominance of M2 type in macrophages in PTB patients compared to healthy volunteers (p < 0.05). Moreover, TNFR2 was highly expressed in M2 macrophages (p < 0.05). Additionally, GRN knockdown was followed by elevated expression of M1 polarization markers TNF-α, iNOS and IL-6 (p < 0.05), decreased levels of M2 polarization-associated factors CD206, IL-10 and Arg1 (p < 0.05), and macrophage polarization towards M1. Subsequently, we found that GRN binds to TNFR2 and that GRN upregulates TNFR2 expression (p < 0.05). In addition, knockdown of GRN elevated M1 polarization marker expression, decreased M2 polarization marker expression, and increased M1 macrophages and decreased M2 macrophages, whereas concurrent overexpression of TNFR2 decreased M1 polarization marker expression, elevated M2 polarization marker expression, and decreased M1 macrophages and increased M2 macrophages.
TNFR2 and GRN are highly expressed in PTB patients and GRN promotes macrophage M2 polarization by upregulating TNFR2 expression.
Keywords
- granulin
- Mycobacterium tuberculosis
- M2 polarization
- macrophages
- tuberculosis
- TNFR2
Pulmonary tuberculosis (PTB) is an infectious disease that poses a serious threat to global public health [1], and mycobacterium tuberculosis (Mtb) is the main pathogen responsible for PTB [2, 3]. Recent research has demonstrated that the polarization state of macrophages plays a key role in the host immune response to Mtb infection [4]. Although M2-type macrophages help to alleviate acute inflammation and tissue damage [5], this anti-inflammatory effect inhibits M1-type macrophages [6]. M1-type macrophages can weaken the host’s defenses against Mtb, leading to the survival and multiplication of Mtb in the host [7], which exacerbates the course of the disease and leads to the development of chronic Mtb infection in the organism [8]. Therefore, balancing the polarization state of macrophages, which can effectively eliminate pathogens while preventing excessive inflammatory damage, is a crucial strategy for treating PTB.
Tumor Necrosis Factor Receptor 2 (TNFR2) is a cell surface receptor [9], whose primary role is to promote cell survival and tissue repair [10]. Studies have shown that the activation of TNFR2 inhibits the bactericidal activity of M1-type macrophages [11], allowing Mtb to persist in infection [12]. Thus, modulating the activity of TNFR2 may enhance the host immune defense against Mtb and alleviate the progression of PTB. In addition, TNFR2 activity is closely associated with Granulin (GRN) levels [13]. GRN is a widely expressed pleiotropic protein involved in different biological processes, including cell proliferation, neuronal development, and wound healing [14]. Previous study has shown that GRN promotes macrophage M2 polarization by inhibiting the secretion of pro-inflammatory cytokines and activating tissue repair by TNFR2 [15]. However, the effect of GRN on TNFR2 and its specific mechanism in M2 polarization and Mtb infection have not been reported.
Therefore, our study aimed to elucidate the potential roles of GRN and TNFR2 in Mtb infection, especially their effects on macrophage polarization status. This study not only helps to deepen the understanding of the mechanism of Mtb infection, but also may provide a theoretical basis for the development of new therapeutic strategies for PTB.
Forty PTB patients who received treatment in the Hospital and 40 healthy volunteers were included in this study. The inclusion criteria for PTB group were: (1) patients receiving anti-tuberculosis treatment in the hospital; (2) pathological confirmation of PTB; (3) no history of other tuberculosis treatment. Inclusion criteria for the normal volunteer group: (1) no history of tuberculosis; (2) no history of major diseases; (3) health examination to confirm good health. The exclusion criteria were: (1) history of mental illness; (2) severe hepatic or renal dysfunction; and (3) coexistence of other types of diseases. All patients or their families/legal guardians as well as healthy volunteers voluntarily signed an informed consent form. The experiment has been approved by the Ningbo Ninth Hospital Medical Health Group Ethics Review Committee (No. NNHM-20230201).
All PTB patients received anti-tuberculosis treatment and the mean age
was (45.3
1
The coding sequences (CDS) of TNFR2 and GRN was cloned into the pcDNA3.1
vector (V79020, Thermo fisher, MA, USA) to construct oe-TNFR2 and oe-GRN,
respectively. The oe-TNFR2, oe-GRN, oe-negative control (NC) plasmids, or si-GRN
(5′-GCCUGACGUCACCAUGACCUG-3′) and si-NC
(5′-UUCUCCGAACGUGUCACGUUU-3′, SIC001, Sigma Aldrich, St. Louis, USA)
were transfected with Lipofectamine 3000 transfection reagent (L3000075, Thermo
fisher, MA, USA) for transfection into RAW264.7 cells. The procedure was as
follows: the RAW264.7 cells were inoculated into 96-well plates at a density of 1
Total RNA was extracted from peripheral serum and RAW264.7 cells of patients in each group using MagMAX Pathogen Nucleic Acid Isolation Kit (A42352, Thermo fisher, MA, USA), and then reversed to cDNA using Reverse Transcription Kit (K1691, Thermo fisher, MA, USA). GAPDH was used as an endogenous reference gene, and the primer sequences were given in Table 1. The PCR experiments were repeated three times with three replicate wells each time, and the absolute relative mRNA content was finally determined by the qPCR algorithm (relative quantification, 2-ΔΔCt method).
| Gene | Organism | Direction | Sequence (5 |
|---|---|---|---|
| TNFR2 | Homo sapiens | F | CACCGGGAGCTCAGATTCTT |
| R | CCGAAAGGCACATTCCTCCT | ||
| Tnfr2 | Mus musculus | F | AAGTGCATGTCCGGGTTAGG |
| R | CCGAGATGACAGAACCCGTC | ||
| Tnf- |
Mus musculus | F | ACCCTCACACTCACAAACCA |
| R | ACCCTGAGCCATAATCCCCT | ||
| Inos | Mus musculus | F | CGCTCTAGTGAAGCAAAGCC |
| R | GCCTAGGTCGATGCACAACT | ||
| Il-6 | Mus musculus | F | GCCTTCTTGGGACTGATGCT |
| R | TGTGACTCCAGCTTATCTCTTGG | ||
| Cd206 | Mus musculus | F | GTCAGAACAGACTGCGTGGA |
| R | AGGGATCGCCTGTTTTCCAG | ||
| Il-10 | Mus musculus | F | GCTCCAAGACCAAGGTGTCT |
| R | AGGACACCATAGCAAAGGGC | ||
| Arg1 | Mus musculus | F | AGCACTGAGGAAAGCTGGTC |
| R | TACGTCTCGCAAGCCAATGT | ||
| GRN | Homo sapiens | F | GTGTGACACGCAGAAGGGTA |
| R | GGGCAGAGACCACTTCCTTC | ||
| Grn | Mus musculus | F | AATGTGAAGGCGAGGACCTG |
| R | GGAGGCCTGAGTAGTGGGTA | ||
| GAPDH | Homo sapiens | F | AATGGGCAGCCGTTAGGAAA |
| R | GCGCCCAATACGACCAAATC | ||
| Gapdh | Mus musculus | F | GAGAGTGTTTCCTCGTCCCG |
| R | ATCCGTTCACACCGACCTTC |
Mononuclear cells in blood were first isolated by density gradient centrifugation. The anticoagulated blood samples were diluted 1:1 in phosphate buffered saline (PBS) and then carefully layered on Ficoll-Paque solution (F4375, Sigma Aldrich, St. Louis, USA) allowed to centrifuge at 400 g for 30–40 minutes. After centrifugation, the interleukocyte layer (monocyte layer) was aspirated, placed in a new centrifuge tube and washed 2–3 times with PBS to remove residues. The isolation process was completed by finally resuspending the monocytes in PBS. After isolation, cells were washed and incubated with human Trustain Fc XTM (422302; BioLegend, San Diego, CA, USA) for 10 min at room temperature to capture the Fc receptor and cells were treated with anti-F4/80-Fluorescein isothiocyanate (FITC) (11-4801-82, Thermo fisher, MA, USA), anti-CD11b-PE-Cy7 (101215, Biolegend, CA, USA) and FVD-eFluor 506 (65-0866-14, Thermo fisher, MA, USA) staining at 4 °C for 30 min. After fixation and permeabilisation, intracellular staining was performed with anti-CD206-PE (12-2061-82, Thermo fisher, MA, USA) and anti-CD86-PE (ab77226, Abcam, Cambridge, UK). FVD-positive cells were considered to be live cells; F4/80 and CD11b-positive cells were identified as macrophages; F4/80-positive, CD11b-positive, and CD206-positive cells were defined as M2-type macrophages; and F4/80-positive, CD11b-positive, and CD86-positive cells were identified as M1-type macrophages [18]. Cells were analysed using a flow cytometer (LSRII; BD Bioscience, Franklin Lake, NJ, USA) and data were analysed using FlowJo software V10 (Tree Star, Ashland, OR, USA).
RAW264.7 cells were lysed using radio immunoprecipitation assay (RIPA)
lysis buffer (ab170197, Abcam, Cambridge, UK) and total protein was collected.
Total protein was determined using the bicinchoninic acid (BCA) Protein
Quantification Kit (P0010, Beyotime, Shanghai, China). The same amount of protein
samples was separated by SDS-PAGE electrophoresis and transferred to PVDF
membranes using a wet transfer method (ab133411, Abcam, Cambridge, UK).
Subsequently, membranes were incubated overnight at 4 °C with the
following primary antibodies: TNFR2 (72337, Cellsignal, Danvers, MA, USA), tumor
necrosis factor-
First, RAW264.7 cells were cultured to a density of 1
RAW264.7 cells were lysed using immunoprecipitation buffer (S7705, Sigma Aldrich, St. Louis, USA), then cell lysates were incubated with agarose beads coupled with immunoprecipitating antibodies GRN (1:70, ab208777, Abcam, Cambridge, UK) and TNFR2 (1:100, ab7369, Abcam, Cambridge, UK) were incubated on agarose beads with IgG (ab172730, Abcam, Cambridge, UK) as a negative control. Unbound proteins were subsequently removed by washing the beads and the immunoprecipitated complexes were analyzed by WB. Bands were developed using ECL (A38554, Thermo fisher, MA, USA) and analyzed using ImageJ software (V1.8.0.112, NIH, Madison, WI, USA).
Statistical analysis was performed using GraphPad Prism9 (Dotmatics,
Boston, MA, USA) software. Comparisons between groups were made using the
Student’s t-test, and one-way ANOVA was used only for three or more groups, with
p
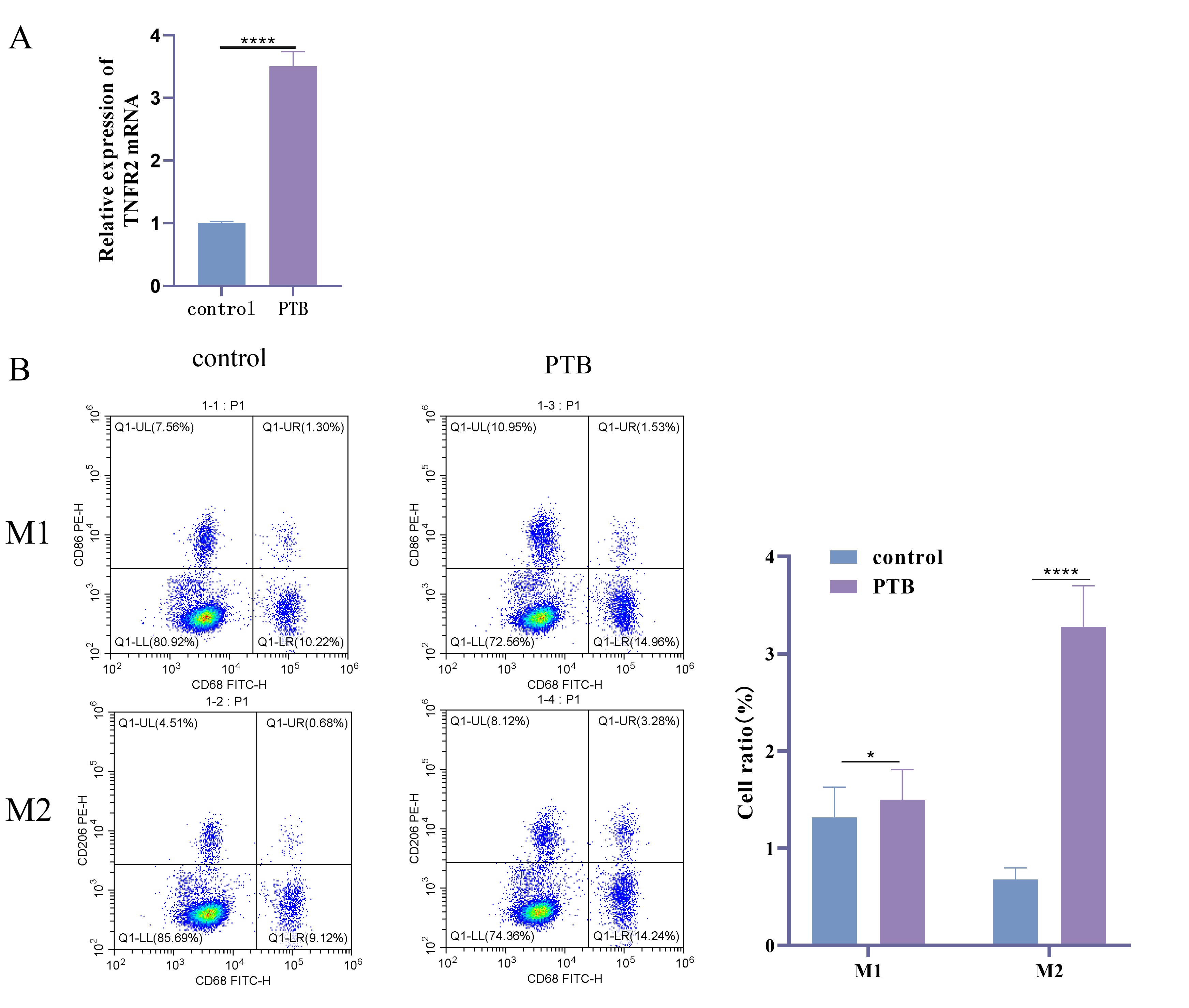
To determine the TNFR2 expression in PTB patients and the polarization
status of macrophages. The expression level of TNFR2 in patients was
detected by RT-qPCR and M1 and M2 type macrophages were analyzed using FC. The
results showed that TNFR2 mRNA expression was significantly elevated in
PTB patients compared with Control group (Fig. 1A, p
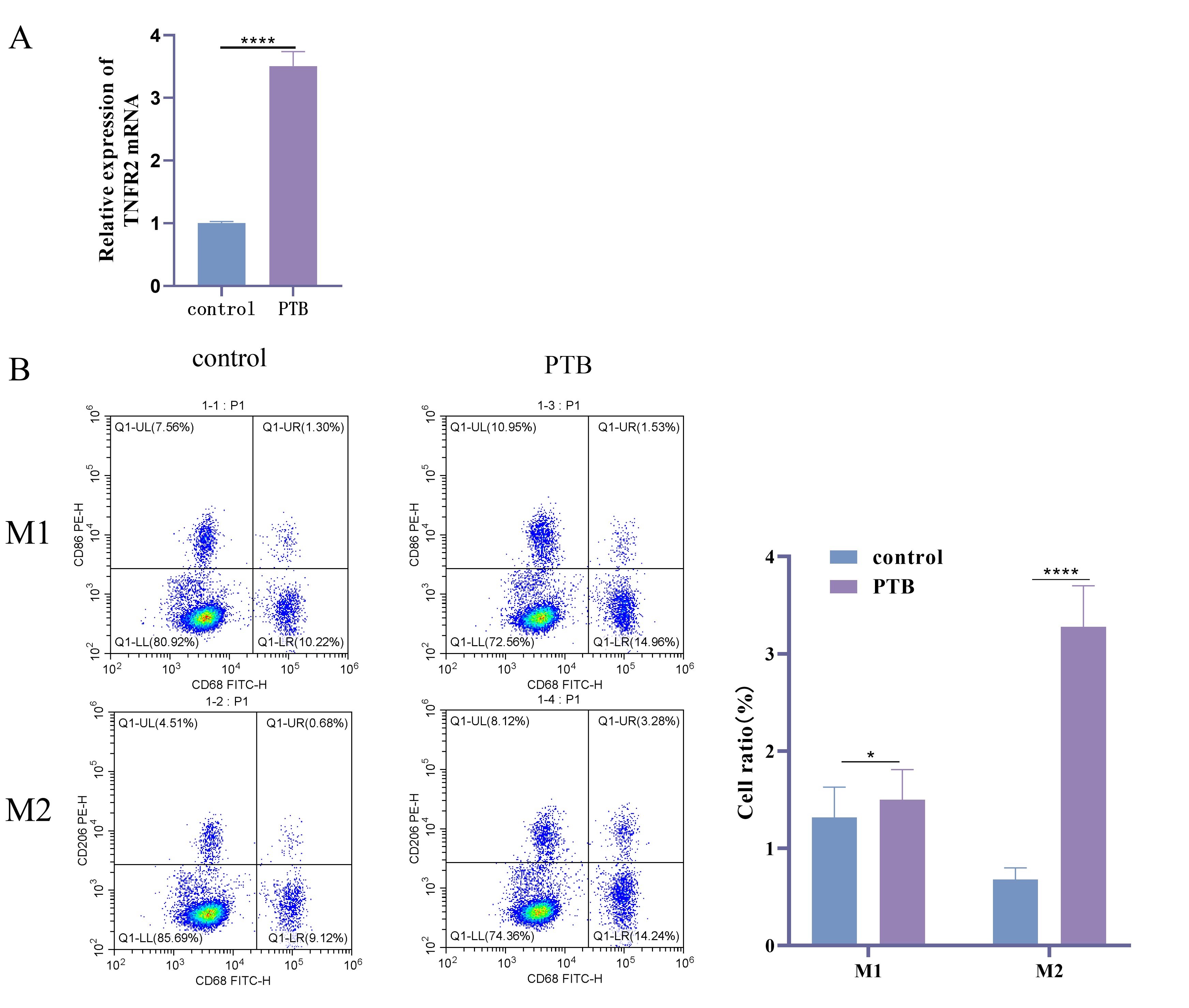 Fig. 1.
Fig. 1.
TNFR2 expression and macrophage phenotype in PTB patients and
healthy volunteers. (A) TNFR2 mRNA expression detected by RT-qPCR. (B)
Using FC to analyze the proportions of M1 and M2 macrophages in the peripheral
blood of PTB patients and healthy volunteers. Specific markers were used to
distinguish between M1 and M2 phenotypes. The data are presented as percentages
of total macrophages. N = 40; *, p
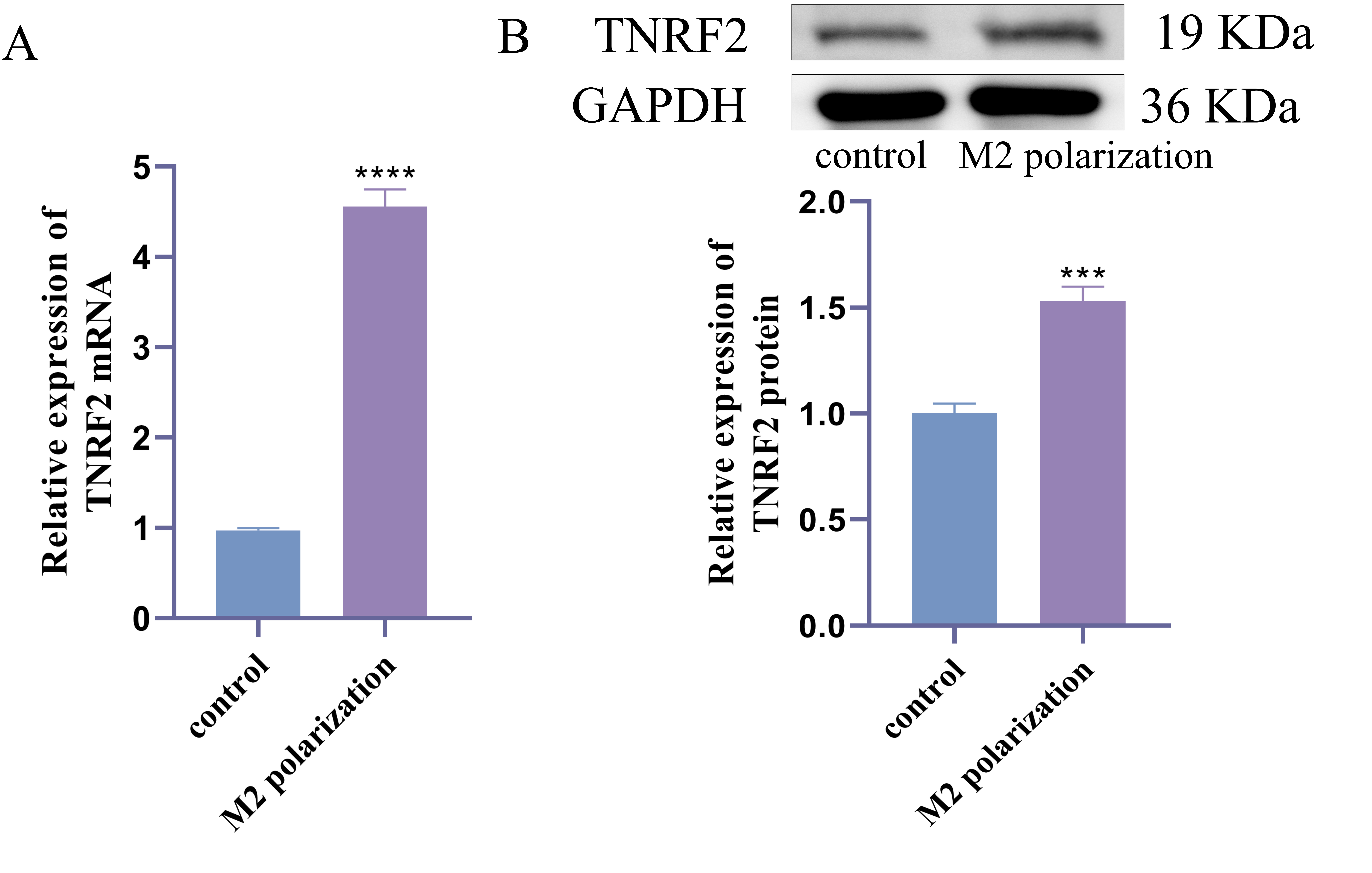
After confirming the expression of TNFR2 in the serum of PTB patients
with M2-polarised macrophages, we performed in vitro experiments. We
induced RAW264.7 into M2 macrophages using IL-4 and examined the expression level
of TNFR2. The results showed that the mRNA expression of Tnfr2 was
up-regulated (Fig. 2A, p
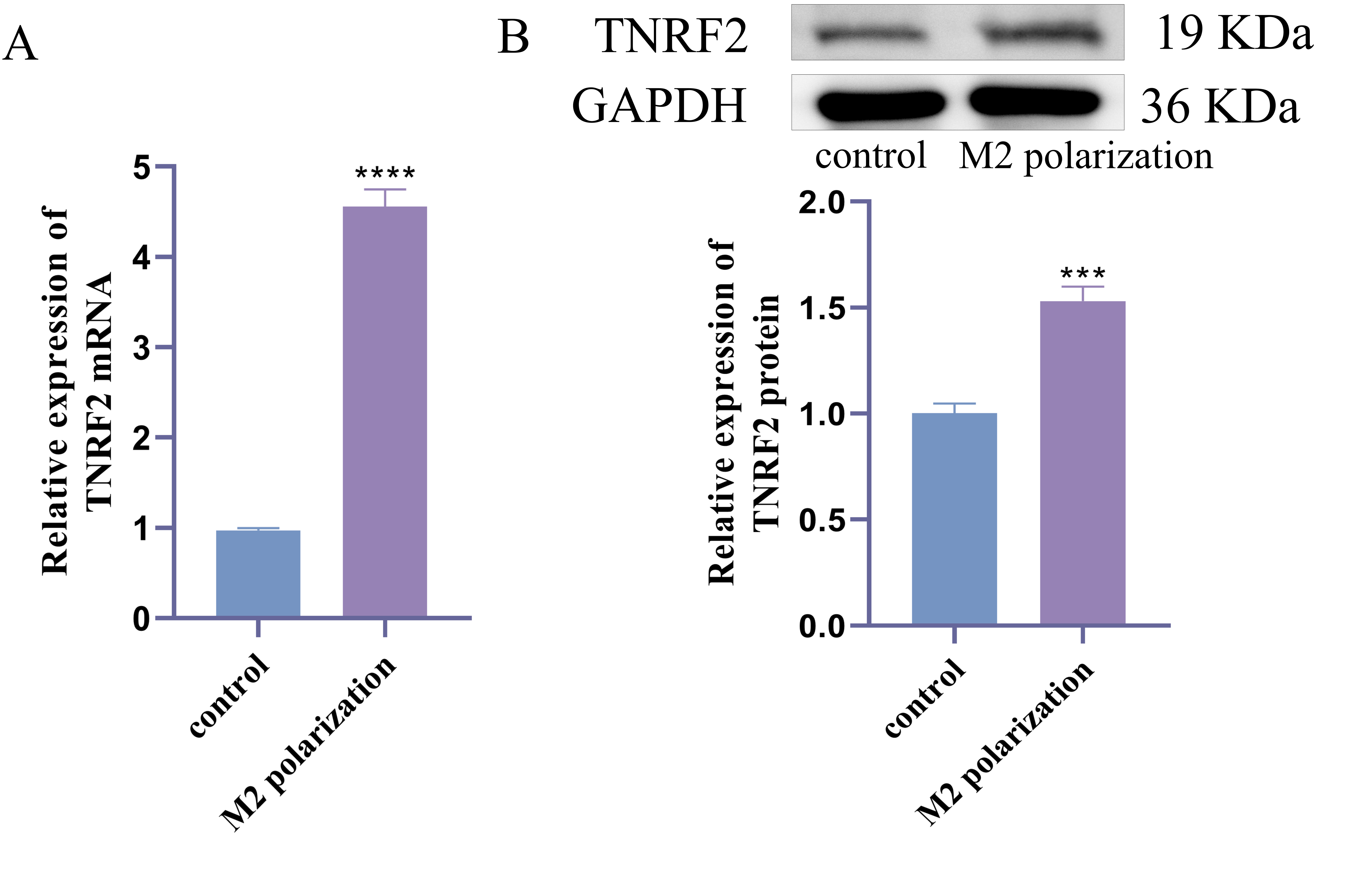 Fig. 2.
Fig. 2.
TNFR2 levels in RAW264.7 cells and M2 macrophages. (A) RT-qPCR
for Tnfr2 mRNA expression in M2 macrophages, (B) WB for TNFR2 protein
expression in M2 macrophages. N = 3; ***, p
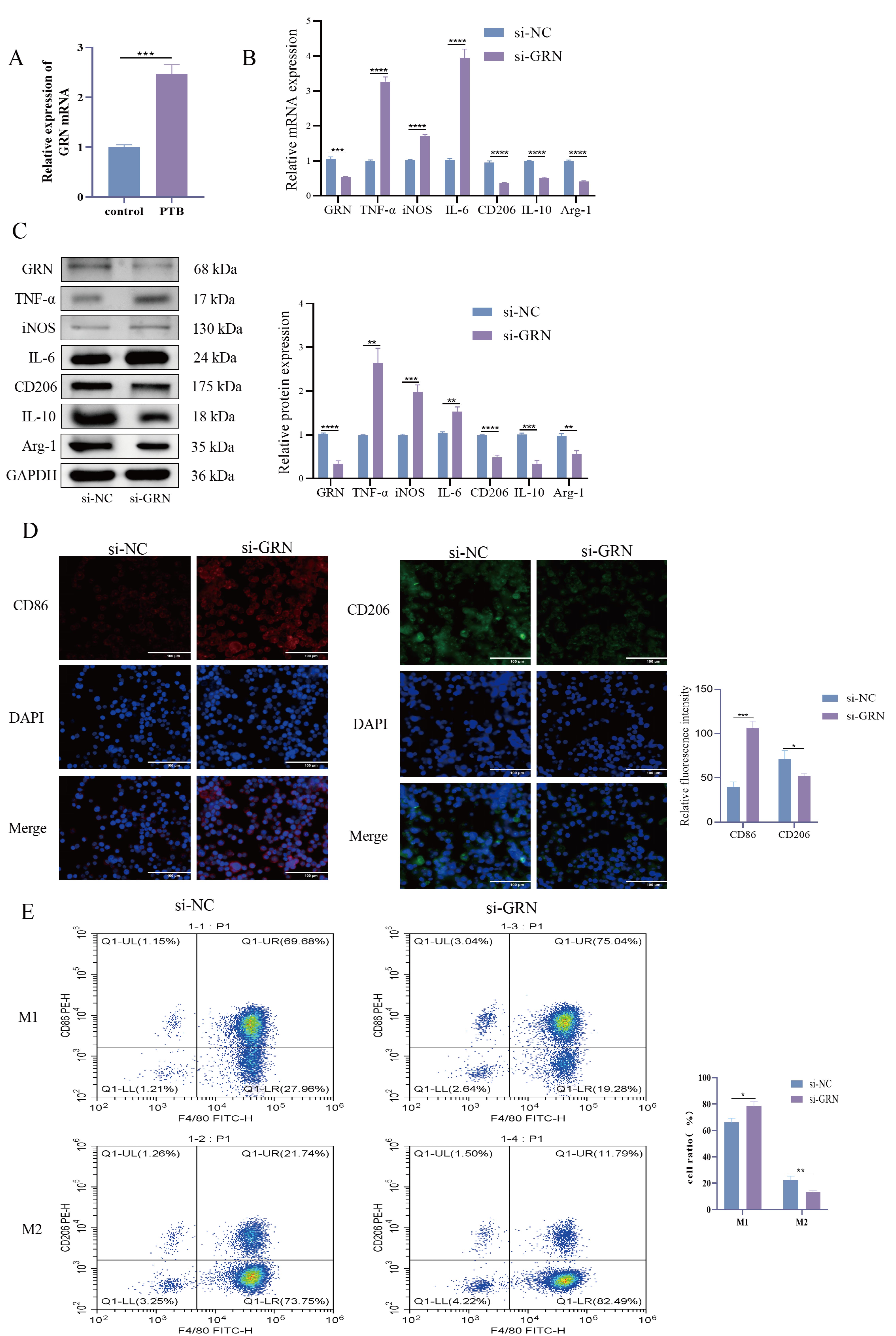
A study has shown that the activity of TNFR2 correlates with changes in
GRN [13]. We found that the expression of GRN was significantly elevated in the
serum of PTB patients (Fig. 3A, p
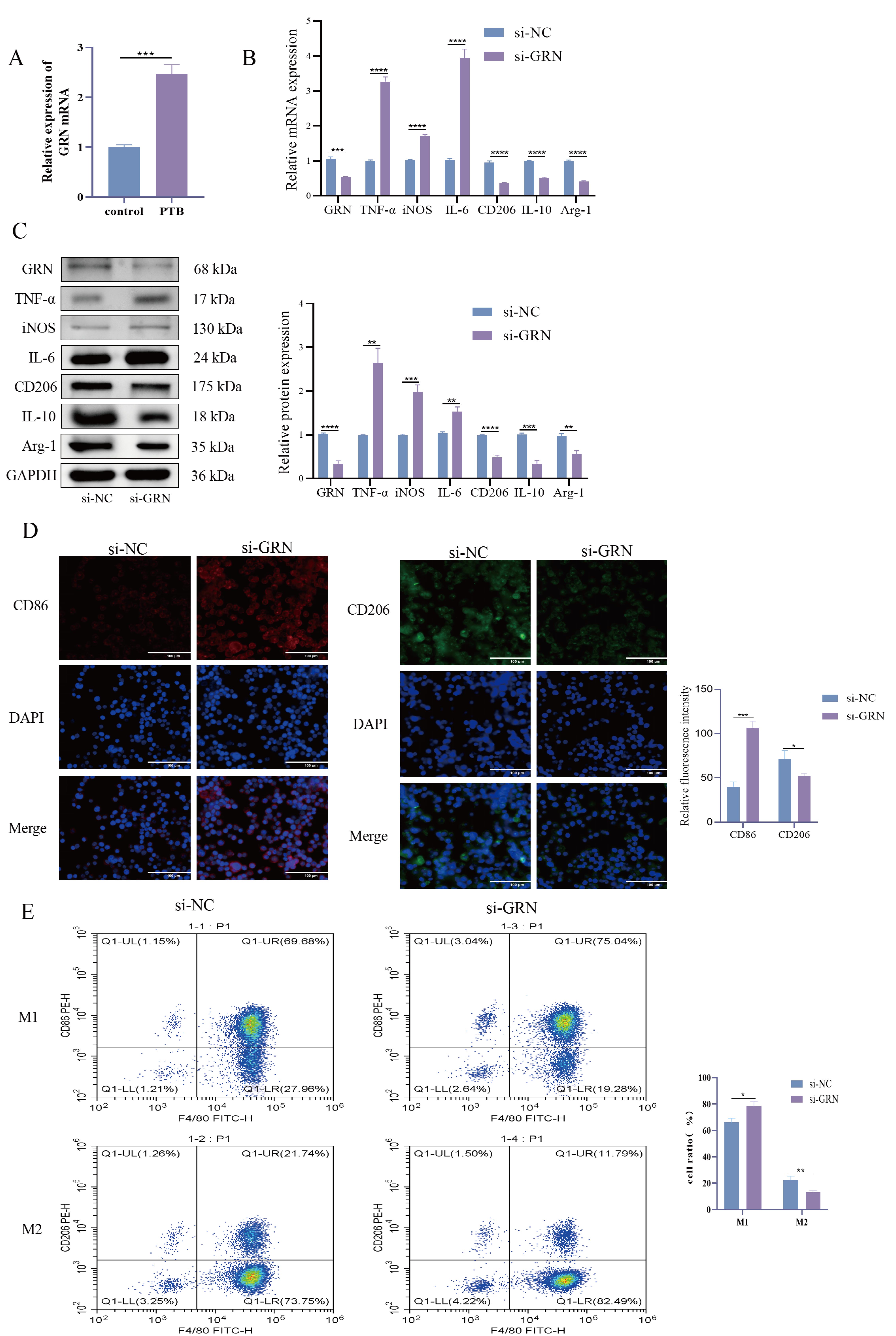 Fig. 3.
Fig. 3.
Macrophage status after GRN knockdown. (A) GRN mRNA
Expression in Serum: RT-qPCR was used to detect the expression levels of
GRN mRNA in the serum of pulmonary tuberculosis (PTB) patients and
healthy volunteers. (B) RT-qPCR to detect the expression of Grn mRNA and
M1 and M2 polarization-related markers in macrophages. Helps to gain insight into
the changes in macrophage polarization state after GRN knockdown. (C) WB
detection of the expression of GRN protein and M1 and M2 polarization related
marker proteins in macrophages. Gain a detailed understanding of the impact of
GRN knockout at the protein level. (D) The fluorescence intensities of CD86 and
CD206 were detected by IF. Measurement of the fluorescence intensity of these
markers provides a visual and quantitative assessment of the polarization status
of macrophages. Magnification 400
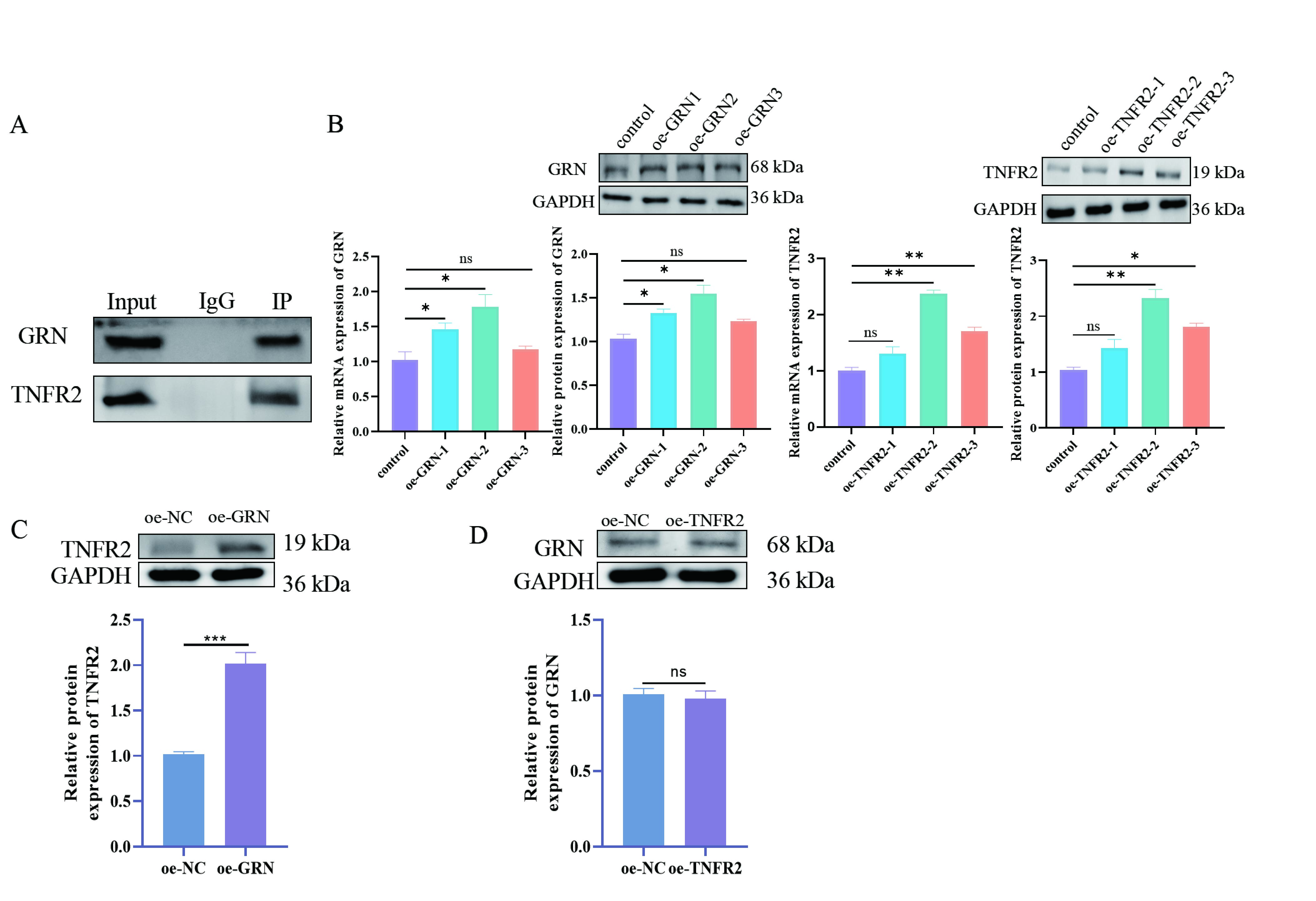
To further explore the interaction of GRN and TNFR2 in macrophage M2
polarization, we examined whether GRN binds to TNFR2 by Co-IP assay. Results
demonstrated that TNFR2 was detected after immunoprecipitation with GRN antibody
and GRN after immunoprecipitation with TNFR2 antibody, but the expression of both
was not detected in the control IgG group, respectively (Fig. 4A), suggesting
that there is a reciprocal binding effect of GRN and TNFR2. Furthermore, we
transfected GRN or TNFR2 overexpressed plasmid into RAW264.7 cells, respectively.
The increased expression of GRN and TNFR2 in the cells indicates successful
transfection (Fig. 4B, p
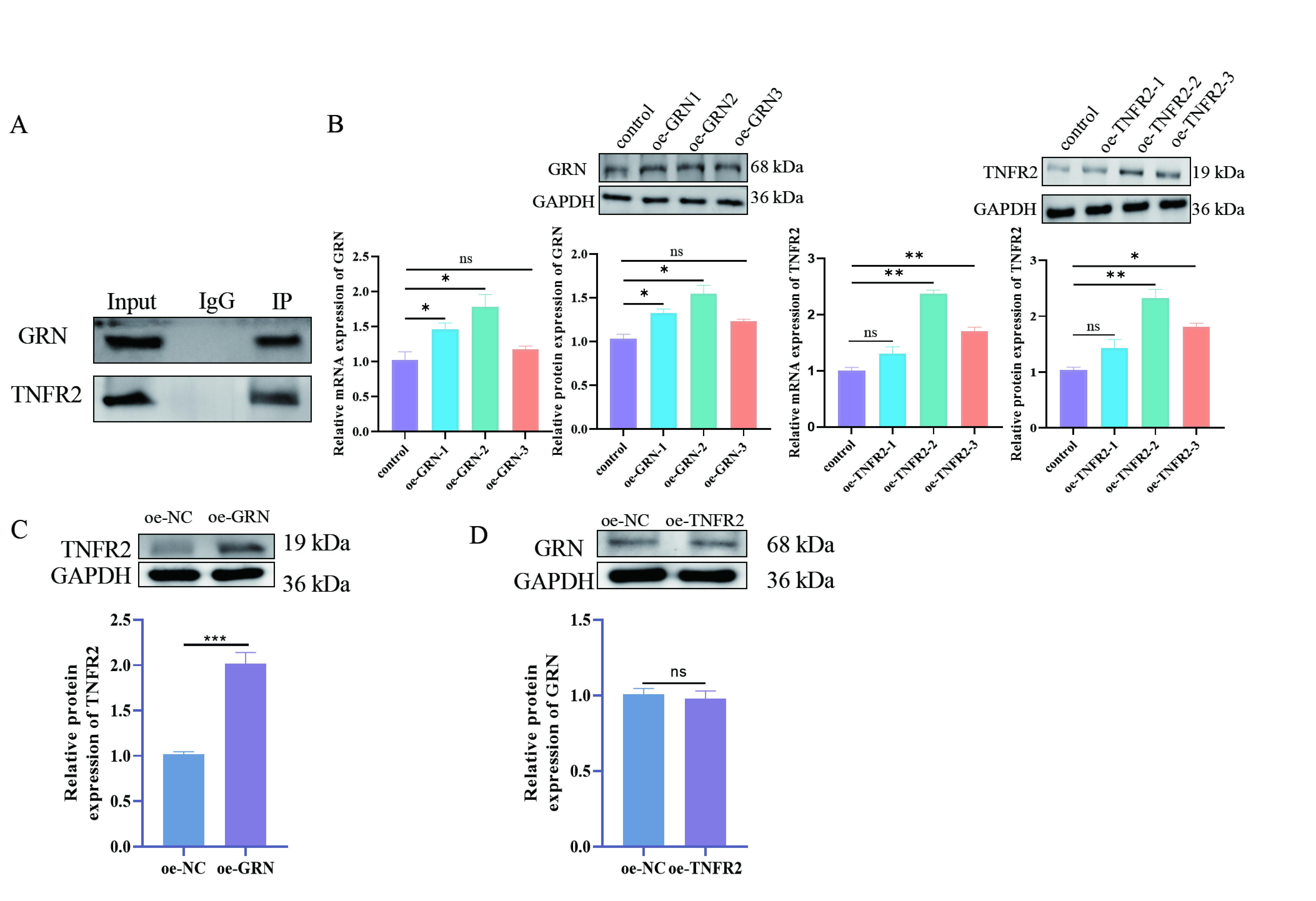 Fig. 4.
Fig. 4.
Validation of the relationship between GRN and TNFR2. (A) Co-IP
combined with WB to detect the interaction between GRN and TNFR2. Protein lysates
from macrophages are immunoprecipitated using antibodies specific for GRN or
TNFR2. The precipitated complexes are then analyzed by WB to detect the presence
of interacting proteins (TNFR2 is detected when GRN is immunoprecipitated and
vice versa). (B) WB and RT-qPCR were performed to detect the expression of GRN
and TNFR2 in RAW264.7 cells. (C) WB were used to detect the expression of TNFR2
in macrophages after GRN overexpression. This experiment was designed to
determine whether increased levels of GRN affect TNFR2 expression, thereby
suggesting a regulatory relationship between the two proteins. (D) WB were used
to detect the expression of GRN in macrophages after TNFR2 overexpression. This
analysis was designed to understand whether upregulation of TNFR2
affects GRN expression, thus further elucidating the interaction between these
proteins. N = 3; ns, p
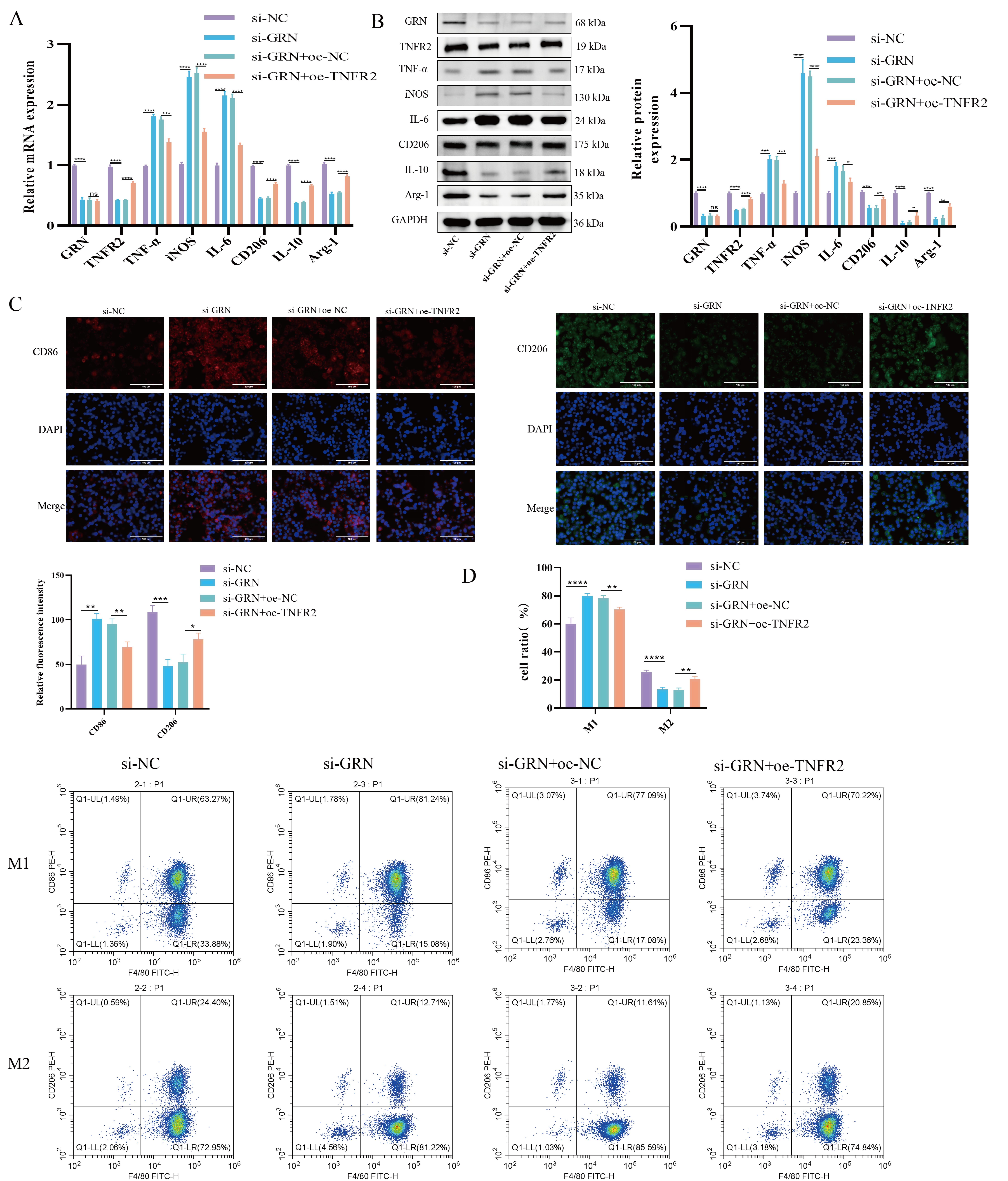
We further overexpressed TNFR2 while knocked down GRN in IL-4-induced
RAW264.7 cells and measured the macrophage M2 polarization level. The results
showed that GRN and TNFR2 protein and mRNA expression was reduced in the si-GRN
group compared to the si-NC group (Fig. 5A,B, p
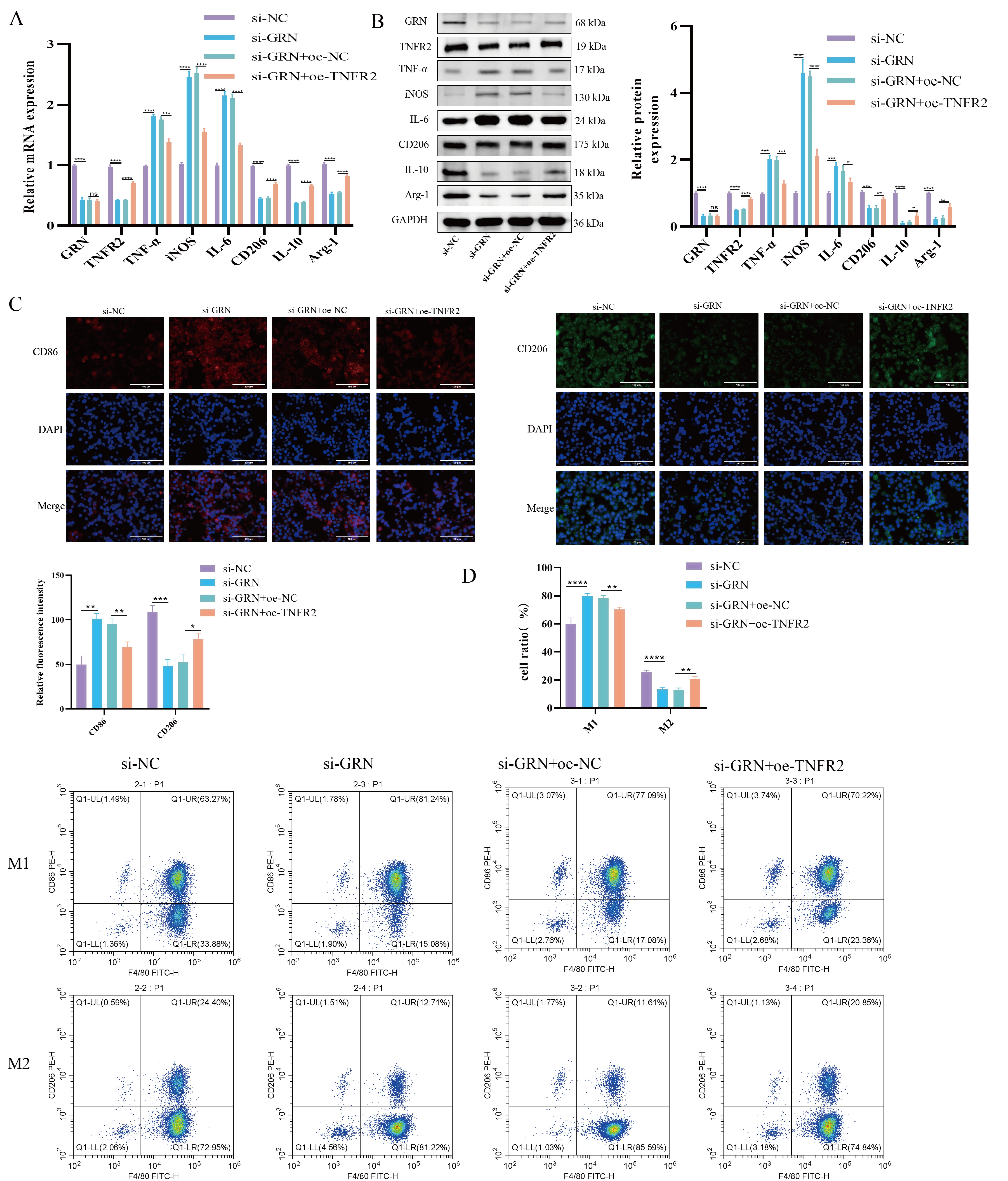 Fig. 5.
Fig. 5.
Expression of indicators and macrophage polarisation when
knocking down GRN and overexpressing TNFR2. (A) RT-qPCR for mRNA expression of
Grn, Tnfr2, and M1 and M2 polarization markers in macrophages.
(B) WB detection of protein expression of GRN, TNFR2, and M1 and M2 polarization
markers in macrophages. (C) The fluorescence intensities of CD86 and CD206 were
detected by IF. Fluorescently labeled antibodies were used to visualize these
markers, and the fluorescence intensity indicates the expression levels and
spatial distribution of the M1 and M2 phenotypes. Magnification 400
PTB is a highly contagious disease caused by Mtb [19] and is the leading bacterial disease causing death worldwide [20]. When the human immune system is dysfunctional [21], the body’s resistance to Mtb is greatly reduced [22]. Mtb is first phagocytosed by macrophages in the alveoli [23]. Macrophages, as the first line of defense of the immune system [24, 25], regulate host immune responses and disease progression through different polarization states [26]. Our study showed that Mtb patients have a significant increase in M2-type macrophages and a decrease in the proportion of M1-type macrophages in their monocytes, which in turn provides favorable conditions for Mtb survival and multiplication in vivo [27, 28]. Past study has shown that Mtb infection induces macrophage polarization to the M2 type and inhibits the bactericidal activity of M1-type macrophages [29], thereby exacerbating the development of PTB [30]. These are consistent with our findings, however, there are few studies related to the mechanisms involved, and perhaps searching for the pathological mechanisms involved may provide new targets for the treatment of PTB.
TNFR2 is a member of the tumor necrosis factor (TNF) receptor family and
functions mainly by binding to TNF-
GRN is a multifunctional protein with important roles in cell growth, inflammation regulation and tissue repair [46]. It has an important role in various diseases such as Alzheimer’s disease [47], rheumatoid arthritis [48] and colorectal cancer [49]. GRN accelerates tissue repair process by promoting cell migration and proliferation [50]. However, the expression and role of GRN in PTB have not been studied. Our results showed that the expression of GRN was significantly elevated in the serum of PTB patients and lower in healthy volunteers. For the first time, we revealed that GRN may have an important role in PTB progression. In addition, GRN has been reported to be able to regulate TNFR2-mediated signaling by binding to TNFR2 [51]. Furthermore, GRN is involved in the regulation of macrophage M2 polarization through TNFR2 in periodontitis [52]. Consistent with these results, the present study also found that GRN could promote M2 macrophage polarization by binding to TNFR2 and upregulating its expression. This suggests that GRN as well as TNFR2 may serve as potential markers of Mtb infection in PTB, which may help in early diagnosis and monitoring of Mtb infection and provide guidance for therapeutic strategies. Furthermore, GRN as well as TNFR2 may serve as targets for intervening in macrophage polarization to the extent that it may also serve as a potential therapeutic strategy in other diseases associated with macrophage polarization. However, more studies are needed to prove these speculations. By detecting serum levels of GRN and TNFR2, the extent and progression of Mtb infection can be assessed to inform clinical decision-making.
TNFR2 and GRN expression was elevated in PTB and M2 macrophages, GRN could bind to TNFR2, overexpression of GRN upregulated TNFR2 expression, overexpression of TNFR2 had no effect on GRN. In addition, high expression of TNFR2 promoted macrophage M2 polarization and attenuated Mtb infection. This study provides a new direction for attenuating Mtb infection and treating PTB.
The data used to support the findings of this study are available from the corresponding author upon request.
BLZ and FW designed the research study. LX and JC performed the research. JZ, RLD and GLM analyzed the data. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
This study has been approved by the Ningbo Ninth Hospital Medical Health Group Ethics Review Committee (No. NNHM-20230201). All patients or their families/legal guardians as well as healthy volunteers voluntarily signed an informed consent form.
Not applicable.
This research received no external funding.
The authors declare no conflict of interest. Guolun Mo is from Beijing STEPPIN Technology Co., LTD., the judgments in data interpretation and writing were not influenced by this relationship.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.