1 Hubei Key Laboratory of Renal Disease Occurrence and Intervention, Medical School, Hubei Polytechnic University, 435003 Huangshi, Hubei, China
2 Department of Critical Care Medicine, Huangshi Hospital of TCM (Infectious Disease Hospital), 435003 Huangshi, Hubei, China
Abstract
Coenzyme A (CoA) is synthesized from pantothenate, L-cysteine and adenosine triphosphate (ATP), and plays a vital role in diverse physiological processes. Protein acylation is a common post-translational modification (PTM) that modifies protein structure, function and interactions. It occurs via the transfer of acyl groups from acyl-CoAs to various amino acids by acyltransferase. The characteristics and effects of acylation vary according to the origin, structure, and location of the acyl group. Acetyl-CoA, formyl-CoA, lactoyl-CoA, and malonyl-CoA are typical acyl group donors. The major acyl donor, acyl-CoA, enables modifications that impart distinct biological functions to both histone and non-histone proteins. These modifications are crucial for regulating gene expression, organizing chromatin, managing metabolism, and modulating the immune response. Moreover, CoA and acyl-CoA play significant roles in the development and progression of neurodegenerative diseases, cancer, cardiovascular diseases, and other health conditions. The goal of this review was to systematically describe the types of commonly utilized acyl-CoAs, their functions in protein PTM, and their roles in the progression of human diseases.
Keywords
- acylation
- acyl-CoA
- cancer
- coenzyme A
- post-translational modification
Coenzyme A (CoA) is a crucial coenzyme that binds with diverse acyl groups to form acyl-CoA, which is involved in numerous essential physiological processes [1, 2]. In 1937, the German-born British biochemist Hans Adolf Krebs (1900–1981) discovered the tricarboxylic acid cycle (TCA cycle), a significant cellular physiological process in energy production [3]. However, certain steps within the TCA cycle require a cofactor, which at the time was unidentified. The German-born American biochemist Fritz Albert Lipmann (1899–1986) carried out extensive studies of this cofactor and successfully isolated CoA from pigeon liver extracts in 1945 [4]. The collective efforts of Krebs and Lipmann led to the discovery of CoA and its pivotal role in cellular metabolism, for which they were ultimately awarded the Nobel Prize in Physiology or Medicine in 1953 [5, 6, 7].
CoA is a vital cofactor found universally in both prokaryotic and eukaryotic cells. Currently, almost all enzymes in the CoA biosynthesis pathway have been identified. Despite sharing similar biochemical functions, prokaryotic and eukaryotic enzymes exhibit significant sequence variations, as revealed by comparative genomics studies [8, 9, 10, 11, 12]. During the logarithmic growth phase of Staphylococcus aureus, CoA contributes to the production of metabolically active thioesters and may also serve as a low molecular weight antioxidant, thereby counteracting oxidative and metabolic stress [13]. In plants, a transferred DNA (T-DNA) mutant of Arabidopsis thaliana is impaired in the penultimate step of the CoA biosynthesis pathway, catalyzed by phosphopantetheine adenylyltransferase (PPAT). This step is critical for plant growth, stress resilience, and seed lipid accumulation [14]. In mammals, CoA acts as a cofactor for over 70 enzymes and participates in a myriad of vital cell physiological processes, including carbohydrate breakdown, fatty acid oxidation (FAO), amino acid decomposition, pyruvate degradation, and the TCA cycle (Fig. 1) [15, 16, 17]. CoA is an essential and irreplaceable molecule in cellular function.
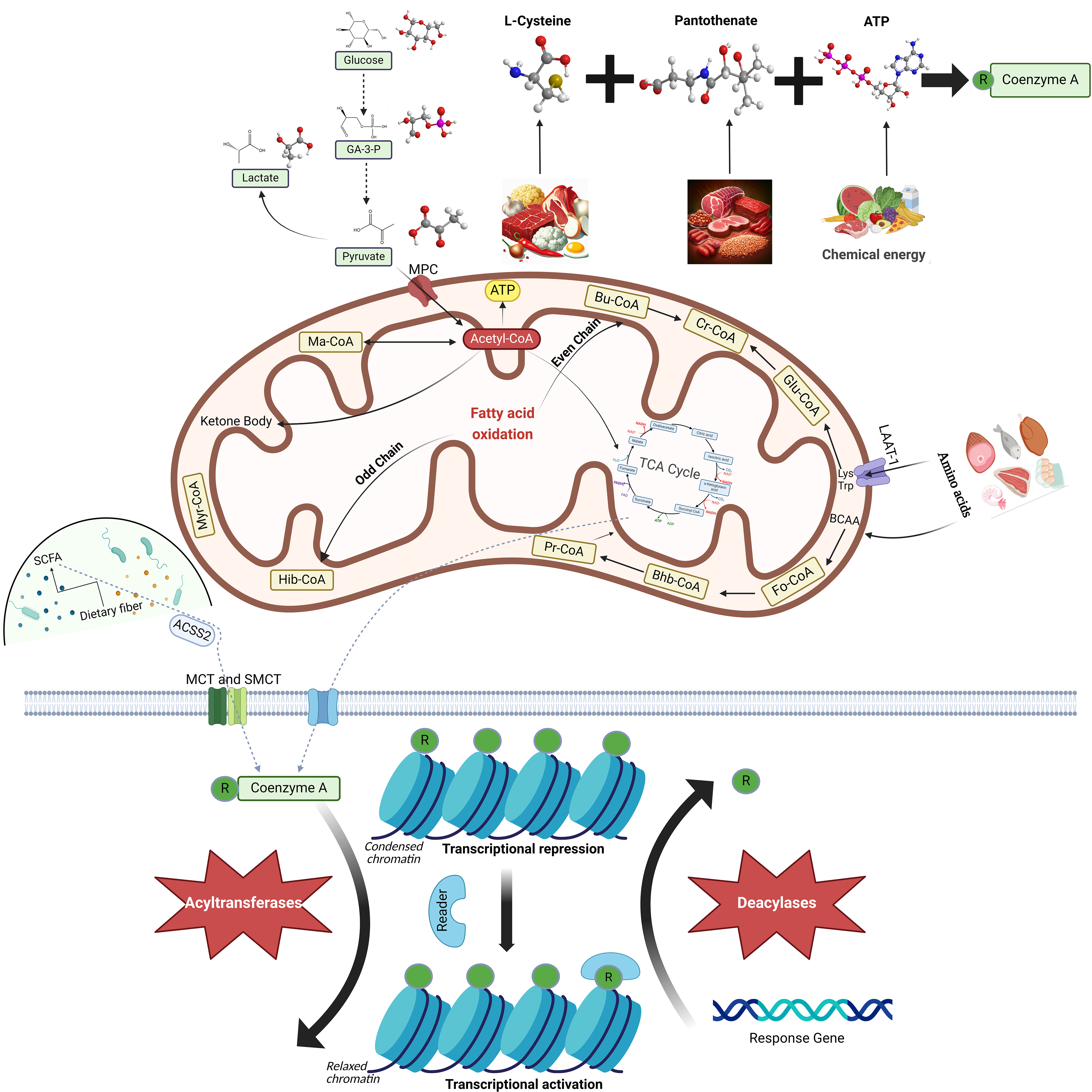 Fig. 1.
Fig. 1.
Biosynthesis, Metabolism, and Function of Coenzyme A (CoA). CoA
is synthesized via a five-step enzymatic reaction involving pantothenic acid
(PAN), L-cysteine, and adenosine triphosphate (ATP). It is sourced from animal
offal, meat, grains, and eggs, and plays a key role in the metabolism of
pyruvate, fatty acids, amino acids, and the tricarboxylic acid (TCA) cycle within
cells. CoA interacts with various acyl groups to form a range of acyl-CoA
molecules under the influence of acyltransferase. Acyl-CoA molecules contribute
to numerous cellular physiological processes. Acetyl-CoA primarily forms through
sugar oxidation in the mitochondria, generating ATP through the
The synthesis of CoA is dependent on three compounds: pantothenate (PAN), L-cysteine, and adenosine triphosphate (ATP) (Fig. 1) [1]. Some bacteria use Cytidine triphosphate (CTP) rather than ATP [18]. Pantothenate, also known as pantothenic acid and vitamin B5, is ubiquitous in nature and serves as a crucial precursor molecule in CoA biosynthesis [19]. Pantothenate kinase (PANK) is the first and rate-limiting enzyme in CoA biosynthesis. PANK phosphorylates pantothenate, resulting in the generation of 4′-phosphopantothenate [20]. Phosphopantothenoylcysteine synthetase (PPCS) facilitates the addition of L-cysteine to 4′-phosphopantothenate, thereby forming 4′-phosphopantothenoylcysteine [21]. Subsequently, phosphopantothenoylcysteine decarboxylase (PPCDC) removes one CO2 molecule from 4′-phosphopantothenoylcysteine, leading to the production of 4′-phosphopantetheine [11]. The addition of an adenylyl group supplied by ATP to 4′-phosphopantetheine and catalyzed by phosphopantetheine adenylyltransferase (PPAT) results in the synthesis of dephospho-CoA [22]. Dephospho-CoA kinase (DPCK) catalyzes the phosphorylation of dephospho-CoA to generate CoA, which is the last step in CoA biosynthesis [23].
CoA thioester derivatives are generated by the formation of thioester bonds with various acyl groups [24]. Acylation involves the introduction of an acyl group and is typically facilitated by acyltransferases [25]. This reaction serves multiple functions within cells. Notably, protein acylation is a significant post-translational modification (PTM) in eukaryotes and exerts regulatory control over protein functionality (Fig. 1) [26, 27].
The acyl-CoA family therefore constitutes a group of vital metabolic factors
that play crucial roles in cellular metabolism, glucose metabolism, fatty acid
synthesis, and protein modification (Fig. 1) [28]. Acyl-CoA can be categorized
into five groups according to the length and structure of the bound acyl group:
(1) short-chain acyl-CoA (number of carbons atoms [Cn]
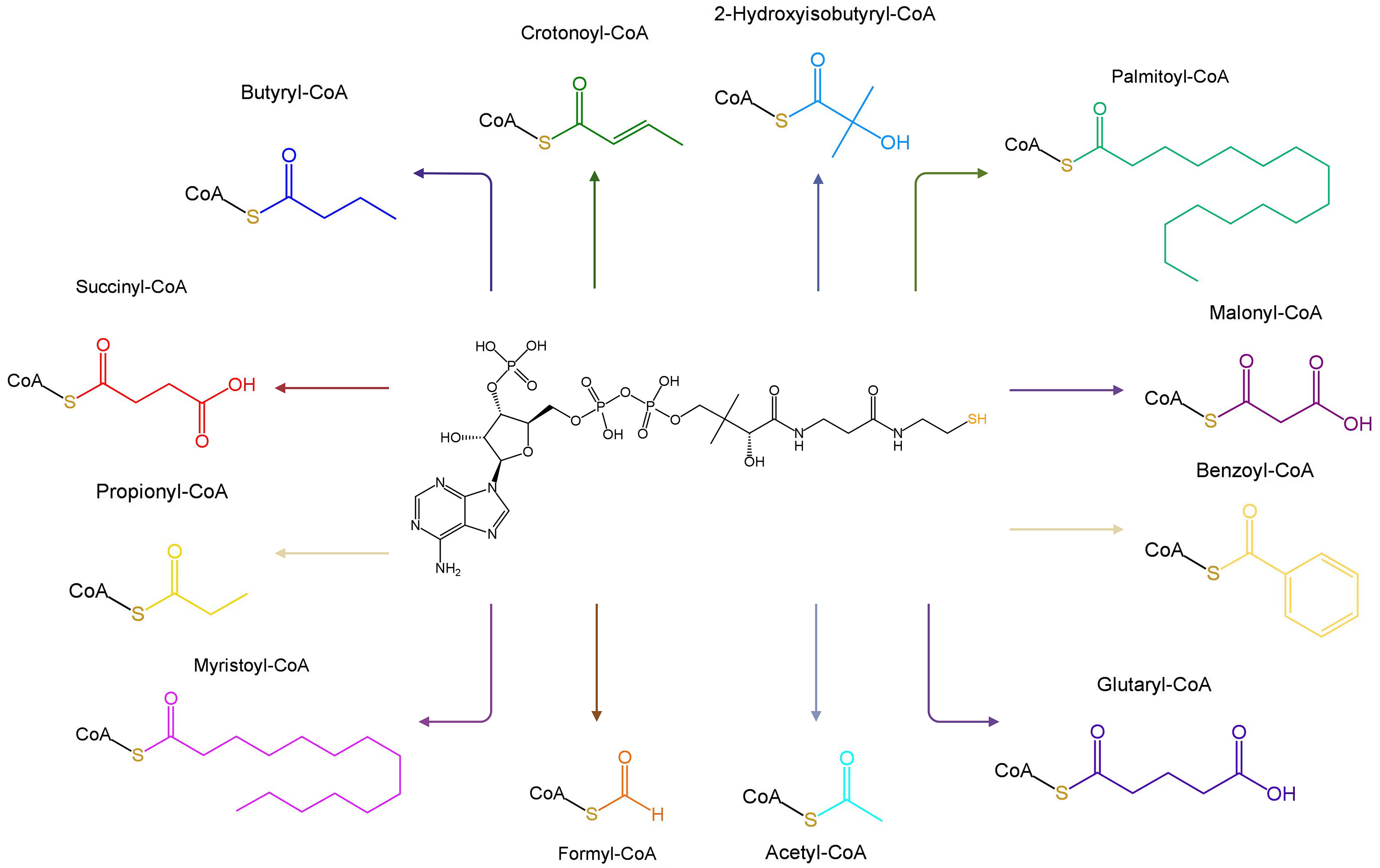 Fig. 2.
Fig. 2.
Molecular structures of Coenzyme A and its 12 common derivatives. Created with BioRender.com.
CoA and its derivatives are predominantly localized in the cytoplasm and mitochondria where they are active participants in the catalytic transformation of various cellular processes [32]. For example, acetyl-CoA carboxylase (ACC) catalyzes the transformation of acetyl-CoA and one bicarbonate molecule into malonyl-CoA, which is involved in fatty acid biosynthesis in many organisms [33].
CoA and its derivatives contribute to approximately 9% of the enzyme reactions in cells [34]. Various acyl-CoA molecules have been identified, including formyl-CoA, acetyl-CoA, propionyl-CoA, butyryl-CoA, malonyl-CoA, succinyl-CoA, glutaryl-CoA, palmitoyl-CoA, myristoyl-CoA, benzoyl-CoA, crotonoyl-CoA, and 2-hydroxyisobutyryl-CoA [35] (Fig. 2). Acyl-CoAs are closely associated with numerous physiological processes, such as the biosynthesis and oxidation of fatty acids, pyruvate oxidation, acetylcholine synthesis, cholesterol biosynthesis, blood lipid regulation, steroid substance synthesis, acyl group transfer, immune activation, and connective tissue formation and repair (Fig. 1) [15, 36, 37].
Multiple acyl-CoA molecules are important in the treatment of cancer, cardiovascular diseases, metabolic disorders, neurological conditions, and inflammatory and infectious diseases (Fig. 3) [38, 39]. Acyl-CoA binding proteins (ACBPs) are highly expressed in glioblastoma multiforme. These proteins maintain a high rate of cell proliferation and promote tumor growth through binding to acyl-CoAs [39]. The above findings highlight the essential roles of CoA and its thioester derivatives in human health.
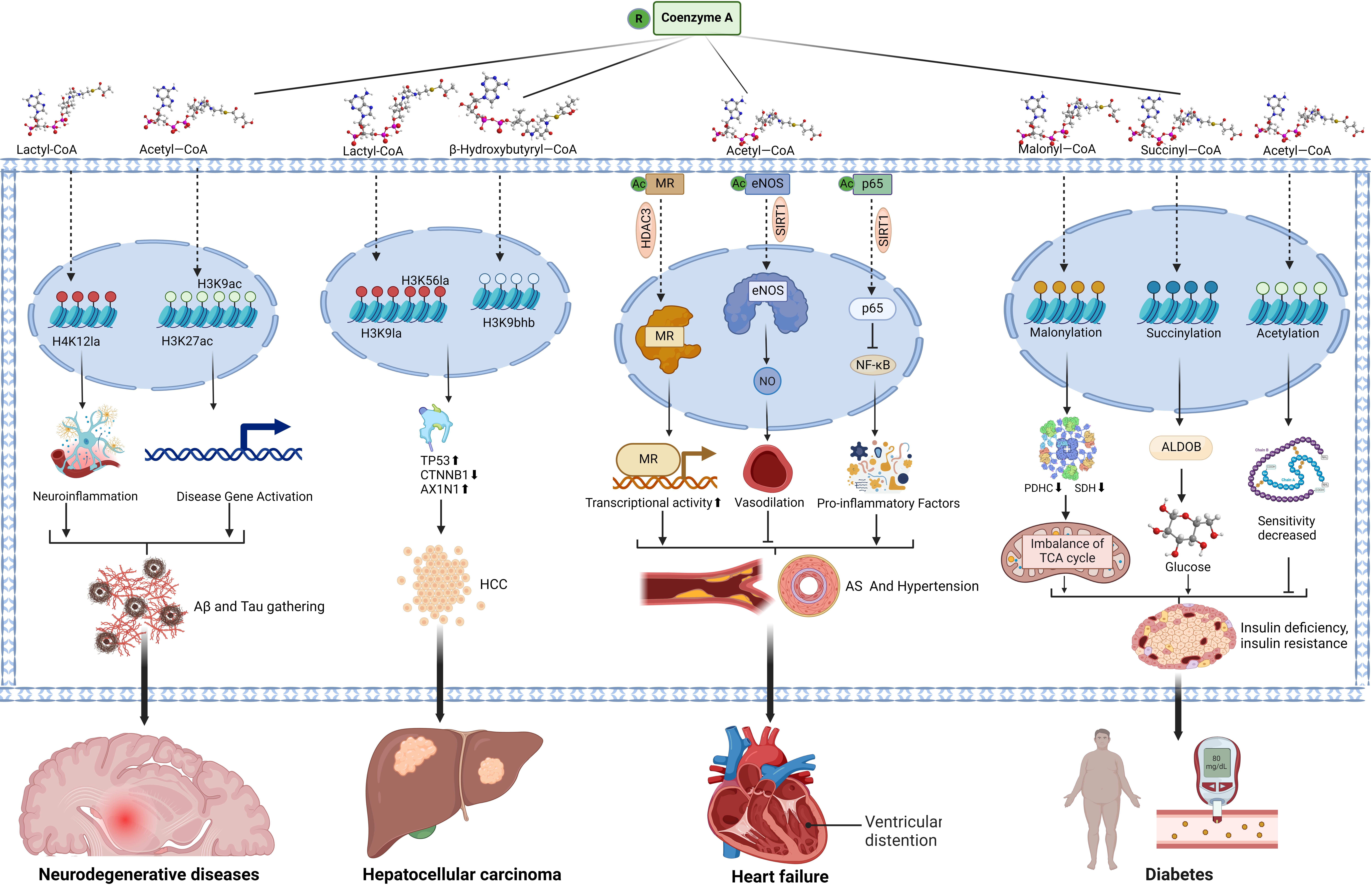 Fig. 3.
Fig. 3.
Coenzyme A and its derivatives in diseases. In
neurodegenerative diseases, lactyl-CoA and acetyl-CoA modulate neuroinflammation
and associated gene expression via histone modification. This increases the
levels of
The aim of this review was to gather information on the discovery and biosynthesis of CoA, the classification of commonly utilized acyl-CoAs, the functions of acyl-CoAs in protein PTM, and the roles of acyl-CoAs in the progression of neurodegenerative diseases, cancer, cardiovascular disease and other health disorders.
CoA plays a crucial role in various biological reactions by encompassing the following key functions: (a) Energy production. Citrate exported from mitochondria is converted to acetyl-CoA and oxaloacetic acid through the action of ATP citrate lyase (ACLY). Acetyl-CoA is transported into the mitochondrial matrix and subsequently enters the TCA cycle to form citrate. Nicotinamide Adenine Dinucleotide (NADH) and Flavine adenine dinucleotide (FADH2) are produced within the TCA cycle, thereby facilitating proton efflux. Following the re-entry of protons through ATP synthase, ATP is generated via oxidative phosphorylation [40]. (b) Fatty acid biosynthesis. Acetyl-CoA undergoes a series of reactions within the cytoplasm that give rise to malonyl-CoA, phospholipids, triacylglycerols, and other lipids. Additionally, acetyl-CoA can be catalyzed by 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) into mevalonate, which subsequently serves as a precursor for cholesterol synthesis [41, 42]. (c) Metabolic regulation. Acetyl-CoA acts as a critical and central regulator of metabolic pathways involved in sugar, fat, and protein modification [43, 44]. (d) Regulation of gene expression. The acylation of diverse histones can modulate the transcription of associated genes [35, 36]. (e) Signaling transduction. Acetyl-CoA has the capacity to transmit signals and to regulate signaling pathways, both intra- and extra-cellularly [43].
CoA can form covalent bonds with 12 distinct acyl groups, leading to a wide range of acylation modifications (Fig. 2). The acyl groups can be categorized as follows: (a) hydrophobic acyl groups, including acetyl, butyryl, benzoyl, crotonyl, palmityl, and myristoyl; (b) negatively charged acidic acyl groups, such as malonyl, glutaryl, and succinyl; and (c) polar acyl groups, including formyl, propionyl, and beta-hydroxybutyryl [35].
Protein acylation has the ability to influence protein localization, functional regulation, and complex formation [35]. Acyl-CoA is commonly employed as an acyl donor in protein acylation [25]. Distinct types of protein acylation play significant regulatory roles in their respective functions [45]. One classical form of protein acylation is PTM, which plays a crucial role in influencing genome function and epigenetic regulation. Consequently, PTM affects gene transcription, DNA replication, and other vital processes (Fig. 1) [45, 46]. The binding of histone acetyltransferase to origin recognition complex subunit 1 (ORC1) is currently understood to catalyze acetylation, propionylation, butyrylation, crotonylation, and other modifications [47]. Lysine acylation includes formylation, acetylation, propionylation, and butyrylation, with each having distinct functions in histone and non-histone proteins [48].
Formyl-CoA donates formyl groups to amino acid residues through iso-peptide bonds. This leads to protein formylation, which is a distinct type of PTM (Fig. 1) [49]. Histone formylation occurs predominantly on lysine or arginine residues situated at the N-terminal of H3 and H4 histones. This modification is intricately associated with chromatin remodeling, gene expression, and cellular dysregulation [50, 51, 52, 53].
Moreover, hindering the formylation of acylated initiating tRNA leads to the creation of a vacant initiation codon. This results from either restricting the formyl donor, or inhibiting the formyl transferase during in vitro translation [54]. In bacterial systems, the essential role of formylated tRNAMet in protein biosynthesis is well recognized. Furthermore, N-terminal formylated proteins play a crucial role in specific metabolic processes [55]. In eukaryotes, N-terminal formylation is primarily restricted to specific mitochondrial proteins, but recent studies have shown its involvement in cytoplasmic N-terminal formylation. This links various cellular stresses to N-terminal-dependent protein degradation [56, 57].
Formylation is a complex and multifaceted biological process that is crucial for maintaining regular cellular function and for adapting to diverse environmental circumstances. It occurs during RNA modification, regulation of protein activity, cell cycle control, intracellular signaling transduction, and the modulation of metabolism. The formylation modification process is a pivotal factor in cell biology, physiology, and the onset of disease.
Lysine acetyltransferase (KAT) is responsible for transferring the acetyl group
from acetyl-CoA to the
Propionyl-CoA acts as a donor for the protein acylating modification. The propionyl group derived from propionyl-CoA is then transferred to protein lysine residues, resulting in a form of protein PTM known as propionylation [69, 70]. Histone propionylation primarily occurs on the N-terminal lysine residues of histone H2A and H2B. The catalytic activity responsible for this modification is attributed to N-terminal acetyltransferases (NATs) [71]. Physiological processes associated with propionylation include chromatin activity and metabolic reactions [72].
The butyryl group derived from butyryl-CoA is transferred to a specific lysine residue to cause protein butyrylation, which is a form of protein PTM [73]. Butyrylation is observed primarily on conserved lysine residues located at the N-terminal region of histones H3 and H4. This modification is closely associated with various physiological processes, including gene transcription, chromatin activity, and metabolic reactions [74, 75]. Butyrylation also plays a significant role in the development of cancer, neurodegenerative diseases, and metabolic disorders (Fig. 1) [76].
Malonyl-CoA mediates the addition of malonyl groups to protein lysine residues.
This results in malonylation, which is another type of protein PTM [77]. During
stimulation by lipopolysaccharide (LPS), glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) undergoes malonylation, leading to its dissociation from tumor necrosis
factor-
Glutaryl-CoA serves as a donor for glutarylation, wherein glutaryl groups are covalently bound to lysine residues. This results in glutarylation, which is a specific type of protein PTM [79]. A mutation in the glutaryl-CoA dehydrogenase (GCDH) gene leads to elevated protein glutarylation and subsequently gives rise to a metabolic disorder known as glutaracidemia type I [79]. Further analysis revealed a significant disparity in protein glutarylation levels between normal men and those diagnosed with asthenospermia, a condition characterized by reduced sperm motility. This disparity suggests a possible link between glutarylation and asthenospermia [80].
Palmitoyl-CoA is involved in the covalent attachment of palmitic acid to protein cysteine residues through a thioester bond. This results in protein palmitoylation, another important form of protein PTM [81]. Palmitoylation is a versatile modification, with a smaller fraction attached to protein arginine and threonine residues. This modification plays a crucial role in regulating protein activity, stability, transport, and protein-protein interactions [82]. Palmitoyl-CoA also plays a pivotal role in regulating the phosphorylation mechanism and acts as an inhibitor of mitochondrial Adenosine Diphosphate (ADP)/ATP transport, thereby exerting a protective effect on mitochondria under ischemic conditions [83].
N-myristoyl transferase (NMT) transfers myristoyl groups derived from myristoyl-CoA to lysine or to N-terminal glycine residues, resulting in protein myristoylation [84]. This protein PTM plays a vital role in enhancing protein adhesion to the cell membrane, as well as facilitating protein interactions within the cell membrane [85]. It is commonly observed at the N-terminal region of various signaling proteins [86]. Myristoylation also significantly influences diverse cellular activities across various diseases. For example, in rheumatoid arthritis, the downregulation of N-myristoyl transferase 1 (NMT1) results in loss of the N-myristoylation modification in AMP-activated protein kinase (AMPK). This leads to reduced localization and activation of mechanistic target of rapamycin complex 1 (mTORC1), and the emergence of pro-inflammatory T cell phenotypes [87]. In plants, the dual lipid modifications of N-myristoylation and S-acylation play a crucial role in facilitating Ca2+ regulation in Arabidopsis kinase. These modifications promote activation of the slow anion channel-associated 1 (SLAC1) anion channel, which is involved in calcium-mediated signaling processes and the regulation of CO2 sensitivity in plant gas exchange [88, 89].
Benzoyl-CoA contributes benzoyl groups to lysine residues, thereby catalyzing the formation of benzoylation, which is a unique protein PTM [90]. A novel benzoylation site, 22 Kbz, was found on histones in HepG2 and RAW cells, making it the sole lysine modification featuring benzene rings [91]. Subsequent investigations highlighted the critical role of YEATS domain-containing 2 (YEATS2), an epigenetic regulator, in modulating the expression of ribosome genes [92]. YEATS2 exhibits selective affinity for benzoylation modifications. Histone acetyltransferase in association with origin recognition complex 1 (HBO1) act as a lysine benzoylation transferase (Kbz) in mammalian cells, targeting predominantly specific lysine residues in proteins. This targeting by HBO1 is essential for various cellular functions, including chromatin remodeling, metabolism, immune response, and tumorigenesis [93].
Succinyl-CoA transfers succinyl groups to protein lysine residues, resulting in succinylation [94]. Following viral infection, succinylation upregulates mitochondrial antiviral signaling protein (MAVS), whereas SIRT5-mediated desuccinylation downregulates MAVS, thus dampening the immune response. This suppression leads to the diminished expression of type I interferon and antiviral genes [95]. Using a mouse model, Yang et al. [96] demonstrated that lead (Pb) exposure reduced lysine acetylation and succinylation in premeiotic, meiotic, and round sperm cells, thereby impairing fertility through the disruption of reproductive processes.
Crotonoyl-CoA transfers crotonoyl groups to lysine residues through histone crotonoyltransferase (HCT), thereby initiating crotonoyl acylation [97]. In chromodomain Y-like (CDYL) transgenic mice, disrupted histone crotonylation leads to diminished sperm cell viability and reduced male fertility [98]. In endodermal cells, induction of the enzyme that produces crotonyl-CoA enhances histone crotonylation levels, thereby stimulating the differentiation of human embryonic stem cells (hESCs) towards an endodermal lineage [99].
The Warburg effect is characteristic of cancer cells and promotes lactate production, which is linked to various intracellular physiological processes including angiogenesis, hypoxia, and immune evasion. While lactate’s role as an energy source and metabolic by-product in cancer cells is well-documented, its non-metabolic functions are less understood. Lactyl-CoA, which donates lactyl groups to histone lysine residues, serves as an epigenetic modifier that directly impacts gene transcription [101]. This epigenetic alteration, known as histone lysine lactylation (Kla), introduces a novel approach for potential cancer treatments through the various roles of lactate [102]. Research on lactylated proteins within gastric cancer Adenocarcinoma Gastric Cell (AGS) cells identified 2375 Kla sites across 1014 proteins, and found that increased Kla levels in gastric tumors were associated with adverse prognosis [103].
In addition, extensive lactylome profiling of hepatitis B virus-related hepatocellular carcinoma (HCC) detected 9275 Kla sites, with 9256 of these being on non-histone proteins. This indicates a widespread presence of Kla that extends beyond histone proteins to influence transcriptional regulation [104].
Protein methylation is a recently uncovered PTM that involves the transfer of a methyl group from methyl-CoA to lysine residues by lysine methyltransferases (KMTs). Histone lysine methylation has been shown to play a pivotal role in the regulation of gene expression, cell cycle control, genome integrity, and nuclear structure [105, 106]. Histone lysine methylation is a ubiquitous PTM and plays a crucial role in the epigenetic regulation of transcription and chromatin dynamics in eukaryotes.
Recent large-scale proteomic studies have revealed significant acylation of mitochondrial proteins that involve the addition of acetyl and succinyl groups. These modifications can occur through enzymatic or non-enzymatic mechanisms [107]. Non-enzymatic acyl protein modifications are likely due to the alkaline nature and high concentrations of acyl-CoA species within the mitochondrial environment (Fig. 1) [108, 109]. Histone non-enzymatic covalent modifications (NECMs) are now recognized as key elements in PTMs that dictate chromatin structure and functionality. NECMs modify histone protein surface topology, thereby influencing their interactions with DNA and chromatin regulators, and also compete with enzymatic PTMs for modification sites [110].
Protein PTMs are pivotal in regulating cell biology and pathological states, with significant implications for health and disease [111]. The various kinds of PTMs are essential cofactors in biochemical reactions within cells and are closely linked to the onset and progression of numerous diseases (Fig. 3). Hence, they present potential strategies and insights for disease therapy.
Pan et al. [112] found that H4K12la positively influences glucose metabolism in the microglia of Alzheimer’s disease, thus offering a new pathway for therapeutic intervention. Nativio et al. [113] reported that excessive increases in H3K9ac and H3K27ac can interfere with chromatin-gene feedback loops to cause transcriptional irregularities that contribute to the onset of Alzheimer’s disease. In the context of Parkinson’s disease, increasing the activity of PTEN induced Kinase 1 (PINK1) enhances the translation of fibrillarin, leading to elevated levels of intracellular CoA and acetyl-CoA (Fig. 3). This process supports mitochondrial autophagy and preserves mitochondrial function [114].
Suppressing the activity of acetyl-CoA carboxylase (ACC) reduces malonyl-CoA production and can effectively diminish de novo lipogenesis (DNL) and curb the proliferation of liver cancer cells [115]. In a study of liver cancer development, Yang et al. [104] reported that lysine lactylation (Kla) affected tumor protein 53 (TP53), catenin beta 1 (CTNNB1), and axis inhibition protein 1 (AXIN1), thereby promoting the proliferation of HCC cells (Fig. 3).
Furthermore, Huang et al. [116] revealed that SIRT6 acts as a tumor
suppressor by regulating the deacetylation of H3K9ac and H3K56ac, thus inhibiting
HCC cell proliferation. Additionally, Zhang et al. [117] found that
metastasis-associated 1 family member 2 (MTA2) can inhibit
In colorectal cancer, Hepatitis B Virus (HBV) pre-S2 trans-regulated protein 3
(HSPC111) phosphorylates ACLY. This facilitates the synthesis of acetyl-CoA,
which then increases the level of histone acetylation to subsequently promote
tumor cell growth and metastasis [118]. The alpha-ketoglutarate dehydrogenase
(
Melanoma cells are specifically dependent on the activity of glutaryl-CoA dehydrogenase (GCDH). Inhibition of GCDH leads to the glutarylation of nuclear factor erythroid 2-related factor 2 (NRF2), resulting in melanoma cell death and the suppression of tumor growth [120]. Under conditions of oxidative stress, succinate-CoA ligase ADP-forming subunit beta (SUCLA2) plays a significant role in mediating the K311 succinylation of glutaminase by regulating succinyl-CoA levels. This metabolic regulatory process results in increased levels of NADPH and glutaminase, and hence the promotion of tumor cell proliferation [121]. The utilization of CoA acylation mechanisms to hinder tumor cell proliferation may therefore be a feasible approach in cancer treatment [122].
Acetyl-CoA and malonyl-CoA can interconvert through the enzymatic actions of acetyl-CoA carboxylase (ACC) and malonyl-CoA decarboxylase (MCD) [123]. In cardiovascular diseases, inhibition of MCD elevates malonyl-CoA levels in the heart, which subsequently reduces the expression of carnitine palmitoyltransferase-1 (CPT-1) and limits mitochondrial fatty acid uptake. This process improves cardiac energy metabolism and presents a novel therapeutic strategy for ischemic heart disease [124, 125].
Using a mouse model, Takada et al. [126] showed that decreased succinyl-CoA levels impair mitochondrial oxidative phosphorylation (OXPHOS) and function, thereby increasing the risk of heart failure. Elevated aldosterone levels during heart failure increase the expression of mineralocorticoid receptor (MR). This in turn raises the blood pressure, which further exacerbates the risk of heart failure [127]. Lysine deacetylases (KDACs), also known as histone deacetylases (HDACs), regulate targeted gene expression via histone deacetylation. HDAC3 is a member of Class I HDACs and exerts deleterious effects on the heart. It facilitates the deacetylation of MR, thereby enhancing its transcriptional activity and leading to elevated blood pressure and the exacerbation of heart failure [128]. Concurrently, MR can activate the innate immune system, which then promotes atherosclerosis [129] (Fig. 3).
Couto et al. [130] found that upregulating the expression of GTP cyclohydrolase 1 (GCH1) can mitigate heart failure by enhancing the bioavailability and sensitivity of nitric oxide (NO) through its interaction with endothelial nitric oxide synthase (eNOS), thus decreasing the level of reactive oxygen species (ROS). Sirtuin 1 is an NAD-dependent class III histone deacetylase that deacetylates and activates eNOS [131]. eNOS stimulates the production of endogenous nitric oxide (NO), which governs blood pressure, smooth muscle relaxation, and vasodilation through peripheral nitrergic nerves [128, 132]. Additionally, SIRT1 plays a pivotal role in regulating vascular remodeling [133] (Fig. 3).
SIRT1 deacetylation activates p65, which in turn inhibits TNF-
The accumulation of acylated proteins impairs mitochondrial function and contributes to the aging process [107]. Mitochondrial lysine deacetylase Sirtuin 3 (Sirt3) can reverse such modifications, offering protection against conditions such as diabetes, obesity, and aging. Thioesterase glyoxalase II (Glo2) acts upstream of Sirt3 to reduce protein N-acetylation. This limits protein S-acetylation and prevents subsequent lysine N-acetylation, which in turn prevents protein dysfunction during acetyl-CoA accumulation [138].
In diabetes mellitus, elevated levels of certain metabolic factors indirectly lead to increased malonyl-CoA content in the body. Hypothalamic accumulation of malonyl-CoA and inhibition of carnitine palmitoyltransferase-1 (CPT1) are linked to reduced food intake in mice. In contrast, decreased levels of hypothalamic malonyl-CoA result in increased food consumption and weight gain. Although the precise mechanisms that underlie the effects of malonyl-CoA remain elusive, it has been suggested they may involve an increased level of long-chain acyl-CoA (LCAC) due to CPT1 inhibition by malonyl-CoA [125]. In type 2 diabetes mellitus (T2D), insulin inhibits lipolysis in white adipose tissue (WAT), leading to reduced acetyl-CoA levels in the liver and decreased activity and flux of pyruvate carboxylase. This mechanism was verified in mice with disrupted insulin signaling and in those lacking adipose triglyceride lipase, underscoring WAT-derived hepatic acetyl-CoA as a critical regulator of hepatic glucose production (HGP) by insulin. This connection also links it to the inflammation-induced hepatic insulin resistance observed in obesity and T2D [139, 140] (Fig. 3).
Individuals with myelodysplastic syndrome and ring sideroblasts (MDS-RS) have mutations in the splicing factor 3B subunit 1 (SF3B1), leading to protein loss and subsequently to reduced CoA and succinyl-CoA levels. This defect impedes erythroid differentiation, affects heme biosynthesis and erythropoiesis, and ultimately contributes to the excessive iron accumulation observed in MDS-RS patients [141].
Coenzyme A (CoA) is a crucial coenzyme that is found extensively in organisms and plays key roles in various metabolic pathways, including those of glucose, lipids, and amino acids [142] (Fig. 1). The sulfhydryl group in the structure of CoA enables the formation of acyl-CoA derivatives with diverse acyl groups (Fig. 2) [143]. These acyl-CoA molecules fulfill numerous physiological functions within cells, including roles in energy metabolism, cellular signaling, and epigenetic regulation [144].
Recent research on acyl-CoA species has moved beyond acetyl-CoA to novel variants such as lactoyl-CoA [101]. The acylation of CoA is intricately linked to two essential physiological functions: cellular heredity and metabolism [62]. The focus is now increasingly shifting towards the acylation of non-histone proteins, which plays a significant role in major biological processes [145]. The complex process of protein PTM through CoA acyl groups is a pivotal mechanism in cellular function that warrants further research. Simultaneously, studies on acyl-CoA are advancing and have enriched our understanding of its many contributions to cellular processes.
Importantly, CoA and its derivatives are deeply implicated in a variety of diseases, including neurodegenerative diseases, cancer, cardiovascular diseases, and metabolic disorders, where they play vital roles in crucial physiological processes within cells [146] (Fig. 3). Continued research into CoA and its thioester derivatives is expected to reveal significant therapeutic targets and strategies for disease diagnosis and treatment.
Ac4C, N4-acetylcytidine; ACC, Acetyl-CoA carboxylase; ACBPs, Acyl-CoA binding proteins; ACLY, ATP-citrate lyase; AD, Alzheimer’s disease; ADP, Adenosine Diphosphate; AGS, Adenocarcinoma Gastric Cell; AMPK, AMP-activated protein kinase; ATP, Adenosine Triphosphate; AXIN1, Axis inhibition protein 1; BDH1,
JX, ZY, YZ, MZ, and YFZ conceived the review. JX and YFZ supervised the review. All authors wrote the draft. JX revised the manuscript. All authors read and agreed to publish the paper. All authors have participated sufficiently in the work to take public responsibility for appropriate portions of the content and agreed to be accountable for all aspects of the work in ensuring that questions related to its accuracy or integrity.
Not applicable.
Not applicable.
This review was supported by Key scientific research projects of Hubei Polytechnic University (23xjz08A).
The authors declare no conflict of interest.
During the preparation of this work, the authors used ChatGpt in order to check for spelling and grammatical errors throughout the text. ChatGPT was also used to refine any incorrect expressions. After using this tool, the authors reviewed and edited the content as needed and took full responsibility for the content of the publication.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.



