1 Department of Histology-Embryology, School of Medicine, Aristotle University of Thessaloniki, 54124 Thessaloniki, Greece
†These authors contributed equally.
Abstract
The neural crest (NC), also known as the “fourth germ layer”, is an embryonic structure with important contributions to multiple tissue and organ systems. Neural crest cells (NCCs) are subjected to epithelial to mesenchymal transition and migrate throughout the embryo until they reach their destinations, where they differentiate into discrete cell types. Specific gene expression enables this precise NCCs delamination and colonization potency in distinct and diverse locations therein. This review aims to summarize the current experimental evidence from multiple species into the NCCs specifier genes that drive this embryo body axes segmentation. Additionally, it attempts to filter further into the genetic background that produces these individual cell subpopulations. Understanding the multifaceted genetic makeup that shapes NC-related embryonic structures will offer valuable insights to researchers studying organogenesis and disease phenotypes arising from dysmorphogenesis.
Keywords
- neural crest cells
- genes
- epithelial-mesenchymal transition
- animal models
- model organisms
- congenital anomalies
The neural crest (NC) is a specific multipotent population of embryonic cells. It arises from each side of the neural plate and specifically from between the neural and non-neural ectoderm [1]. Neural crest cells (NCCs) undergo migration throughout the embryo by a process known as epithelial-mesenchymal transition (EMT) and differentiate into specific types generating a diverse array of cells [2]. NCCs serve as precursors to a wide range of tissues, that are scattered all throughout the embryonic body. They contribute to many structures in the cranial area, most prominently, to the facial and pharyngeal bones and cartilage, while also taking part in dental development and the morphogenesis of sensory placode derived structures [3, 4, 5]. Additionally, NCCs contribute to the shaping of the cardiac outflow tract, while they also give rise to sympathetic and sensory neurons, melanocytes, neuroendocrine cells and contribute to the enteric nervous system.
NC can be divided into several functional domains. In vertebrates, the cephalic or cranial NC originates from the diencephalon to the third somite, playing a role in the formation of bones, cartilage, and connective tissues in the head and neck regions [6]. Additionally, they contribute to the development of pigment cells, localized in the cranial region. Subsequent to cranial NC is the vagal region, a subset of which is the cardiac NC, extending from the otic level to the fourth somite. This region participates in the formation of the heart’s outflow tract [3]. The truncal NC, located caudally to the fourth somite, gives rise to peripheral nervous system (PNS) sympathetic ganglia and enteric ganglia, pigment cells (dorsolateral migration), and the endocrine cells found in the adrenal medulla (ventral migration) [7]. Lastly, the sacral NC is responsible for forming the enteric nervous system (ENS), with a contribution from vagal NC.
Unique groups of distinctly expressed transcription factors and proteins orchestrate the differentiation of cells into specialized forms and functions [8, 9, 10]. In this review, an attempt is made to deleneate the key genes that participate in the acquisition of NCCs’ terminal composition, from rostral-to-caudal region specifiers while presenting miscellaneous derivatives. The data collection aims to present a robust categorization of all the “master genes” implicated in developmental morphogenesis across a plethora of species, with a perspective to address potentional alterations as a causal effect to observed NC-derived malformations.
NCCs interact with one another while on the move, navigating towards specific locations with their target points being scattered throughout the whole embryonic body. Once they reach their intended destinations, they develop into distinct, specialized cell types, morphing later into the distinct organs that we know of [11]. Being divided in cephalic, cardiac, truncal, and enteric NC, the NC plays a role in developing and contributing to a vast range of systems, such as craniofacial and neck structures, the heart’s outflow tract and the enteric nervous system. Here we are going to describe the genes involved in the morphogenesis of these systems.
In the cranial region NCCs play a significant role in forming the majority of the cartilage and bone structures in the skull, contributing in the facial and pharyngeal skeleton [6]. NCCs located at the front of the cranial region contribute significantly to the formation of the frontonasal bones and the membranous bones of the skull. On the other hand, NCCs situated in the posterior part of the cranial region fill in the pharyngeal arches [12]. An interesting scheme that is worth mentioning is the different molecular mechanisms that drive stem cells into entering the cranial rather than the trunk NC family. In particular, ets proto-oncogene 1 (ets-1) is of high importance in the distinction between these two cell fates, as it binds to an enhancer region in the forkhead box d3 (Foxd3) gene, which is a NC marker, and is required for the differentiation into cranial NC [13]. On the other hand, the same enhancer region needs the Zic1 gene in order to form trunk cell progenitors. The same regulatory pattern is observed in sex determining region Y (SRY) - box transcription factor 10 (sox10) gene expression in the cranial region [14, 15]. During the process of delamination in chicken and mice, another crucial difference in proteins that take part is observed, with cranial NC requiring solely sheath interacting protein 1 (Sip1) [7, 13, 16, 17].
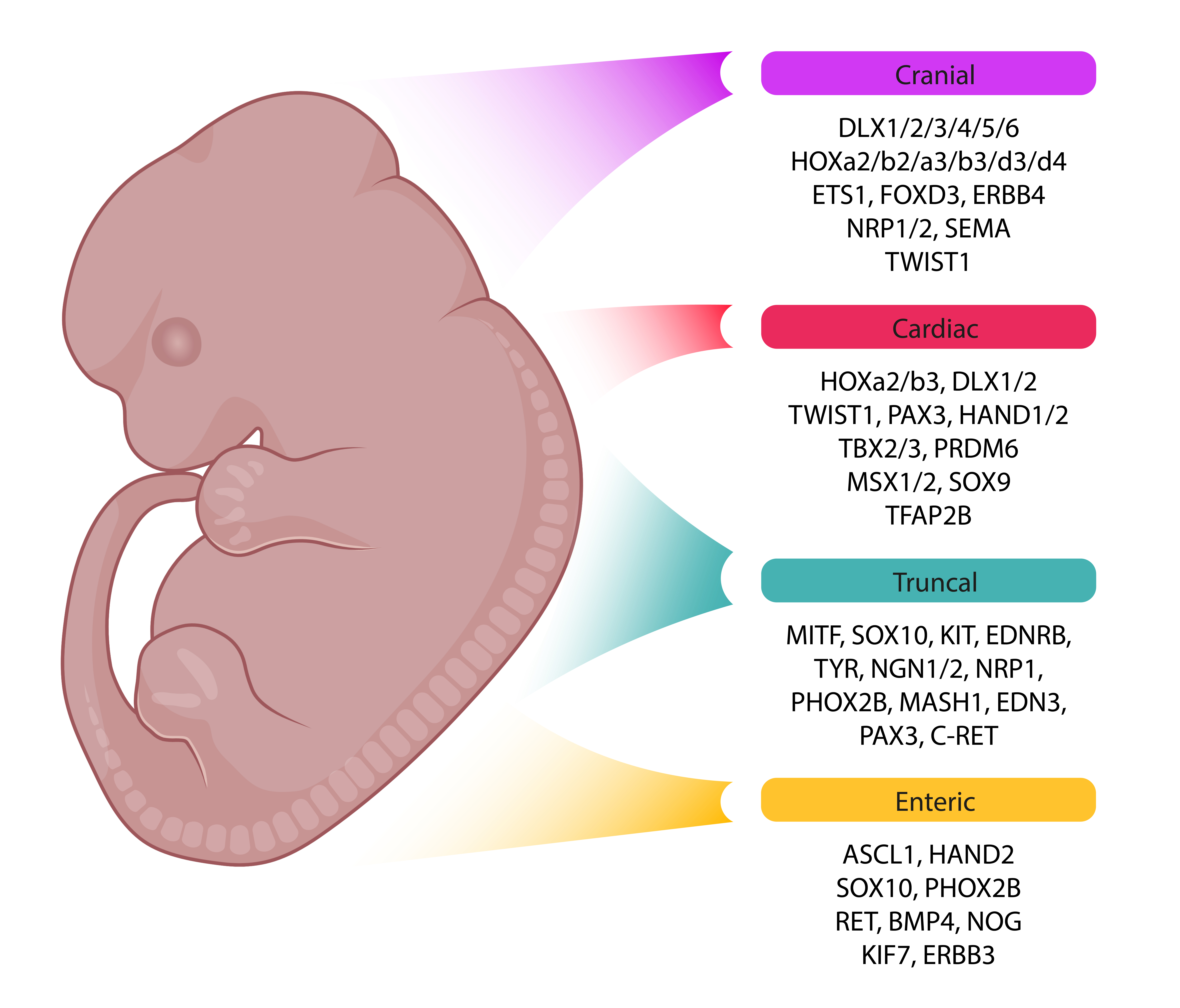
Starting from the hindbrain, the migratory pathways of NCCs are organized into distinct streams, with regions lacking NCCs found laterally of the third and fifth rhombomeres, which are the segmented compartments of the hindbrain (r3/5) [18]. The most common model of migration includes three streams starting from three regions of the hindbrain. In Xenopus, the first stream travels around the optic area and enters the first arch, the second stream enters the second arch, while the third stream extends to both the third and fourth arches [19]. Environmental cues guide these streams along specific routes. Specifically, the mesenchyme adjacent to r3 and r5 inhibits NCC movement, obstructing the entry of NCCs into r3 and r5, partially through the actions of erb-b2 receptor tyrosine kinase 4 (Erbb4), which is expressed in r3 and r5 [20, 21]. Apart from the repulsive interactions between Eph tyrosine kinase receptors and their ephrin ligands (Eph), the repulsive interactions between the transmembrane neuropilin (Nrp) receptors and their secreted semaphorin (Sema) ligands, also contribute to the shaping of the migratory pathways of NCCs [6, 19, 22, 23, 24]. In zebrafish, the receptors nrp2a and nrp2b are present in migrating NCCs heading towards the first, second and third pharyngeal arches (PA1-3). The ligands sema3f and sema3g are found in areas without NCCs, specifically lateral to r3 and r5 [22]. Also, C-X-C motif chemokine receptor 4a (Cxcr4a) is present in these NCCs, while its receptor, stromal cell derived factor 1b (Sdf1b), lies in the endoderm of the pharyngeal arch. In chicks, Sema3a and Sema3f are expressed in regions r1, r3, and r5 [23]. In chicks, migrating NCC streams express EphA3, EphA4, EphA7, EphB1, EphB3, and ephrin B2, while cells surrounding the streams of cranial NCCs express ephrin B1 and EphB2 [25]. In mice, Nrp2 and Nrp1 are present in migrating NCCs targeting PA1-2 and PA2, respectively. Sema3f is present in the caudal midbrain, and both in r3 and r5, while Sema3a is found in r3, where NCCs are absent [24, 26]. In Xenopus, Eph tyrosine kinase receptors and their ephrin ligands, also, play here a crucial role. Genes expressed in migrating cranial NCCs include EphA4 and EphB1. These genes are active in the migrating NCCs of the third arch, and in the third and fourth arches, respectively, as well as in the adjacent mesoderm [19]. The ligand ephrin-B2, which interacts with these receptors, is expressed in the nearby NC and mesoderm of the second arch, even before migration begins. The coordinated expression of EphA4/EphB1 receptors and ephrin-B2 is crucial for limiting the mixing of NCCs from the second and third arches and guiding the third arch NC to its destination. Specifically, EphA4 is expressed in regions r3 and r5 of the Xenopus embryo’s hindbrain, as well as in NCCs adjacent to r5 that are destined for the third arch. NCCs expressing EphA4 migrate into the third branchial arch. In contrast, EphB1 expression is observed in a broader domain compared to EphA4, encompassing NCCs migrating toward both the third and fourth branchial arches, as well as mesoderm [19]. After NC migration, cells with high levels of EphB1 expression are confined to the fourth and more caudal arches [27]. Thus, a distinct boundary exists between the NCCs of the second and third arches, as cells expressing EphA4 and EphB1 are restricted from migrating into the second arch. Similarly, the presence of ephrin-B2 in the mesoderm of the second arch might contribute to proper migration by restricting the entry of third arch NCCs [19]. In Xenopus EphA2 is expressed in r4 and also contributes in the distinction of the third and fourth arch NC streams [28]. The most prominent NC genes are depicted in Fig. 1.
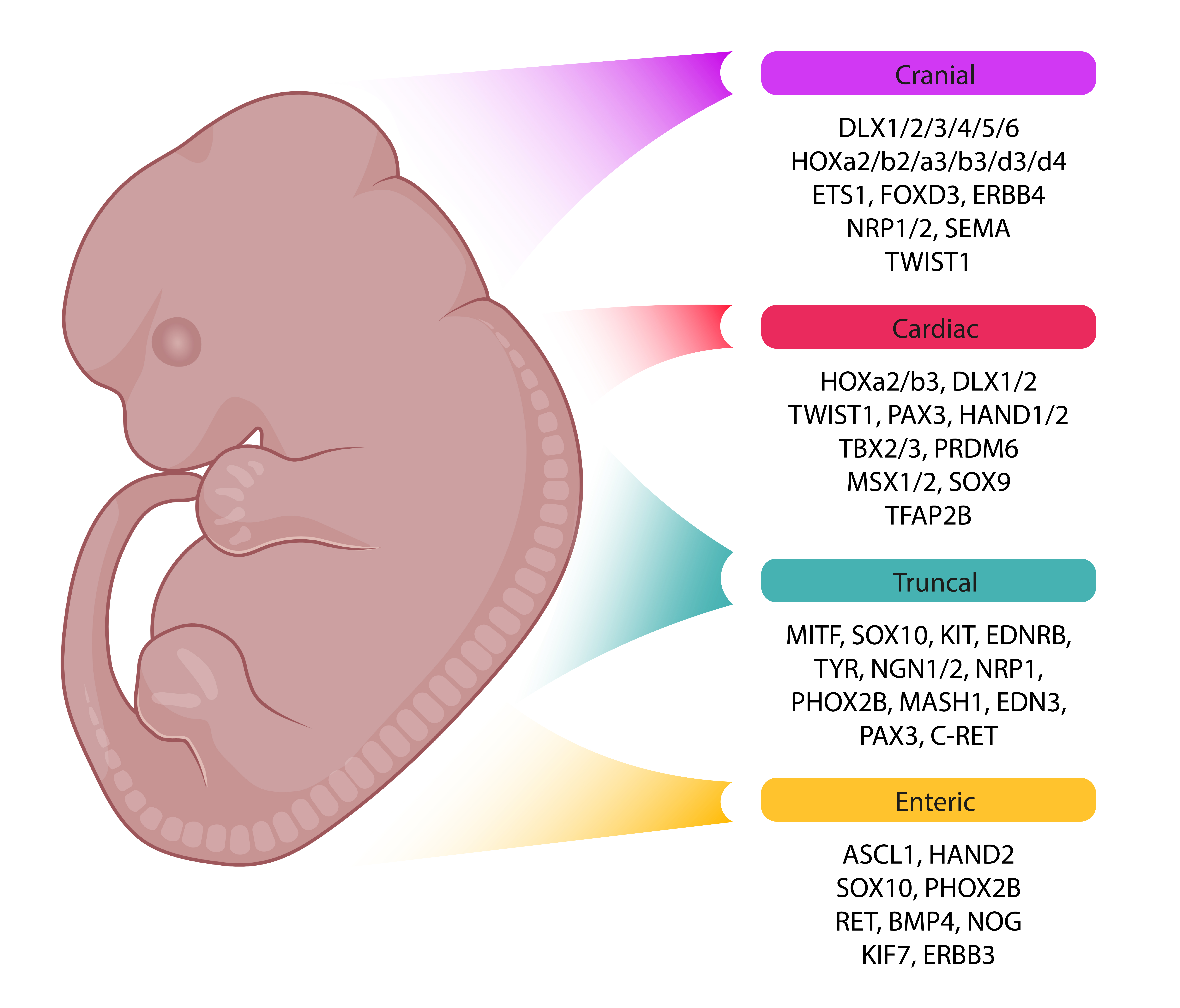 Fig. 1.
Fig. 1.Key genes expressed in NC subdivisions of the vertebrate embryo. Each color depicts a group of genes expressed in the corresponding region; magenta: cranial NCC, red: cardiac (part of vagal) NCC, cyan: truncal NCC, yellow: enteric (part of vagal and sacral) NCC. Abbreviations: NC, neural crest; NCC, neural crest cell; ASCL, achaete-scute family basic helix-loop-helix (bHLH) transcription factor; BMP, bone morphogenetic protein; DLX, distal-less homeobox; EDN, endothelin; EDNR, endothelin receptor; ERBB4, erb-b2 receptor tyrosine kinase 4; ETS 1, ETS proto-oncogene 1; FOX, forkhead box; HAND, heart and neural crest derivatives; HOX, homeobox; KIF, kinesin family member KIT, KIT proto-oncogene; MITF, melanocyte inducing transcription factor; MSX, MSH homeobox; NC, neural crest; NCC, neural crest cell; NGN1/2, neurogenin-1/2; NRG, neuregulin; NRP, neuropilin; PAX, paired box; PHOX, paired like homeobox; PITX, paired like homeodomain; PRDM, PR/SET domain; RET, Ret proto-oncogene; SEMA, semaphorin; SOX, Sex determining region Y-box transcription factor; TBX, T-box transcription factor; TFAP, transcription factor AP; TWIST, twist family bHLH transcription factor.
Notably significant among cranial NCCs in mouse embryos are chondrocytes. The transcription factor that plays a key role in the differentiation of NCCs into chondrocytes is Sox9 [29, 30], which, except for its capacity to regulate its own expression, is strongly affected by WNT signaling in mouse embryos. In accordance with SoxD, it activates a cascade including other genes such as Agc1 and Col2a1. Agc1 encodes a cartilage proteoglycan that is a marker for the differentiation of NCCs into chondrocytes. The procedure of chondrogenesis relies upon Agc1 and Col2a1, which determine the function and morphology of NC derived chondrocytes. Specifically, in the beginning of chondrogenesis, these genes are upregulated in chondrocytes, however, their production is decreased when terminal differentiation is achieved. Regulation of these two marker genes in chondrocytes requires the presence of both Sox9 and Sox5/6. In mesenchymal cells and NCCs that are going to differentiate into chondrocytes, such marker genes are detected in low levels, as Sox5/6 are missing. This prevents Sox9 to bind to chondrocyte enhancers and kickstart their transcription. During the process of differentiation, Sox5/6 are induced and Sox9 can then bind to the enhancer site. This interaction drives crest derived progenitors into acquiring the fate of chondrocytes [31]. On the other hand, an antagonist transcription factor of Sox9 that suppresses its activity is Osr1, which is found extensively in soft tissues [32]. Moreover, in mice Sox9 is present in a number of different precursors of the myoskeletal system including tendon, ligament, bone and cartilage [33, 34]. The determination of these destinies relies on nearby signaling molecules and transcription factors, such as such as scleracxis, osterix and Runx2 [35, 36]. Initially, osteoblastic precursors separate from the Sox9 group, followed by the differentiation of tendocytes and ligamentocytes [36]. On the other hand, skeletal muscle development requires other genes and transcription factors [37, 38, 39, 40].
In human development, prechondrocytes originating from NCCs, differentiate into chondrocytes and then secrete matrix molecules that will finally form cartilage. Between prechondrocytes and chondrocytes different marker genes seem to be expressed, revealing the transcriptional change that takes place during the process of embryonic chondrogenesis. Particularly, in the initial stages of development genes like SOX5/6/9, forkhead box P4 (FOXP4), NKX 3-2, GDF5, PCDH8/10, PTCH1 and bone morphogenetic protein receptor 1 beta (BMPR1B) are transcriptionally active [41, 42, 43]. These genes have, also, been detected in mice and chick embryos as well. As prechondrocytes matured into periarticular chondrocytes, there was a gradual reduction in the expression of various mesenchymal genes such as NCAM1 (encodes CD56), MCAM (encodes CD146), CDH2 (encodes N-cadherin), CD24, and, to a smaller degree, ALCAM. From these molecules, CD146 is implicated in the process of EMT during early stages of embryonic development, as it contributes to the invasiveness and migration of NCCs. CD166 is another gene that serves the role of a surface marker mostly in the perichondrium. Accordingly with CD146, it enhances both the invasive and migratory character of mesenchymal cells [44, 45, 46]. Thus, CD146 and CD166 are gradually decreased as prechondrocytes differentiate into chondrocytes that are immotile terminal differentiated cells. In resting immature chondrocytes at later stages of chondrogenesis, bone morphogenetic protein receptor 1 beta (BMPR1B) is highly expressed in the cells on the surface of articular cartilage, along with leukemia inhibitory factor receptor subunit alpha (LIFR). LIFR is thought to preserve the superficial progenitors of cartilage. These receptors are detected in growth plates after birth, but only for the period that the process of chondrogenesis is active. In mice, BMPR1B is required for chondrogenesis. In terminal differentiated chondrocytes transforming growth factor b1 and 2 (TGF-b1, TGF-b2) and leukemia inhibitory factor (LIF) are highly expressed [47].
In the context of frontal bone development, the ectomesenchyme originating from
cranial NCCs is the exclusive source responsible for populating the initial
structure of the frontal bone and its underlying dura mater [48]. During this
process, TGF plays a vital role as a major factor, promoting the proliferation of
NCCs in both the orbital and lateral aspects of the developing frontal bone, even
though TGF receptor type II (Tgfbr2) is not required for the survival of
osteoblast progenitors [49]. Within the cranial NC-derived ectomesenchyme and
cranial sutures, the presence of Tgfbr2 has been identified, demonstrating its
significant involvement in human skull development [50]. TGF
During the early stages of craniofacial development, the Fibronectin Leucine-Rich Transmembrane (Flrt) gene family proteins are crucial for NC migration and craniofacial morphogenesis. In mouse embryos, Flrt2 is initially highly active in the cranial NCCs, while Flrt3 exhibits heightened expression in the midbrain with an anterior boundary at the border between midbrain and forebrain [58]. Subsequently, both genes are expressed in the developing pharyngeal region. Specifically, Flrt2 is expressed in the NC-derived mesenchyme located medially in the developing frontonasal region, closely associated with the expression of Fgfr2 [59]. Additionally, Flrt2 expression is present in the vomero-nasal organ, mandibular primordia, and the posterior sections of the secondary palatal shelves. In contrast, Flrt3 expression was more limited, observed in the mesenchyme beneath the ectoderm of the medial nasal process and beneath the mandibular primordium, with its expression being aligned with the expression of Bmp4 [60].
The terminal position of cranial NCCs is largely determined in pre-migratory cells by genes belonging in the homeobox (HOX) family. This variety of genes has been excessively studied in Drosophila. A gradient of the expression of these genes is present along the anterior-posterior axis in the pharyngeal arches [61, 62]. First of all, the most commonly expressed, homeobox a2 (Hoxa2), is active in a very broad region. This region stops at the border of the first and second rhombomeres (r1 and r2). However, NCCs originating from r2 and moving into the first pharyngeal arch (PA1) do not possess any active Hox genes, as in PA1 Hox genes are not expressed [6]. As a result, the mandibular process has a morphogenetic programme characterized by the absence of these genes’ expression. Moving downwards, Hoxa2 is active in PA2 along with Hoxb2 in a smaller degree, while in PA3 and PA4, Hoxa3/b3/d3 expression is present, with Hoxd4 expression appearing in the last of the pharyngeal arches [6]. When it comes to the dorsal-ventral axis, the latter is organized by distal-less homeobox (Dlx) genes, which are six in mammals. In PA1, Dlx1 and Dlx2 are active in both the upper (maxillary) and lower (mandibular) jaw structures, covering the area closer to the dorsal part of the embryo. Dlx5 and Dlx6 genes, however, are specifically active in the lower jaw and their expression extends near the future joint area between the maxilla and the mandible [55, 63]. The domains where Dlx3 and Dlx4 genes are expressed are even more restricted, limited to the farthest tip of the lower jaw structure, inhabiting the ventral region of the pharyngeal arches. In the first pharyngeal arch, Dlx5/6 stimulate and/or sustain the activity of various genes crucial for the formation of the lower jaw structure. These genes include Dlx3/4, heart and neural crest derivatives 1/2 (Hand1/2), Alx homeobox 3/4 (Alx3/4), paired like homeodomain 1 (Pitx1), gastrulation brain homeobox 2 (Gbx2), and Bmp7 and Evf2 [6, 64]. Crucial genes are listed in Table 1 (Ref. [6, 13, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 47, 55, 56, 57, 58, 59, 60, 63, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97]).
| NC subdivisions | Genes | Species they are studied in | References |
| Cranial | EPHA4/EPHB1 | Chicken, Xenopus | [19, 25, 27, 28] |
| DLX1/2/3/4/5/6 | Chicken, Mouse | [55, 56, 63] | |
| HOXa2/b2/a3/b3/d3/d4 | Chicken, Mouse, Zebrafish, Xenopus | [6] | |
| ETS1 | Chicken | [13] | |
| FOXD3 | Chicken | [13] | |
| ERBB4 | Mouse | [20, 21] | |
| NRP1/2 | Mouse, Zebrafish | [22, 24, 26] | |
| SEMA | Chicken, Mouse, Zebrafish | [22, 23, 24, 26] | |
| TWIST1 | Mouse | [57] | |
| FLRT2/3 | Mouse | [58, 59, 60] | |
| Cardiac | HOXa2/b3 | Mouse | [6, 65] |
| DLX1/2 | Mouse | [6, 65] | |
| TWIST1 | Mouse | [6, 65] | |
| PAX3 | Mouse | [66, 67, 68, 69] | |
| HAND1/2 | Mouse | [6, 65] | |
| TBX2/3 | Mouse | [65, 70] | |
| PRDM6 | Mouse | [71, 72] | |
| MSX1/2 | Mouse | [6, 65, 68, 69] | |
| SOX9 | Mouse | [71, 72] | |
| TFAP2B | Mouse | [71, 72] | |
| Truncal | EDNRB | Mouse | [73] |
| EDN3 | Mouse | [47] | |
| PAX3 | Mouse | [74, 75] | |
| SOX10 | Mouse | [74] | |
| C-RET | Mouse | [74] | |
| PHOX2B | Mouse | [76] | |
| MASH1 | Mouse | [77] | |
| NGN1/2 | Mouse, Xenopus | [78, 79, 80, 81, 82] | |
| NRP1 | Mouse | [79, 83, 84] | |
| MITF | Mouse, Zebrafish | [85, 86] | |
| SOX10 | Mouse, Zebrafish | [85, 86, 87] | |
| KIT | Mouse, Zebrafish | [88, 89] | |
| EDNRB | Mouse | [90] | |
| TYR | Mouse, Zebrafish | [85, 91] | |
| Enteric | ASCL1 | Mouse | [92] |
| HAND2 | Mouse | [93] | |
| SOX10 | Mouse | [93] | |
| PHOX2B | Mouse | [93] | |
| RET | Mouse | [93] | |
| BMP4 | Mouse | [94] | |
| NOG | Mouse | [95] | |
| KIF7 | Mouse | [96] | |
| ERBB3 | Chicken, Mouse | [78, 97] |
An integral aspect of the cranial NCC, is its contribution in the development of the sensory placodes, such as the eye and the ear. These structures are formed from pre-placodal ectoderm, thickenings of the ectoderm in the area of the head, which collaborate with the NC to produce the cranial sensory part of the nervous system [98, 99]. Before NC migration begins, both the NCCs and placodes are situated in neighboring areas of the lateral ectoderm. The NCCs are found on both sides of the neural plate, while the placodes partially envelop both the neural plate and the NC [100]. In Xenopus laevis, NCCs go through EMT and display collective cell migration. Meanwhile, placodes positioned in the lateral ectoderm release C-X-C motif chemokine ligand 12 (CXCL12), a widely recognized chemoattractant [101, 102]. NCCs, which express Cxcr4, the primary CXCL12 receptor, migrate towards the latero-ventral regions due to chemotaxis guided by its ligand. CXCL12 activates Rac1, which plays a crucial role in the extension of cellular protrusions and the formation of focal adhesions in cephalic NCCs, and thus enhances attachment between cells and the extracellular matrix [101, 103]. In this whole interaction, matrix metalloproteinase MMP14, which is expressed in Xenopus NCCs [104], might also play a crucial part, through its effect on cleaving the matrix fibronectin [103].
2.1.1.1 Eye
The NC vastly contributes to several structures of the eye, such as the optic cup, lens, ciliary body, iris, cornea, trabecular meshwork, and the primary vitreous [105, 106]. A specific group of cranial NCCs migrates and populates the periocular mesenchyme participating in the formation of the eye [107]. TGF signaling aids the NCCs in their migration, guiding them towards the periocular mesenchyme, and controls ocular development. Regulated by TGF signaling, is the expression of Pitx2 and forkhead box C1 (FoxC1), which produce transcription factors [108, 109]. PITX2 contribution to eye development regards the morphogenesis of structures of the anterior segment of the eye. In chick embryos, Sema3A present in the lens placode restricts the movement of neuropilin 1 expressing NCCs, preventing their migration into the periocular region. This restriction continues until a particular subset of NCCs decreases their Nrp1 expression [110]. Moreover, paired box 6 (PAX6) is one of the necessary genes required to initiate human eye formation and lens differentiation [111]. Its homologous gene eyeless in Drosophila can alone initiate eye development [112]. Furthermore, it has been shown that in zebrafish embryos that two separate NC streams enter the optic cup with pax6 being a necessary guide for their proper migration.
2.1.1.2 Ear
The embryological origin of the ear can be traced back to all three germ layers. The outer ear is formed from the ectoderm, the middle ear is derived from the mesenchyme of the first and second pharyngeal arches, while the inner ear develops from the vesicles of the otic placodes. NCCs originating from the hindbrain and populating the mandibular area of the first pharyngeal arch form Meckel’s cartilage, as well as the middle ear bones incus and malleus [113, 114]. NCCs are also involved in the morphogenesis of the inner ear by contributing to the cochleovestibular ganglion. During the formation and growth of the inner ear, NCCs migrate from r4 to the otocyst, where they undergo specialization into glial cells of the cochleovestibular ganglion and intermediate melanocytic cells of the cochlear stria vascularis. This process is greatly dependent on SOX10, mutations of which cause Waardenburg syndrome, in which deafness is present [115, 116]. Moving into PA2, NCCs form the middle ear bone stapes and contribute to a section of the hyoid bone. Precise gene expression domains are essential for the development of these bones. In mice, in PA1, no Hox family gene is expressed. Additionally in the area where the incus is formed, Dlx1/2 expression is present, whereas in the area where the malleus is formed Dlx1/2, Dlx5/6 are expressed. In the PA2 derivatives, where Hoxa2 is the only member of its family to be expressed in cranial NCCs [117], the formation of the stapes depends on the expression of Hoxa2 as well as the expression of Dlx1/2. Hoxa2 is also a defining factor in the external ear formation [6]. A known regulatory factor that interacts with Hoxa2 is the super enhancer region called HIRE1/2, which mediates cell interactions that occur selectively in cranial NCCs from PA2, but not in those from PA1 [117].
When it comes to the formation of teeth, the latter are formed by the dental lamina, an ectoderm thickening situated in the first PA. The mechanism of the dental formation in mammals is well studied as part of the morphogenesis of the first PA. Cranial NC cells play a crucial role in the formation of mammalian teeth. They serve as the exclusive source of mesenchyme capable of supporting tooth development [5]. These cells not only contribute to most dental tissues but also form the periodontium. The process of tooth development is carefully orchestrated through interactions between cranial NCCs and the oral epithelium [118]. NCC identity regarding dental pattern specification is most likely not determined before the NCCs reach the mandible mesenchyme, but it is determined by their position and the secreted signals of the mandible [119]. The first pharyngeal arch is characterized by the absence of Hox family gene expression [6]. Although mammalian tooth development is confined to specific regions, namely the first arch and frontonasal process, where Hox genes are not expressed, the formation of dental organs in mice appears to occur independently of the Hox patterning program [120, 121]. As an example, Hoxa2 overexpression in the area does not disturb the normal formation of the teeth [121]. However, different genes are expressed in each specific area of the mandible. Ectomesenchymal cells in different areas exhibit unique developmental capabilities and will give rise to different types of teeth, thus the different gene patterns [119]. In the proximal molar mesenchyme, Barx homeobox 1 (Barx1) and Dlx1/2 genes are active, whereas in the distal incisal mesenchyme, MSH homeobox 1/2 (Msx1/2) and Alx4 are expressed. In the former, Barx1, Dlx1 and Dlx2 orchestrate the formation of molars, while in the latter, Msx1, Msx2, and Alx4 contribute to the development of incisor morphology [5]. The patterning of the mandible mesenchyme from proximal to distal mirrors the patterning of the overlying oral epithelium. Proximal mesenchymal markers are stimulated by Fgf8, which is secreted by the proximal epithelium [122], while distal mesenchymal markers are induced by Bmp4, secreted by the distal epithelium [123]. Along the rostrocaudal axis, LIM homeobox 7 (Lhx7) expression in the rostral mesenchyme, regulated by Fgf8, is complementary to Goosecoid (Gsc) expression in the caudal mesenchyme.
NCCs originating from the occipital region of the embryo belong in a distinct category, called the cardiac NC. This specific subpopulation initiates its migration with the purpose of forming the septum of the cardiac outflow tract and constructing parts of the aortic arches [3]. Along their route, these cells are observed moving from the occipital neural tube, invading the third, forth and sixth branchial arch and later settling into the aortic sac and aortopulmonary outflow tract [124, 125]. They exhibit characteristics of both cranial and trunk NCCs, thus the need for their classification in a separate category. Some basic genes expressed in NCCs, while they still reside at the pharyngeal arches, are common with genes expressed in cranial NCCs and include Hoxa2, Hoxb3, Dlx1, Dlx2 and Twist1 [6, 65]. NCCs destined to differentiate into cardiac NCCs express Tbx2 and Tbx3, which are T-box genes taking part in cardiac development. Later, when these cells arrive at the heart to create its outflow tract, they express not only Tbx2/3, but also Hand1/2, Msx1/2, Gata3, forkhead box f1 (Foxf1) and Isl1/2. At an even later point, when cardiac NNCs form the outflow tract and the pharyngeal arch arteries by transitioning into smooth muscle cells, smooth muscle cell genes such as Acta2, transgelin (Tagln), myosin light chain 9 (Myl9), myosin heavy chain (Myh9), Rgs5 and Cnn1 are expressed [65]. Apart from T-box transcription factor genes 2 and 3 (Tbx2/3), the gene Tbx1, which also produces a T-box transcription factor, shows minimal expression in cardiac NCCs. However, it exhibits robust expression in nearby cells of the pharyngeal arches and is crucial for the proliferation and terminal differentiation of cardiac NCCs [70]. A transmembrane receptor known as Roundabout (Robo) is present on cardiac crest cells and is crucial for the final stage of their migration, as it binds with its Slit ligand, a glycoprotein released into the extracellular space. Slit serves as a navigational signal, helping cardiac NCCs reach their destination [126]. When genes involved in Slit-Robo signaling are mutated, valve abnormalities and septal defects arise [126].
One of the most prominent genes expressed in the cardiac NC is Pax3. In mouse embryos, Pax3 expression is expressed in early NC progenitors and is a marker for cardiac NCCs and essential to their migration to the developing heart [66]. In wildtype mouse embryos, Pax3 expression is at first undetectable, but it became significantly strong during the point when cardiac NCCs enter the cardiac outflow tract, and then, subsequently, the expression decreased. Cardiac NCCs invade the developing heart around that point of Pax3 expression. As a result, it can be hypothesized that cardiac NCCs in mice express Pax3 at least to the point where they enter the cardiac outflow tract [67]. Downstream of Pax3, Msx2 is expressed and its repression is regulated by Pax3. However, the related gene Msx1, which has been found active in other tissues, has not been found to be expressed in cardiac NCCs [68]. Other cardiac NC markers are the genes Hoxa-3, CrabpI, Prx1, Prx2, and c-met which were found to be expressed at cells moving through the third, fourth and sixth branchial arches and acquiring their final position in the aortic sac and the cardiac outflow tract [127]. However, all these genes are not expressed at the same level or at the same time points throughout the cardiac NCCs and some of them might also be associated with other types of NC that dwell on localized areas. In more detail, Hoxa-3 and Prx1 are expressed at the first and second branchial arches [128], CrabpI is expressed only at the first arch at the same time, while Pax3 is not expressed at all at the first and second branchial arch [69].
Semaphorin signaling is also a recurring factor for cardiac morphogenesis,
making its first appearance already from the post delamination stage for cardiac
NCCs, as it helps them to find their way from the dorsal aorta to the development
area of the embryonic heart and to navigate through the pharyngeal arches [129].
It also is required for cardiac outflow tract septation. In mice, plexinA2
is present in cardiac NCCs as they leave the neural tube. This gene is expressed
in the NC, while it still resides in the neural tube, in cells giving rise to the
outflow tract of the heart and the aortic arches and also along the migration
path connecting the two [130]. Neuropilin 1, which is present in cardiac NCCs
migrating, Neuropilin 2 and other plexin genes have been implicated in being
active in the cardiac NC. However, their additional expression in tissues
surrounding it, prevents us from reaching a definitive conclusion as to if they
can be characterized as marker genes for the cardiac NC [129]. Sema3C and
plexinA2, which are present in the roof plate of the neural tube and the
compacted mesenchyme of the cardiac outflow endocardial cushions, serve as an
appealing directional cue for cardiac crest migration and is found in the cardiac
outflow tract [129]. Moreover, necessary signals spanning from the early to
the final steps of cardiogenesis are provided and mediated by bone morphogenetic
proteins (BMPs) of the TGF family and specifically BMP1, BMP2, BMP4 and BMP7
[131, 132, 133, 134]. When migration is completed, cardiac NCCs come together and condense
to shape the aorticopulmonary septum. In mice, the TGF
A notable marker and morphogenetic gene is PR/SET domain gene 6 (PRDM6). The latter functions as a histone methyltransferase in smooth muscle cells, controlling histone modification and gene expression, acting as a transcriptional repressor and enabling other genes [71]. It plays a crucial role in cardiac development, particularly in the migration of cardiac NCCs pre and post EMT. H4K20 monomethylation, a modification mediated by PRDM6, is essential for cell cycle progression and NCC proliferation. In mice, the expression of PRDM6 is notable in specific cardiac regions like the cardiac outflow tract and the ductus arteriosus, where the smooth muscle cells are derived from cardiac NCCs [71, 72]. In cardiac NCCs, PRDM6 is vital for the closure of the ductus arteriosus and the formation of compacted myocardium. It acts as a regulator for various genes involved in cardiac NCC specification, including Wnt1, transcription factor AP 2b gene (Tfap2b), and Sox9. Prdm6 serves as a central hub and coordinates the detachment, migration, and differentiation of specific cardiac NCCs, a process dependent on the Wnt1 pathway. Prdm6 maintains the balance between Bpm4 and Wnt1 signals allowing the NCCs to leave the neural tube and progress through their cell cycle. Downstream of Wnt1, Tfap2b and Sox9, which are NC specifiers, are positively regulated by Prdm6 and in turn cause the expression of contractile proteins Myh11 and Tagln, which are crucial for differentiation into vSMC [71]. Lastly, some other examples of genes that are typically active in mice premigratory or migrating cardiac NCCs, as well as in tissues located near the pathways of migration are endothelin receptor A (EdnrA), neurofibromin 1 (Nf1), Cx43, Foxc1 and Foxc2 [137, 138, 139]. The last two genes belong to the forkhead family transcription factors and are implicated in mice in the morphogenesis of the arch arteries, the outflow tract and the second heart field [140].
The trunk NCCs originate in the lower part of the embryo and migrate through three distinct routes: a dorsolateral pathway between the ectoderm of the embryo and the somites, a ventrolateral pathway between the somites and a ventromedial pathway between the neural tube and the posterior sclerotome [141]. The trunk NC contributes to the formation of the peripheral nervous system, including the sympathoadrenal system and the enteric ganglia, while also giving rise to melanocytes. The most important genes expressed in the truncal NC derivatives are listed in Table 2 (Ref. [47, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 142, 143]).
| Truncal derivatives | Genes | Species they are studied in | References |
| Sympathoadrenal | NGN1/2 | Mouse, Xenopus | [78, 79, 80, 81, 82] |
| NRP1 | Mouse | [79, 83, 84] | |
| PHOX2B | Chicken, Mouse | [76, 142, 143] | |
| MASH1 | Chicken, Mouse | [142, 143] | |
| Enteric ganglia | EDNRB | Mouse | [73] |
| EDN3 | Mouse | [47] | |
| PAX3 | Mouse | [74, 75] | |
| SOX10 | Mouse | [74] | |
| C-RET | Mouse | [74] | |
| PHOX2B | Mouse | [76] | |
| MASH1 | Mouse | [77] | |
| Melanocytes | MITF | Mouse, Zebrafish | [85, 86] |
| SOX10 | Mouse, Zebrafish | [85, 86, 87] | |
| KIT | Mouse, Zebrafish | [88, 89] | |
| EDNRB | Mouse | [90] | |
| TYR | Mouse, Zebrafish | [85, 91] |
The NC gives rise to various neuronal lineages, from the sympathoadrenal system to the enteric ganglia. These neural derivatives of the NC share common expression of some gene markers and transcription factors. As far as the formation of glial and neuronal products is concerned, a majority of genes and signaling cascades seem to be involved. Apart from its significant role in melanocyte formation, SOX10 is a crucial gene that is responsible for the differentiation of NCCs into neuronal and glial progenitors [144, 145]. This can be proved by noting the implications that follow mutations in SOX10 gene, which can lead to tumors with neural origin, like schwannomas or malignant peripheral nerve sheath tumors [146]. MEFC2, another gene that is the direct transcriptional target of SOX10, seems to play a significant role in neuronal development in humans, with its mutations being associated with implications including epilepsy, seizures and intellectual disability [147]. Additionally, the signaling molecule neuregulin determines the fate of peripheral glia while BMP is responsible for differentiation of NCCs into sympathetic neurons [148].
In mice embryos, much discussion has been raised about the distinction between sympathetic and parasympathetic ganglia. The development of noradrenergic characteristics in sympathetic precursor cells begins once they have migrated near the dorsal aorta and is defined by the production and secretion of tyrosine hydroxylase (TH), an enzyme required in catecholamine synthesis [149]. The initiation of sympathetic neuron differentiation is characterized by BMPs production [150], then followed by the induction of multiple transcription factors such as paired like homeobox 2b (Phox2b), Phox2a, Insm1, achaete-scute family bHLH transcription factor (Ascl) 1, Gata2/3, Hand2, Sox4 and Sox11 [151]. Research in chick embryos unveiled a step-by-step initiation of gene activity, commencing with Phox2b and Ascl1, succeeded by Hand2, Phox2a, and Gata2 and last but not least genes that are specific for certain cell subtypes [152]. In the process of parasympathetic ganglia formation, which also depends on Ascl1 and Phox2b expression [153], most cells adopt a cholinergic identity, denoted by the presence of choline acetyltransferase and vesicular acetylcholine transporter. Essential for the acquisition of noradrenergic or cholinergic fate is the homeobox transcription factor HMX1. In adrenergic neurons, it conserves the level of TH through the suppression of Tlx3 and Ret and activation of Trk1. The opposite cascade that starts with Hmx1 repression is responsible for the differentiation of cholinergic neurons [154].
2.3.1.1 Sympathoadrenal
The autonomic nervous system consists of chromaffin cells found in the adrenal medulla, the sympathetic chain, the organ of Zuckerkandl, and the rest of the prevertebral ganglia. This system is made up of both cranial and trunk NCCs. The former originate above somite five and give rise to structures like sensory, sympathetic, and possibly parasympathetic ganglia, while the latter are formed under somite four and contribute to the development of sympathetic and sensory neurons as well as adrenal chromaffin cells. In particular, common precursor cells give rise to both sympathetic neurons and chromaffin cells of the medulla, while the differentiation between these two cell types happens after delamination [155]. Along with the formation of the sympathoadrenal system, also occurs the formation of melanocytes, with the only difference between them being that melanocyte precursors follow a dorsolateral path when leaving their home site. On the contrary, the other group of NCCs that generates chromaffin cells in the adrenal medulla and cells in the sympathetic and sensory nervous systems migrates ventrally through the intersomitic space or within the anterior sclerotome [156]. The formation of sympathetic ganglia, sensory ganglia, and melanocytes mainly occurs in a sequential manner, meaning that delamination and migration time point plays a substantial part in determining the fate of these common precursors [157]. In greater detail, early migrating NCCs are primarily destined for neuronal and glial roles, whereas NCCs that migrate later tend to become melanocytes. The sympathoadrenal (SA) NCCs are a distinct population of NCCs, deriving between the 18th and 24th somite in most species gathering around the large vessels of the abdomen, where they take their final ganglial form, with a subset of them venturing further down to construct the adrenal medulla and acquiring chromaffin cell characteristics [7, 151]. During their descent, in chickens, the Eph/ephrin family plays a role in promoting ventral migration [158], while in mice, Sema3A/Nrp1 signaling serves a similar function, meaning that these two pairs of ligands and receptors direct the path that SA NCCs will follow. NCCs that will form the sympathetic ganglia express Nrp1 but not Nrp2, that seems to be distinctive of cells belonging to the sensory nervous system [79, 83, 84]. Cells that will become sensory neurons of the dorsal root ganglia (DRGs) express in premigratory level and during their ventral migration neurogenin-1/2 (Ngn1/2), which direct them towards this phenotype [79, 80, 81, 82]. Additionally, ErbB/Neuregulin signaling pathway is important, apart from its role in migration of trunk NCCs, for their differentiation into DRG as well as sympathetic ganglia [78].
The anatomical formation most involved in the procedure of SA cells’
differentiation, is the dorsal aorta. This vesicle acts as factory responsible
for the production of BMPs, including BMP2, BMP4 and BMP7 which are usually
secreted by endothelial cells of the dorsal aorta in chicks [142]. Due to these
molecules, the expression of Phox2a/b, Mash/Cash1,
Insm, Hand2 and GATA2/3 is increased. The BMPs are
also essential for the activation of SDF1 and neuregulin 1
(NRG1) in the mesenchyme surrounding the aorta, which also are
attractive ques, subsequently influencing the direct control of SA cell
morphological development. Afterwards, SDF1 and NRG1 function as chemical signals
that attract SA precursors, which directionally migrate towards those secretion
sources [148, 155, 159]. In chick embryos specifically, in some of the ventrally
migrating NCCs heading towards the dorsal aorta the receptor CXCR4 is present.
This guides them towards SDF1 rich areas [160, 161]. AP2 transcription factors
also characterize the ventrally migrating NCCs. However, a specific link between
their expression and the survival of sympathoadrenal NCCs has not been described
[79]. The receptor that is necessary for the binding of BMP molecules is Alk3,
which is present in premigratory NCCs. The presence of the transcription factor
Phox2b is crucial for the sustenance of sympathetic precursors. Its expression is
triggered by BMP signaling and is regulated by Alk3. WNT signaling is
another regulatory mechanism that takes part in the distinction between sensory
and sympathetic neurons. Specifically, canonical WNT signaling, through direct
activation of
2.3.1.2 Enteric Ganglia
The enteric ganglia is subset of the ENS, that is derived from truncal, sacral and vagal NC. A trio of genes, Pax3, Sox10, and c-RET, plays a vital role in the development of the NC in the intestine and the enteric ganglia in particular. Precursor cells that express Pax3 are fated to form enteric ganglia, a morphogenesis event that Pax3 is crucial for [74]. It is typically present in NCCs before they migrate, but its expression decreases as the cells migrate [66]. This gene can activate the expression of c-RET, the second gene in this triad. Pax3 collaborates with the transcription factor Sox10 to initiate the transcription of c-RET. In the context of enteric ganglia development in the intestine, the c-RET tyrosine kinase receptor is indispensable, as it is required for their viability and multiplication [75]. Pax3 and Sox10 together significantly increase endogenous c-RET expression. For their synergistic activation a specific 45-bp element within the c-RET enhancer is required. This enhancer contains adjacent binding sites for Pax3 and Sox10 [74]. All enteric ganglia precursors also express the c-RET receptor, necessary for their survival and proliferation, which binds with its ligand, glial cell-derived neurotrophic factor (GDNF) [163, 164]. The latter is produced by surrounding mesenchymal cells of the intestine. Apart from Sox10, several other transcription factors, such as Phox2b [76], and Mash1 [77], regulate various aspects of enteric NC differentiation and proliferation. On the other hand, connexin 43 (Cx43) promoter directs expression in specific regions, including the dorsal neural tube, dorsal root ganglia, cardiac NC, and other NC-derived tissues [165]. The endothelin B receptor (EDNRB) and endothelin 3 (EDN3) are also essential for distal hindgut ganglia formation [166]. Ednrb is typically expressed in enteric ganglia and surrounding mesenchyme [73].
This cell population is formed mostly by differentiation of truncal NCCs, through a process mediated by a vast number of different genes and signaling cascades. Melanocytes derived from the trunk NC will migrate towards most parts of the embryonic body. In mouse embryos, a transcription factor that is specific and indicative of melanocyte progenitors is melanocyte inducing transcription factor gene (Mitf) [85]. This marker gene is responsible for the survival of melanocyte progenitors, by regulating antiapoptotic factors like Bcl12. It also contributes to cell multiplication, by controlling INK4A/p16, p21, Tbx2, and cyclin-dependent kinase (CDK) 2, and cell differentiation. Tha latter is achieved through its effect on activating genes such as Aim-1, Dct, Tyrosinase related protein 1 (TYRP1), Tyrosinase (TYR), Mart1, Silver/Pmel17 and Mc1r [167]. In zebrafish embryos, nacre is a counterpart of mitf. This molecule is responsible for the production of a transcription factor which controls the transcription of genes involved in the pigmentation process, like trp-1. The referred regulation is achieved through WNT signaling [108]. Considering the major contribution of Mitf in melanocyte development, it has to be mentioned that additional transcription factors are required for the full melanocyte differentiation. Specifically, in mice Sox10 is necessary for the expression of Dct/Trp2, whose transcription product is required for the proper synthesis of eumelanin. Eumelanin is the basic type of melanin, produced by tyrosine through the process of melanogenesis. The interaction of Sox10 with Dct/Trp2 can be both direct and indirect, through the mediation of Mitf [91, 168]. However, recent examination of this pathway in zebrafish proposes a different scenario, indicating that sox10 might actually have a negative effect on the expression of melanogenic enzymes. In this hypothesis mitf acts as a repressor of sox10 [87]. In Zebrafish the distinct role of sox10 is the regulation of mitf in a linear way, without affecting the transcription of other genes. The direct correlation between SOX10 and the activation of MITF can be noted in human crest cells too [86]. It can be easily concluded that mutations in SOX10 are directly associated with dysregulated melanocytic differentiation, leading in neuroectodermal tumors, like melanoma [4]. Pax3 is another important gene involved in the pigmentation procedure in ascidian embryos. Firstly it increases the survival rate of melanocyte precursors and secondly it activates Mitf promoter along with Sox10. However, the hypothesis of the immediate control of Mitf needs further investigation [169, 170]. On the other hand, an antagonist in the promotion of melanocyte differentiation induced by mitf, is foxd3, which restrains its transcription. As a result, it prevents crest cells from evolving into melanocytes in zebrafish [171]. An additional gene that is involved in the process of pigmentation during the early stages of mice embryogenesis is Mef2c, along with its transcriptional product MEF2C. MEF2C’s loss of function causes defects in the process of melanogenesis, due to a decrease in the number of differentiated melanocytes. It is responsible for defects in the gut innervation as well [172]. MEF2 protein family plays a significant role in governing the development of the brain, lymphocytes, skeleton, and cardiovascular system in many vertebrates including mice and Drosophila. However, in Drosophila there is only one Mef2 gene, instead of a protein family, which is pivotal for the proper melanogenesis [173]. In mouse embryos, Mefc2c enhancer contains three functional sites that bind to SOX proteins as well as a crucial MEF2 site. This indicates that Mefc2c is directly regulated and dependent on the SOX10 and MEF2 protein family. The transcription factor MEF2C, encoded by the Mef2c gene, has been proven to preserve a differentiating phenotype in melanoblasts. Cosequently, it is on charge of the proper amount of melanoblasts [172].
The major molecular signaling pathways that take part in the procedure of
melanocyte formation include WNT, KIT proto-oncogene (KIT) and EDNRB. To begin
with, canonical Wnt/
The progenitors of ENS are the vagal and sacral NC. Cells from these regions start migrating, multiplying and finally they give rise to both neurons and glial cell products in the developing embryonic gut. ENS consists of operational neural networks consisting of various types of neurons and glia, structured into two linked clusters of ganglia: the myenteric and submucosal plexuses within the gut. Vagal NC, situated between one and seven somites, is responsible for forming the vast majority of the ENS, with its cells colonising the digestive tube in a ‘head to tail’ direction. This cell population is divided into two subtypes; one extending from somites level one through 3 and the second lying between somites four through seven. The first subtype is separated in two smaller groups, the first of which migrates in a dorsolateral direction beneath the ectoderm to populate the pharyngeal arches and the cardiac outflow tract, while the second migrates ventrally and gives birth to the dorsal root and sympathetic ganglia as well as forming part of the ENS by invading into the initial part of the foregut [183]. Crest cells from somite level four through seven move ventrally and end up colonising the gut [184]. According to other subdivisions, in chicken embryos NCCs in the surroundings of somites one and two generate the circumpharyngeal crest as well as precursor cells for Schwann cells. These precursors then populate the vagus nerve, subsequently directing their migration into the esophagus and stomach. They establish themselves along nerves by detecting axonal Nrg1 via the tyrosine kinase receptor ErbB3. ErbB3 is, also, required for the development of the sympathetic nervous system in mouse embryos [78, 97]. On the other hand, crest cells positioned near somites three through seven, take part in the formation of the sympathetic ganglion chain and invade the whole digestive tube [185]. Sacral crest provides approximately 20% of the neurons found in the descending colon and rectum.
To begin, retinoic acid (RA) plays a crucial role in the differentiation of NCCs
into enteric lineage. RA is secreted by the paraxial mesoderm in the space around
somites in quail embryos and interacts with its receptors RAR
Netrins are a significant laminin related protein family, generated by
epithelial cells located in the gut. They have been demonstrated as significant
participants in the development of ENS, contributing in the surveillance and
migration of ENCCs. To begin with, they are being expressed in the developing
intestine and pancreas in mouse and chick embryos. NETRIN-1 is a member of the
family that acts as an attractive source for ENCCs and fosters not only the
growth but also viability of cells originating from enteric crest. It is also
responsible for the axonal growth of neurites. Specifically in order to enhance
neurite extension, NETRIN-1 necessitates co-factors that are generated by cells
within the bowel wall. However, these co-factors do not originate from the crest.
Netrins, also, stimulate the inward migration of neurons from the myenteric to
the submucosal region, with mice missing these protein molecules, lacking the
submucosal as well [194]. Another protein family produced by the epithelial
cells in mouse embryos is hedgehog (Hh) family, crucial for controlling the
accumulation and specialization of the mesenchyme [195, 196]. Hedgehog
proteins, including sonic hedgehog (SHH) and indian hedgehog (IHH), are signaling
molecules which are secreted by the enteric epithelium from the initial phases of
intestinal organ formation. They directly affect BMP4 production, which is
pivotal for the proper ECM formation and prevents the development of smooth
muscle under the enteric epithelium. Moreover, this group of proteins is involved
in the activation of Foxf1 and Foxf2, in the mesenchymal cells
(forkhead transcription factors), which are crucial for the proper production and
formation of the ECM (especially Foxf2). FOXF proteins are responsible
for the induction of BMP4, which in turn inhibits the transcription of
Wnt5a. In mutant mice with loss of Foxf2 alleles, this
imbalance in signaling molecules leads to a decrease in collagen synthesis and
finally in general ECM disintegration. Such deficiency of the ECM disrupts
interactions between confinement molecules and as a result, epithelial cells of
the intestinal villi lose their normal polarisation and become round with
centrally located nuclei. However, cells remain viable and present resilience in
apoptosis due to increased WNT signaling and
Moreover, EDNRB signaling encourages the growth of ENCCs while preventing their transformation into neurons [73, 184]. This effectively keeps them in an undifferentiated and multiplying condition which comes in contrast with GDNF signaling. The latter, drives progenitor cells to differentiate into neurons [163]. ENDRB signaling pathway is not necessary until the migration stream of NCCs invades the region of the cecum. In the balance between neurons and glial cells in mice, Ezh2 is also involved. Ezh2’s expression is increased in ENCCs committed to the glial lineage, where it functions as a transcriptional activator. On the other hand, it is reduced in cells that are going to differentiate into neurons, where it serves the role of transcriptional repressor [197]. Ezh2 is regulated by Kinesin family member 7 (Kif7) [96]. Transcriptional factors detected in glial cell progenitors are Sox10, Erbb3, Fabp7, and Plp1, while those participating in the determination of the neuronal fate include Elavl4, Tubb3, Phox2b and Ret [197, 198].
The procedure of the terminal differentiation of ENCCs encloses different molecules. In particular, ASCL1 and HAND2 are two transcription factors that are also involved in the process of enteric nervous system formation, especially in their terminal differentiation [89]. Specifically, Ascl1 is present in all progenitors of enteric neuronal subtypes in mouse and human embryos (ASCL1). It is required for proper neurogenesis, with its loss being associated with delayed formed and atrophic neurons. But even in its absence, migration rates are not affected. Neuronal development in mutant embryos can be saved by increased expression of Ngn2, a proneural gene. However, Ngn2 cannot replace Ascl1’s function in determining the formation of particular types of the enteric neurons. Consequently, Ascl1 is a necessary transcription factor in the normal differentiation procedure [92]. Another gene involved in the terminal differentiation of ENCCs is Hand2. It is a prerequisite for the differentiation of NCCs into enteric neurons in mice and is present in cells that are undergoing development as both neurons and glial cells. In mice mutants that do not express Hand2, crest-derived precursors usually populate the Hand2-deficient gut, and its presence is not necessary for the initial activation of Phox2B, Ret or Sox10. However, its expression is essential for the final stage of differentiation of neurons located in the gut [93]. Additionally, in mutant mice lacking Hand2, notable reductions are observed in the counts of cholinergic and nitrergic neurons. Nitric oxide synthetase (NOS), which is necessary for the formation of NO, is missing in these mutant embryos, demonstrating its downstream dependance on Hand2. NOS offers guidance for the migration of particular groups of cells [199]. Since cells producing two crucial neurotransmitters in the enteric nervous system, nitric oxide (NO) and acetylcholine (ACh), tend to develop early in the developmental process, the substantial decrease in both of these cell types in the progenitors of neural endopeptidase cells implies that neurotransmission would likely be adversely impacted.
When NC genes are altered, NCCs’ migration is disrupted and characteristic phenotypic changes occur. These phenotypes are thoroughly studied using various animal models that help elucidate the complexity of gene pathways and reveal the distinct phenotypic alterations that occur when these pathways are compromised. Zebrafish, Xenopus, avian, and mouse embryos each offer unique insights into the complexities of NC migration and differentiation. Xenopus, an adaptable vertebrate model, has been instrumental in studying various neurocristopathies through diverse experimental methodologies. Its large eggs allow for early-stage injections, facilitating accessibility throughout embryonic development. Additionally, the transparency of Xenopus tadpoles in later stages enables imaging akin to zebrafish larvae, thus enabling the monitoring of differentiated cells originating from the neural crest [200].
The avian embryo has been pivotal in elucidating cell migration and histogenesis since the inception of the quail-chick marking system in 1969. Through this method, chimeras generated by replacing specific embryonic regions of one species with those of another enable the investigation of the behavior and destiny of grafted NC cells. By substituting fragments of the neural fold or tube between chick and quail embryos along the neural axis, migration pathways and fates of neural crest cells exiting at different levels of the neural tube can be discerned [201]. Although mice models are valuable for studying NC pathways in mammalian species, they have limitations. For instance, null mice lacking key NC genes sometimes do not exhibit a failure of neural crest cell induction similar to non-mammalian organisms [202]. Nevertheless, the applications of all these animal models contribute to deciphering the “master genes” of neural crest cells and their morphogenetic fate.
The pathological changes due to NC-related gene alterations that are observed in these animal models are categorized under the umbrella term “Neurocristopathies”, which encompasses a broad spectrum of disorders arising from issues during migration and differentiation of NCCs. Mutations in EphA2 led to abnormal migration of cranial NC and loss of the distinction between third and fourth arch streams [28]. Regarding FGF signaling, mice mutants with Fgfr2 deficiency display reduced osteoblast differentiation [51]. FGFR2 is also implicated in syndromic craniosynostosis [203]. Multiple craniofacial defects with the most prominent of them being agenesis of frontal bones and absence of parietal and interparietal ossification is exhibited in Dlx5 mutant embryos, with this gene being involved in the pathogenesis of frontonasal dysplasia. Absence of Twist1 function leads to anomalies in the cranial mesenchyme, hindrance in the closure of the neural tube in the cranial area, and various other anomalies, including Saethre–Chotzen syndrome [204]. The temporary deactivation of Hoxa2 from the stage before migration to the later stages after migration consistently results in the complete alteration of skeletal structures in the second arch to resemble those of the first arch, as seen in Hoxa2-deficient mutants [205]. Embryos with Dlx1/2 mutations have impaired development of the upper jaw and the hinge region, while in brain development Dlx1/2 mutations are linked to abnormal striatal subventricular zone formation and differentiation [63, 206]. Loss of Dlx5 leads in proximal lower jaw anomalies, profound deformities in the vestibular system, postponed ossification in the cranial vault, irregular bone development and abnormalities in the hinge region [55] Tricho-Dento-Osseous syndrome is likely connected to Dlx5 mutations. The simultaneous inactivation of both Dlx5 and Dlx6 at the same time results in a lower jaw that mirrors the upper jaw [207, 208].
When it comes to placode formation, mutated mice with loss of TGF
Various heart abnormalities are also observed. Tbx1 deactivation leads to diminished NC cell presence in the heart and the maturation and differentiation of cardiac NCCs is hindered, due to a decrease in MAPK signaling and an early activation of BMP signaling [65]. Tbx1 mutations are linked to Di George syndrome, CHARGE syndrome as well as Velocardiofacial syndrome [65, 210]. Mouse embryos lacking either Sema3C or PlexinA2, are not able to properly arrange NCCs in the cardiac outflow tract [69, 211]. Moreover, the absence of Semaphorin 3C leads to the disruption of the aortic arches’ morphogenesis, while neuropilin containing receptors interact with vascular endothelial growth factor (VEGF) signaling [3, 212]. BMP1 inactivation results in the inability to separate the outflow tract and a lack of proper development of smooth muscle in the heart. Similarly, impairing BMP receptor 1A in NCCs also leads to defects in the outflow tract [131]. Loss of SMAD4 results in several cardiac defects and extensive cell death in cardiac NCCs and NCCs populating the pharyngeal arches [213, 214], while its disruption causes cardiac hypertrophy [215]. Loss of Alk2 led to impaired cardiac NC migration and several cardiac and craniofacial defects [71, 216]. Loss of Prdm6 in cardiac NCCs leads to a disruption in the pre-EMT stage, causing NCCs to stay in the dorsal neural tube. Moreover, loss of Prdm6 in mice can result in thinner myocardial walls and subsequent heart failure [72].
Loss of Mitf or Sox10 leads to pigmentation defects in mice and both genes are implicated in the pathogenesis of Waardenburg syndrome [86]. Disabling Mef2c within mice NC leads to diminished activation of genes associated with melanocytes during their development, causing a notable decrease in. This occurs as a consequence of impaired cellular differentiation and a decreased number of melanocytes [172]. Alk3 deletion in embryos does not influence migration of neural progenitors towards the dorsal aorta but it weakens their ability to form aggregates, leading to cardiac abnormalities and high fatality rate. The absence of Phox2b, which is connected to Hirschprung disease, might explain the absence of sympathetic nervous system (SNS) precursors in Alk3 knockout embryos [217].
In the current study, we attempted to organize specifier genes according to location and individual cell type basis with extension to related dysmorphogenesis in animal models. The NC contributes to craniofacial and placode development, with HOX family genes’ and DLX family genes’ guidance being the most prominent. In cardiac development, TBX2/3, PAX3 and PRDM6 are crucial, while in enteric development, ASCL1, HAND2 and RET are key genes. When it comes to distinct cell types, neural-crest derived neuronal lineages are characterized by NGN1, NRP1/2, PAX3, SOX10 and C-RET expression. KIT, MITF and SOX10 have important roles in melanocytes formation, SOX family genes are vital for chondrocyte fates. Additionally, we aimed to illustrate distinct genetic alterations that result in phenotypic examples across various species. Through such examination, a deep understanding of the fundamental processes involved in abnormal morphogenesis is acquired. Nonetheless, more systematic approaches should be implemented to achieve such a result. Understanding the intricate processes involved in NC cell migration and differentiation requires a roadmap of molecular cues and genetic drivers [218, 219]. In this pursuit, animal models serve as essential tools for unraveling complex gene pathways.
The multipotency of NCCs presents intriguing avenues for research and potential medical applications. NCCs use can serve as a mean to tissue and organ repair and regeneration while potentionally aiding in the treatment of congenital forms of NC-related diseases [220, 221]. Loss-of-function techniques, namely dominant-negative constructs, antisense morpholinos, and RNA interference (RNAi) have been preferred methods for transiently disrupting neural crest development in chick, Xenopus, and Zebrafish embryos. Miscellaneous technologies such as Cre/loxP, TALENs, Flp/FRT and newer ones like CRISPR-Cas9 are widely employed in generating knockout animals. However, challenges persist in NCC research, as these methods alone may not fully elucidate the underlying gene networks and interactions [222]. Moreover, variability in existing NC induction models poses further complications. Contemporary technologies, e.g., next-generation sequencing (NGS), proteomics and transcriptomics, may offer an alternative view into congenital disease progression and mechanisms of gene interactions [223]. Moreover, the knowledge aquired will accelerate the rise of “personalized medicine” and “precision medicine” [221]. Through these promising approaches, early diagnosis can be achieved and specialized interventions catered to each patient can be designed, producing individualized therapeutic solutions.
AAA, abdominal aortic aneurysm; ACH, acetylocholine; ALX, ALX homeobox; ASCL, achaete-scute family bHLH transcription factor; BARX, BARX homeobox; BMP, bone morphogenetic protein; BMPR1B, bone morphogenetic protein receptor 1 beta; CDK, cyclin-dependent kinase; CX, connexin; CXCL12, C-X-C motif chemokine ligand 12; CXCR4, C-X-C motif chemokine receptor 4; DLX, distal-less homeobox; DRG, dorsal root ganglia; DV, dorsal-ventral; ECM, extracellular matrix; EDNR, endothelin receptor; EMT, epithelial mesenchymal transition; ENCC, enteric neural crest cell; ENS, enteric nervous system; EPH, ephrin; ERBB4, erb-b2 receptor tyrosine kinase 4; ETS, ETS proto-oncogene; FGF, fibroblast growth factor; FLRT, fibronectin leucine-rich transmembrane; FOX, forkhead box; GBX, gastrulation brain homeobox; GDNF, glial cell derived neurotrophic factor; GSC, goosecoid; HAND, heart and neural crest derivatives; HH, hedgehog; HOX, homeobox; IHH, Indian hedgehog; KIT, KIT proto-oncogene; LHX, LIM homeobox; LIFR, LIF receptor subunit alpha; MITF, melanocyte inducing transcription factor; MMP, matrix metalloproteinase; MPNST, malignant peripheral nerve sheath tumor; MSX, MSH homeobox; MYH, myosin heavy chain; MYL, myosin light chain; NC, neural crest; NCC, neural crest cell; NEP, neuroepithelial; NF, neurofibromin; NO, nitric oxide; NOS, nitric oxide synthetase; NGN1/2, neurogenin-1/2; NRG, neuregulin; NRP, neuropilin; PA, pharyngeal arch; PAX, paired box; PHOX, paired like homeobox; PITX, paired like homeodomain; PNS, peripheral nervous system; PRDM, PR/SET domain; RA, retinoic acid; ROBO, roundabout; R3/5, rhombomere 3/5; SA, sympathoadrenal; SDF, stromal cell derived factor; SEMA, semaphorin; SHH, sonic hedgehog; SIP, sheath interacting protein; SMAD, SMAD family member; SNAP, synaptosome associated protein; SOX, SRY-box transcription factor; TAGLN, transgelin; TBX, T-box transcription factor; TGFBR2, TGF receptor type II; TH, tyrosine hydrosylase; TNC, tenascinC; TFAP, transcription factor AP; TGF, transforming growth factor; TYR, tyrosinase; TYRP, tyrosinase related protein 1; TWIST, twist family bHLH transcription factor; vSMC, vascular smooth muscle cell; ZIC1, ZIC family member 1.
SAK and DC collected, analysed and reviewed the information, wrote and edited the manuscript; ID reviewed and assisted in the analysis; SG, MEM and SM provided advice on the review; PT conceptualized and designed the review. All authors contributed to editorial changes in the manuscript. All authors contributed to the drawing of charts and figures. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Not applicable.
Not applicable.
This research received no external funding.
The authors declare no conflict of interest. Given his role as Guest Editor, Paschalis Theotokis had no involvement in the peer-review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to Margarita M Ivanova and Viviana di Giacomo.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.