1 Shandong Branch of National Vegetable Improvement Center, Institute of Vegetables, Shandong Academy of Agricultural Science, 250131 Jinan, Shandong, China
2 College of Life Science, Qufu Normal University, 273165 Qufu, Shandong, China
3 Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, 200032 Shanghai, China
4 Horticultural Science and Engineering, Shandong Agricultural University, 271002 Taian, Shandong, China
5 College of Life Science, Shandong Normal University, 250061 Jinan, Shandong, China
†These authors contributed equally.
Abstract
Background: DELLA protein is a crucial factor which played pivotal roles in regulating numerous intriguing biological processes in plant development and abiotic stress responses. However, little is known about the function and information of DELLA protein in Chinese cabbage. Methods: Using 5 DELLA gene sequences in Arabidopsis Thaliana as probes, 5 DELLA genes in Chinese cabbage were identified by Blast search in Chinese cabbage database (Brassica database (BRAD)). The National Center for Biotechnology Information (NCBI), ExPaSy, SWISS-MODEL, DNAMAN, MEGA 11, PlantCARE were used to identify and analyze the DELLA gene family of Chinese cabbage. Gene expression was analyzed by quantitative real-time polymerase chain reaction (qRT-PCR). The function of BraA10gRGL3 was verified by overexpression and phenotypic analysis of BraA10gRGL3 and yeast hybrid. Results: In this study, 5 BraDELLAs homologous to Arabidopsis thaliana were identified and cloned based on the Brassica database, namely, BraA02gRGL1, BraA05gRGL2, BraA10gRGL3, BraA06gRGA and BraA09gRGA. All BraDELLAs contain the DELLA, TVHYNP, and GRAS conserved domains. Cis-element analysis revealed that the promoter regions of these 5 DELLA genes all contain light-responsive elements, TCT motif, I-box, G-box, and box 4, which are associated with GA signaling. Transcriptome analysis results proved that the expression of BraA02gRGL1, BraA05gRGL2, and BraA10gRGL3 in Y2 at different growth stages were lower than them in Y7, which is consistent with the phenotype that Y7 exhibited stronger stress tolerance than Y2. It is worth emphasizing that even through the overexpression of BraA10gRGL3-Y7 in Arabidopsis resulted in smaller leaf size and lower fresh weight compared to the wild type (WT) Arabidopsis: Columbia, a stronger response to abiotic stresses was observed in BraA10gRGL3-Y7. It indicated that BraA10gRGL3-Y7 can improve the stress resistance of plants by inhibiting their growth. Moreover, the yeast two-hybrid experiment confirmed that BraA10gRGL3-Y7 can interacted with BraA05gGID1a-Y7, BraA04gGID1b1, BraA09gGID1b3-Y2, and BraA06gGID1c, whereas BraA10gRGL3-Y2 cannot interact with any BraGID1. Conclusions: Collectively, BraDELLAs play important role in plant development and response to abiotic stress. The differences in amino acid sequences between BraA10gRGL3-Y2 and BraA10gRGL3-Y7 may result in variations in their protein binding sites, thus affecting their interaction with the BraGID1 family proteins. This systematic analysis lays the foundation for further study of the functional characteristics of DELLA genes of Chinese cabbage.
Keywords
- Brassica rapa
- DELLA protein
- evolution
- stress tolerance
- growth and development
Chinese cabbage (Brassica rapa L. ssp. pekinensis) is one of the most important leafy vegetables, belongs to the genus Brassica of the cruciferous family [1], is originated from China. It is an economically important winter vegetable, that is widely consumed and cultivated globally, especially in Asia [1, 2]. Due to its high mineral nutrients, vitamins and crude fiber leads to fundamental part of health diet [3]. However, Brassica rapa is extremely sensitive to various environmental influences, which can cause significant reductions in yield [4]. To survive environmental stresses, plants have evolved physiological and biochemical resistance mechanisms, that respond at the molecular, cellular, and physiological level [5, 6]. Plant hormones can regulate the growth and development of plants by integrate various internal and external signals, to help them cope with abiotic stresses [7, 8].
Gibberellin (GA) is not only an important regulatory factor in plant growth and development, but also an important regulatory factor in plant response to stress. GA signal transduction process limits plant growth under cold, salt, and osmotic stress, and suggested an important role in response to abiotic stress [9]. DELLA protein is an integrated factor in a variety of hormone signals involved in environmental signaling pathway to regulate responses and is a negative regulator in the GAs metabolic pathway belonging to the plant-specific GRAS family [10]. The DELLA protein has a highly conserved C-terminal sequence but a large variation of N-terminal [11]. In the entire amino acid sequence, the DELLA protein includes a total of 8 conserved domains, which are divided into three parts, the N-terminal DELLA, VHYNP, the Poly S/T/V (serine, threonine, valine rich region in the middle part) [12, 13, 14]. Numerous DELLA genes (GAI (GA-INSENSITIVE)), RGA (REPRESSOR OF GA1-3), RGL1 (RGA-LIKE1), RGL2 and RGL3 is highly conserved in different plants, and have been cloned in a variety of plants, such as Arabidopsis thaliana, rice, barley, tomato, and cucumber [15, 16]. These findings suggested that RGA and GAI are involved in flower induction and inhibition of plant vegetative growth [17] and RGL2 inhibits seed germination [18]. In addition, RGA, RGL1 and RGL2 have a synergistic regulatory effect on flower development [19]. Furthermore, RGL3 enhances plant resistance to adversity [20].
Previously, five DELLA proteins were identified in Arabidopsis, GAI, RGA, RGL1, RGL2 and RGL3, which are shown to inhibit plant growth [16]. DELLA proteins can interact between GA signaling pathway and CBF1-dependent cold signaling pathway CBF1 (C-repeat/dehydration response element binding factor1), while CBF cold response pathway and plant cold acclimation [13]. The transcription factor CBF1/DREB1b, enhanced responds to low temperature, and increased the expression levels of GA2ox3 and GA2ox6 in the GA synthesis pathway, thereby reducing the content of endogenous GA, stabilizing the DELLA protein, and improving the cold tolerance of plants [21, 22]. When the gibberellin receptor protein GID1 is not bound to GA, its N-terminal extension (N-Ex) has a relatively flexible structure, and in the presence of GA signal, the conformation of N-Ex starts to change, and GA tightly binds to the GID1 protein, forming the GA-GID1 complex. Phosphorylation of the EL1 (Earlier flowering 1) protein and the SPY (Spindly) protein can activate the activity of the DELLA protein, making it easier for the DELLA protein to bind to the GA-GID1 complex and form a more stable trimer, known as GA-GID1-DELLA [23, 24]. This trimer can be polyubiquitinated by a specific ubiquitin E3 ligase complex (SCFSLY1/GID2) and subsequently degraded by the 26S proteasome, resulting in a GA response [25]. This was designed to identify and characterize DELLAs protein family in B. rapa for abiotic stress tolerance [26, 27].
DELLA proteins are not only involved in the GA responses but also integrate plant responses to various environmental signals (like light, temperature, moisture, and salt) and hormone signals (such as jasmonic acid, and auxin) [28]. DELLA proteins are beneficial for plants under stress, as they inhibit certain plant responses. The higher the DELLA protein content, the more tolerant the plant is to the environment [29]. By responding to external signals, DELLA proteins can regulate numerous plant processes, including seed germination, seedling growth, floral organ formation, fruit development, and plant senescence. Leaf elongation in seedlings depends on the proteasome-mediated derepression of DELLA [30]. LlDELLA1 in Lupinus luteus contributes to flower and pod development, with expression levels fluctuating during various stages of growth and reproduction [31]. DELLA plays a crucial role in grain development, the expression of the DELLA gene in tomatoes and Arabidopsis induces parthenocarpy [32]. Following the overexpression of the peach Della family member, PpeDGYLA, in Arabidopsis, the plants exhibited a dwarf phenotype and displayed insensitivity to GA [33]. The DELLA protein in rice can competitively inhibit the GID1-NGR5 interaction. This response is crucial for nitrogen-regulated plant growth and development. It also explains the increased tillering observed in green revolution varieties [34]. The maize WRKY28 interacts with the DELLA protein D8, influencing embryonic layer formation. Additionally, it plays a role in regulating shade avoidance and overall plant structure [35]. Moreover, DELLA proteins play a crucial role in various processes such as alleviating seed dormancy, promoting flowering, extending the flowering period, enhancing fruit quality, slowing plant aging, and controlling the synthesis of secondary metabolites. Consequently, the findings on DELLA proteins hold extensive application potential in managing plant growth, development, resistance, and disease resilience.
Until now, the knowledge regarding DELLA, the evolutionary relationship and functional characterization in B. rapa is poorly understood. In this study, we conducted the identification of family members of DELLA in Chinese cabbage, delved deeply into their physicochemical properties, gene structure, protein domains, distribution of conserved motifs, phylogenetic tree, cis-regulatory element analysis, inter-species collinearity analysis, gene expression pattern analysis. Furthermore, it provides preliminary validation of their functions through gene editing of Arabidopsis. We aimed to provide a theoretical basis for further studying the biological functions and practical applications of the DELLA genes in Chinese cabbage.
Chinese cabbage of high generation inbred line “Y2” and “Y7” were used in
this study. Heathy seeds were selected and germinated in a glass petri dish
containing a little water. Transplanted the budding seedlings into pots (4
seedlings per pot) containing a growth medium having vermiculite and peat (3:1)
and grown in the greenhouse at 20
For salinity and drought treatments, plants were treated with 200 mM NaCl (salt)
and 30% (w/v) PEG6000 (polyethylene glycol) (dry) and 200 mM Mannitol. For heat
and cold treatments, plants were treated in control growth chamber at 38
In previous studies, the information of DELLA in Arabidopsis thaliana [16], Brassica oleracea [36], Oryza sativa [37], Zea mays [38], Triticum aestivum [39] and Brassica napus [40] have been reported. The whole genome sequences, protein sequences and gene annotation files of Brassica rapa and Brassica oleracea were downloaded from the Brassica database (BRAD, https://db.cngb.org/brassica/#/). The whole genome sequences, protein sequences and gene annotation files of Arabidopsis thaliana, Oryza sativa, Zea mays, Triticum aestivum and Brassica napus were downloaded from the Ensembl Plants. We perform Blast in the online website BRAD to obtain the DELLA homologous sequences of Brassica rapa using the AtDELLA genes as template.
The motif of DELLA protein sequences was analyzed with MEME online tools
(http://meme-suite.org/tools/meme). The molecular masses of DELLA proteins were
calculated using the Compute pI/Mw tool of ExPaSy
(http://web.expasy.org/compute_pi/). The number of amino acids, molecular weight
(MW), and theoretical isoelectric point (pI) were computed using the ProtParam
tool (https://web.expasy.org/protparam/). SWISS-MODEL
(https://swissmodel.expasy.org/) are used to predict the secondary and tertiary
structure models of BraDELLA proteins. Protein structure was predicted on NCBI
database (NCBI Conserved Domain Search (nih.gov), https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), (E-value
All the DELLA protein sequences in Brassica rapa, Brassica oleracea, Brassica napus, Arabidopsis thaliana, Zea mays, Triticum aestivum and Oryza sativa were used to perform phylogenetic. Multiple sequence alignments were conducted by ClustalW in MEGA 11 with default parameters. The alignment result was then used to construct a phylogenetic tree based on the neighbor-joining (NJ) method of MEGA 11. The parameters of NJ analysis were as follows: bootstrap method done 1000 times for statistical testing, model with JTT + G, and 50% sites coverage cutoff.
The promoter sequences (2000 bp upstream of ATG) of five BraDELLA genes were extracted from genome sequences of Brassica rapa by TBtools software (version 1.098696, Chengjie Chen, South China Agricultural University, Guangzhou, China) [41]. The cis-elements in promoter region were predicted using the PlantCARE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
The synteny of DELLA genes among the Arabidopsis, Brassica oleracea, Brassica napus, Brassica nigra, B. rapa was visualized by the One-Step MCScanX function of TBtools software. The Dual Systeny Plot for the MCScanX function of TBtools software was used to visualize the synteny.
Total RNA was isolated from each sample using the Plant RNA Extraction Kit
(DP432, Tiangen Biochemical Technology Beijing Co., Ltd., Beijing, China). cDNA
synthesis was carried out using a PrimeScript™ RT reagent kit with
a gDNA Eraser (RR037B, Takara, Dalian, China). qRT-PCR was performed using a SYBR Green Master mix (Takara, Dalian, China) on
an IQ5 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The qRT-PCR
primers for the BraDELLA genes and Actin gene are listed in
Supplementary Table 1. Three replicates of each sample were conducted to
calculate the average cycle threshold (Ct) values. The relative expression level was calculated by
2
The BraA10gRGL3 cDNA of Y2 and Y7 were cloned into pCAMBIA1307 vector NcoI and EcoRI sites under the control of the 35S promoter. The recombinant plasmid was transformed into A. tumefaciens strain GV3101 to incubated 2 days at 28 °C on the Yeast Extract Peptone Broth Medium (YEP) plate with kanamycin and rifampicin. The resulting constructs were named BraA10gRGL3-Y2 (OE-Y2) and BraA10gRGL3-Y7 (OE-Y7), and the sequences of the primers used are given in Supplementary Table 1. The floral dip method was used to transform Arabidopsis. The inflorescences were immersed in the transformation medium for 30 s, followed by one day of light avoidance cultivation, and then normal cultivation between the cultivations. After one week, the same method was used to immerse the inflorescences again. Approximately one month later, the transformed Arabidopsis seeds were harvested. The received T0 generation Arabidopsis seeds were grown on 1/2MS plates containing kanamycin (50 µg/mL) and hygromycin (50 µg/mL) for about 10 days. The plants with good growth and green leaves were considered positive seedlings. They were then transplanted into a culture medium (vermiculite:soil = 3:1) for further growth and individually numbered. The seeds harvested next were considered the T1 generation. The same process was repeated until obtaining the T3 generation homozygous line which was used for phenotypic analysis.
The Matchmaker Gold Yeast Two-Hybrid system (Clontech) was used to perform the yeast two-hybrid (Y2H) screen. To verify the interaction between Bra10gRGL3 and GID1 in yeast, the CDS of GID1 were cloned into the pGADT7 vector and the CDS of Bra10gRGL3 were cloned into the pGBKT7 vector using the NdeI and SmaI sites; then, the pGBKT7-Bra10gRGL3-Y2-GID1 and pGBKT7-Bra10gRGL3-Y2-GID1 pairs were separately transformed into yeast strain Y2H Gold and grown on SD/-Leu/-Trp medium. The positive transformants were further detected on SD/-Leu-Trp-His-Ade medium as described in the Yeast Protocols Handbook (Clotech, PT3024-1). The interaction between pGADT7-T7 and pGBKT7-53, and between pGADT7-T7 and pGBKT7-Lan were regarded as a positive and negative control, respectively. Primers used for construction are listed in Supplementary Table 1.
Five DELLA genes were identified in Chinese cabbage reference genome Brapa_genome_v3.0 (BRAD, http://www.brassicadb.cn/#/) after homologous sequence alignment with DELLA genes from Arabidopsis (At1G14920; AtGAI, AT2G01570; AtRGA, AT1G66350; AtRGL1, AT3G03450; AtRGL2, AT5G17490; AtRGL3). For further analysis, online database NCBI (http://www.ncbi.nlm.nih.gov/Structure) and ExPASy (http://web.expasy.org/protparam) were used for additional confirmation DELLA and GRAS domains. Referring to the naming method of the Arabidopsis DELLA gene family and further based on the relative position of the five genes on the chromosome, they were renamed BraA02gRGL1 (BraA02g017030), BraA05gRGL2 (BraA05g041910), BraA10gRGL3 (BraA10g022510), BraA06gRGA (BraA06g040430), and BraA09gRGA (BraA09g023210).
The basic properties including the length of protein sequence, gene locus, theoretical isoelectric point (pI), amino acid [39], molecular weight (MW) and chromosome localization (Chr. No) were analyzed to further characterize the BraDELLA proteins (Table 1). The predicted protein lengths varied from 488–573 amino acids, which is quite similar to DELLA proteins in Arabidopsis (512 to 588 amino acids), B. oleracea (507 to 576 amino acids), and B. napus (507 to 579 amino acids), but different from O. sativa (626 amino acids), T. aestivum (621 amino acids), and Zea mays (631 amino acids). Evolutionary analysis suggested that BraDELLA genes are located on different chromosomes, A02, A05, A06, A08 and A09, with various sequence lengths from 1467 to 1722 bp and molecular weight from 53.89 to 62.51 KDa in Chinese cabbage. Besides, DELLA protein sequences were similar to predicted isoelectric point (pI) values (ranging from 4.83 to 5.58). The isoelectric point (pI) value of DELLA protein of B. napus, ranges from 4.72 to 5.58 and B. oleracea, ranges from 4.73 to 5.52, indicate that these proteins are highly acidic in nature (Table 1). Five BraDELLA genes distributed on five chromosomes (Fig. 1).
| Group | Gene name | Gene ID | Chr. No | Gene locus | AA | pI | Mw (kDa) | Arabidopsis Orthologues | AtAA | ||
| ORF | Start | End | |||||||||
| III | BraA06gRGA | BraA06g040430.3C | A06 | 1722 | 22952357 | 22954078 | 573 | 5.58 | 62.51 | AT1G14920 (AtRGA2) | 534 |
| III | BraA09gRGA | BraA09g023210.3C | A09 | 1644 | 13719647 | 13721290 | 547 | 5.33 | 59.96 | AT2G01570 (AtGAI) | 588 |
| II | BraA02gRGL1 | BraA02g017030.3C | A02 | 1467 | 882873 | 8830203 | 488 | 5.09 | 53.89 | AT1G66350 (AtRGL1) | 512 |
| I | BraA05gRGL2 | BraA05g041910.3C | A05 | 1578 | 4919233 | 4920810 | 525 | 4.83 | 57.66 | AT3G03450 (AtRGL2) | 548 |
| I | BraA10gRGL3 | BraA10g022510.3C | A10 | 1578 | 4919233 | 4920810 | 525 | 4.83 | 57.66 | AT5G17490 (AtRGL3) | 524 |
ORF, Open reading frame; pI, Isoelectric point; Mw, molecular weight.
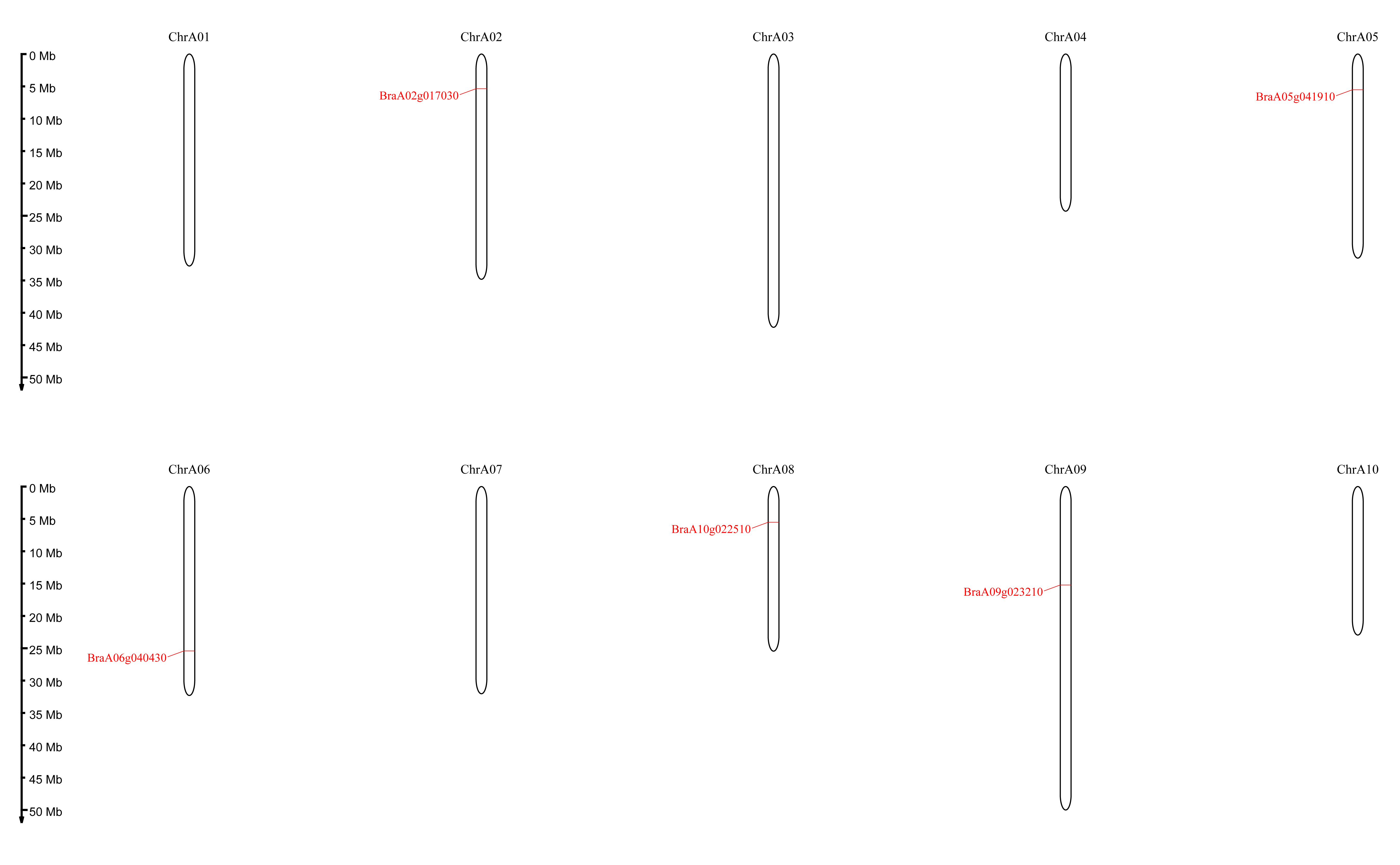 Fig. 1.
Fig. 1.Locations of the BraDELLA genes on the chinese cabbage chromosomes. The chromosome number is indicated at the top of each chromosome representation.
To explore the phylogenetic relationship of DELLA family genes in Brassica rapa [42], Brassica oleracea (Bo), Brassica napus [43], Arabidopsis thaliana (At), Zea mays [7], Triticum aestivum [44], and Oryza sativa (Os). A neighbor-joining (NJ) tree was constructed using the full-length protein sequence alignments of the identified 5 BraDELLAs, 5 BoDELLAs, 5 BnDELLAs, 5 AtDELLAs, 1 ZmDELLAs, combined with 1 TrDELLAs and 1 OsDELLAs. Based on the phylogenetic analysis, 28 DELLA genes were cluster into four groups, according to the topologies and bootstrap support (Fig. 2). The group Ⅰ contain RGL2 and RGL3 of Bra, Bo, Bn and At. The group Ⅱ holds RGL1 of B. rapa, B. oleracea, B. napus and Arabidopsis. The group Ⅲ includes TrRH1, ZmDWARF8 and OsSLR1. The group Ⅳ consists of RGA and GAI of B. rapa, B. oleracea, B. napus and Arabidopsis. B. rapa DELLA genes were relatively closer to the Arabidopsis. A 100% similarity between BraDELLAs and BnDELLAs was observed. ZmDELLAs, TrDELLAs and OsDELLAs was identified in the same clade which indicated a close relationship.
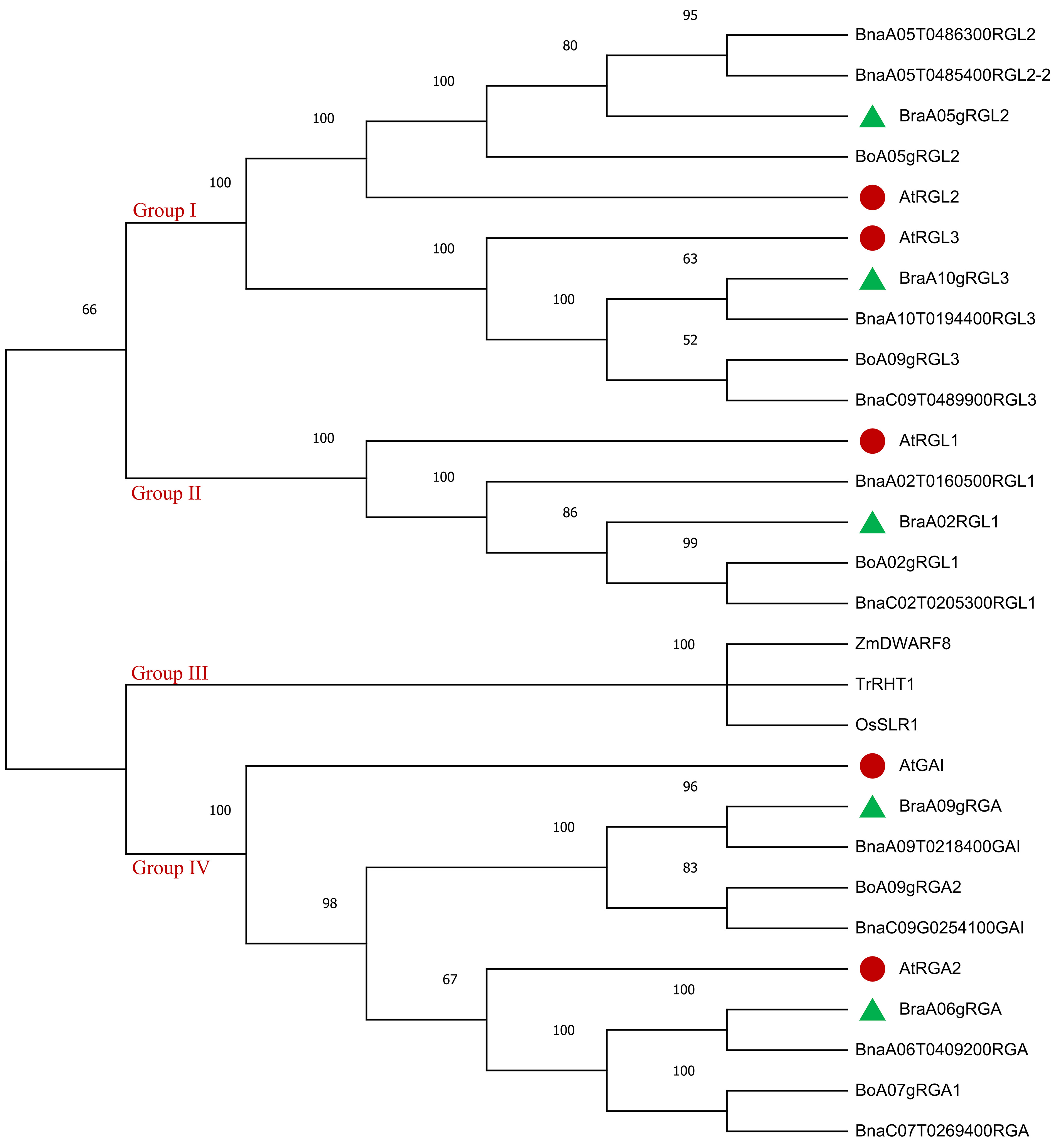 Fig. 2.
Fig. 2.The phylogenetic tree of DELLA proteins from Brassica rapa (the green triangle), Brassica oleracea, Brassica napus, Arabidopsis thaliana (the red circle), Zea mays, Triticum aestivum, and Oryza sativa using the neighbor-joining method. The bootstrap value (1000 replicates) is indicated close to the tree branches.
The secondary protein structure refers to the regular polypeptide chains,
stabilized by hydrogen bonds arrangements, while tertiary protein structure
refers to overall three-dimensional arrangements of its polypeptide chain in
space. The secondary and tertiary structure predicted that BraDELLA proteins has
a complex structure. The alpha (
| Protein name | Secondary Structure | |||
| Alpha helix (%) | Extended strand (%) | Random coil (%) | Beta turn (%) | |
| BraA02gRGL1 | 47.34 | 17.01 | 26.23 | 9.43 |
| BraA05gRGL2 | 48.32 | 12.08 | 33.07 | 6.53 |
| BraA10gRGL3 | 50.29 | 13.90 | 28.38 | 7.43 |
| BraA06gRGA | 43.98 | 18.50 | 30.72 | 6.81 |
| BraA09gRGA | 47.90 | 15.72 | 30.72 | 6.40 |
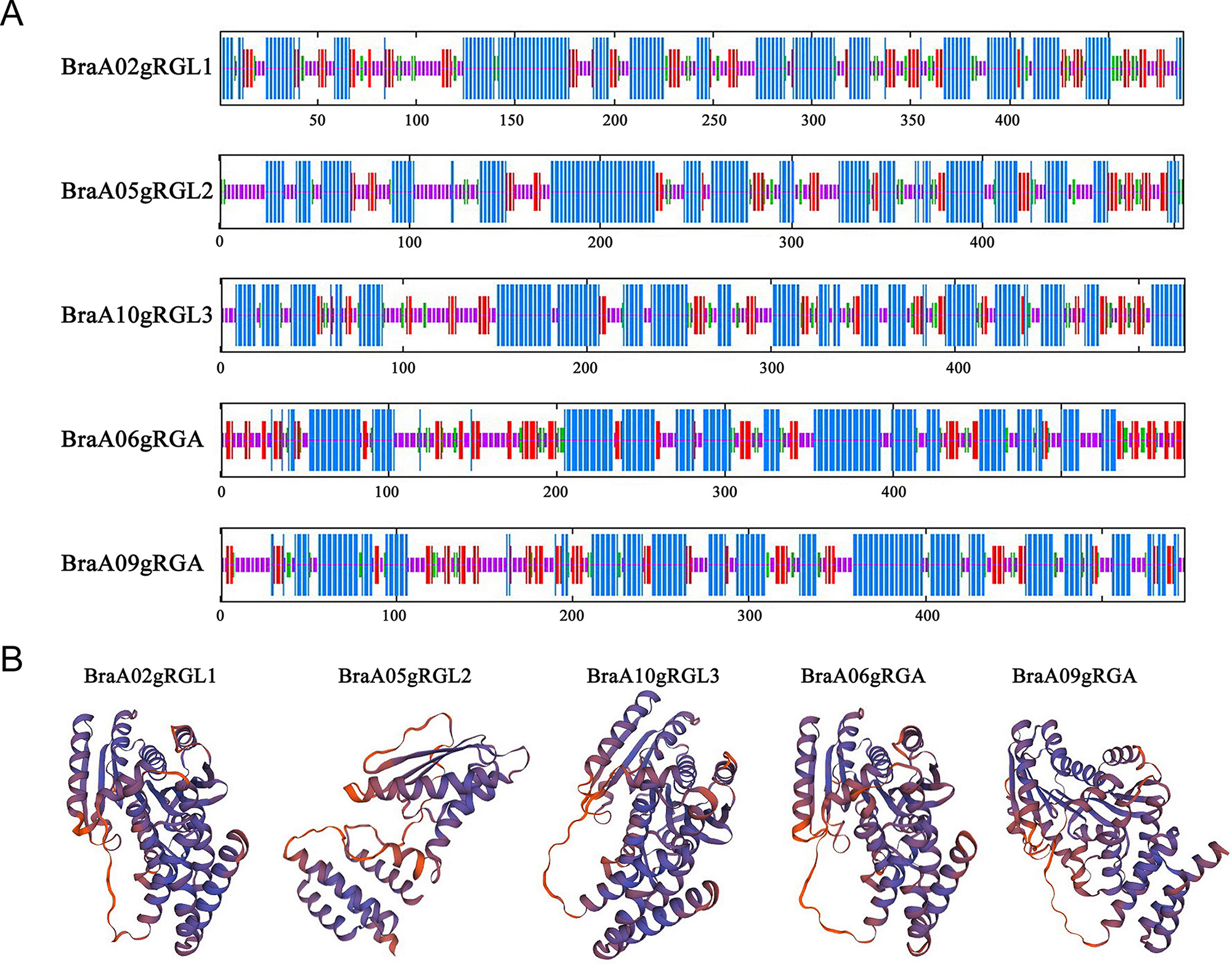 Fig. 3.
Fig. 3.Secondary structure and tertiary structure prediction of BraDELLA proteins. (A) Secondary structure prediction structure of BraDELLAs. (B) Tertiary structure prediction of BraDELLAs.
To further study the structural characteristics of DELLAs, we grouped the DELLAs of B. rapa, B. oleracea, B. napus, Brassica juncea and Arabidopsis. All the DELLAs were divided into three groups (Fig. 4A). Their conserved motifs were analyzed using MEME online software, and 12 conserved motifs were found (Fig. 4B). The results showed that motifs distribution among these BraDELLAs are similar, but the number of the motif among different genes varied greatly, ranging from 3 to 12. Among them, BnaA02g12260D contains at least 3 motifs, BniB05g046370 contains 7 motifs, and BnaC05gRGL2 contains 8 motifs. According to sequence alignment, motif 6 and motif 8 are DELLA domain and TVHYNP domain, respectively (Fig. 4C, which are two conserved domains of DELLA family. In addition, BnaA02g12260D does not have motif 8 and BniB05g046370 does not have motif 6, which may be caused by base loss during tandem repeat. From the perspective of domain distribution, DELLA domain and GRAS domain exist stably at the N-terminal and C-terminal of BraDELLAs, but compared with AtDELLAs, an obvious mutation in the BraDELLAs domain were found. DELLA domain of BraA09gRGA is D-E-L-L-A but others are D-E-L-L-X (Fig. 5).
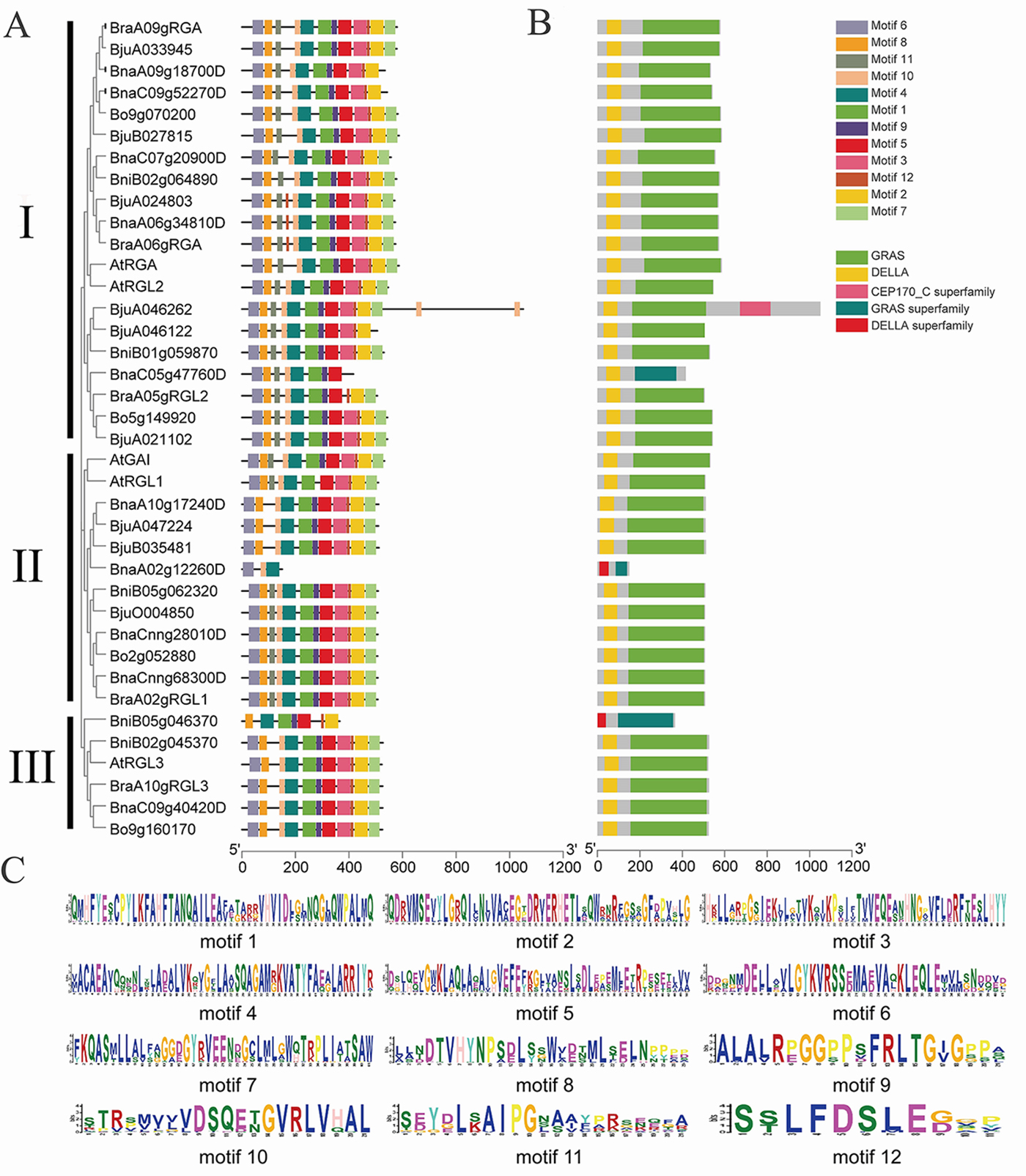 Fig. 4.
Fig. 4.Motif composition analysis of DELLAs. (A) The group of
different DELLA genes. (B) The 12 motifs are distributed in different
DELLAs (p-values
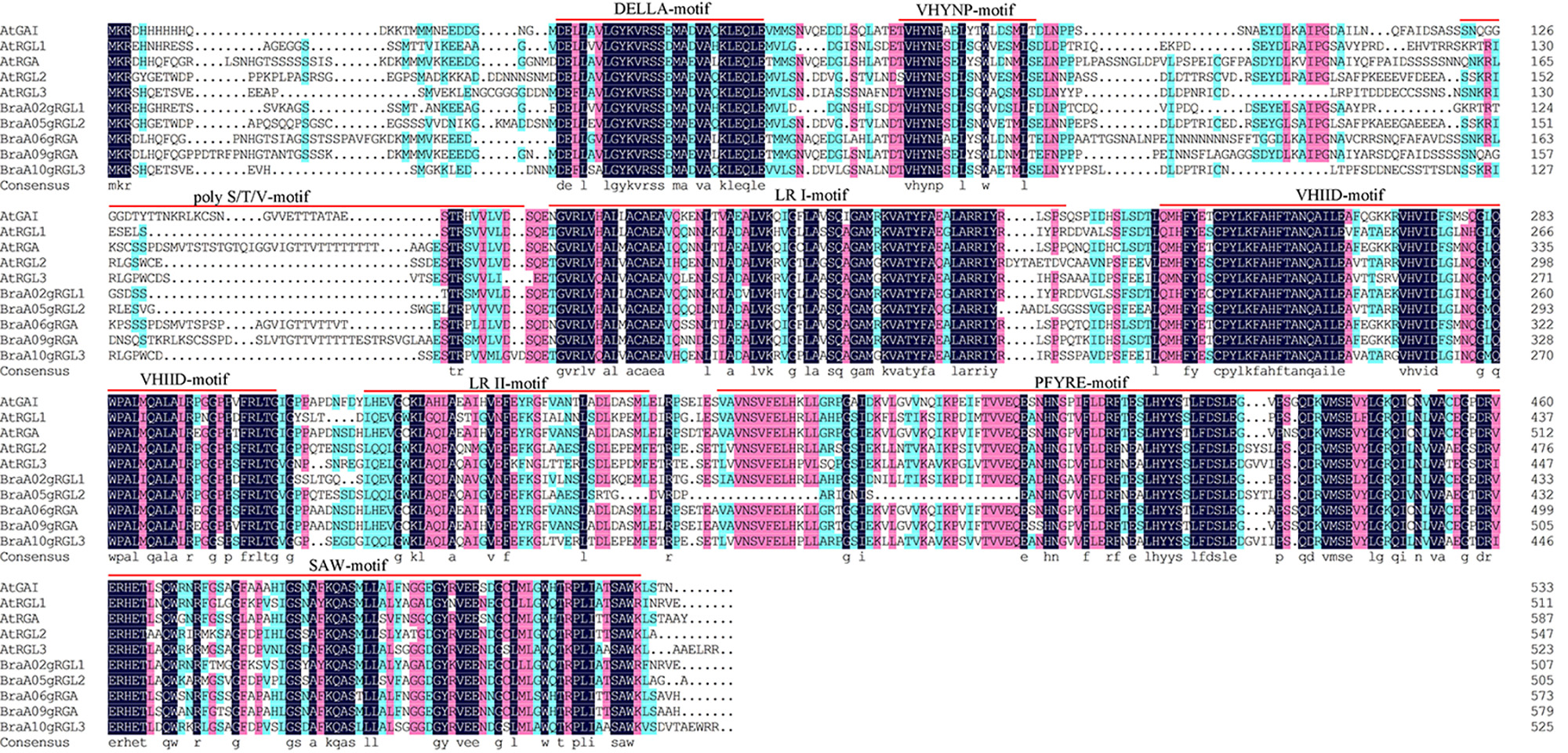 Fig. 5.
Fig. 5.Multiple sequence alignment of the protein sequence of Arabidopsis thaliana and Brassica rapa.
To further infer the phylogenetic mechanisms in the BraDELLAs family, we constructed the syntenic maps of B. rapa, B. oleracea, B. nigra, B. napus, and Arabidopsis (Fig. 6). A total of 5 DELLA genes in Chinese cabbage showed collinear relationship with 5 DELLA genes in Arabidopsis, 4 DELLA genes in cabbage, 4 DELLA genes in B. nigra, and 6 DELLA genes in B. napus, respectively. In addition, 4 DELLA genes in cabbage showed collinear relationship with 5 DELLA genes in Arabidopsis, 5 DELLA genes in B. nigra, 6 DELLA genes in B. napus. 6 DELLA genes in B. napus showed collinear relationship with 6 DELLA genes in B. rapa, B. oleracea, B. nigra and Arabidopsis. This indicated that the DELLA genes family in cruciferous crops has high homology, and the evolution of DELLA genes family experienced similar pathways between different species.
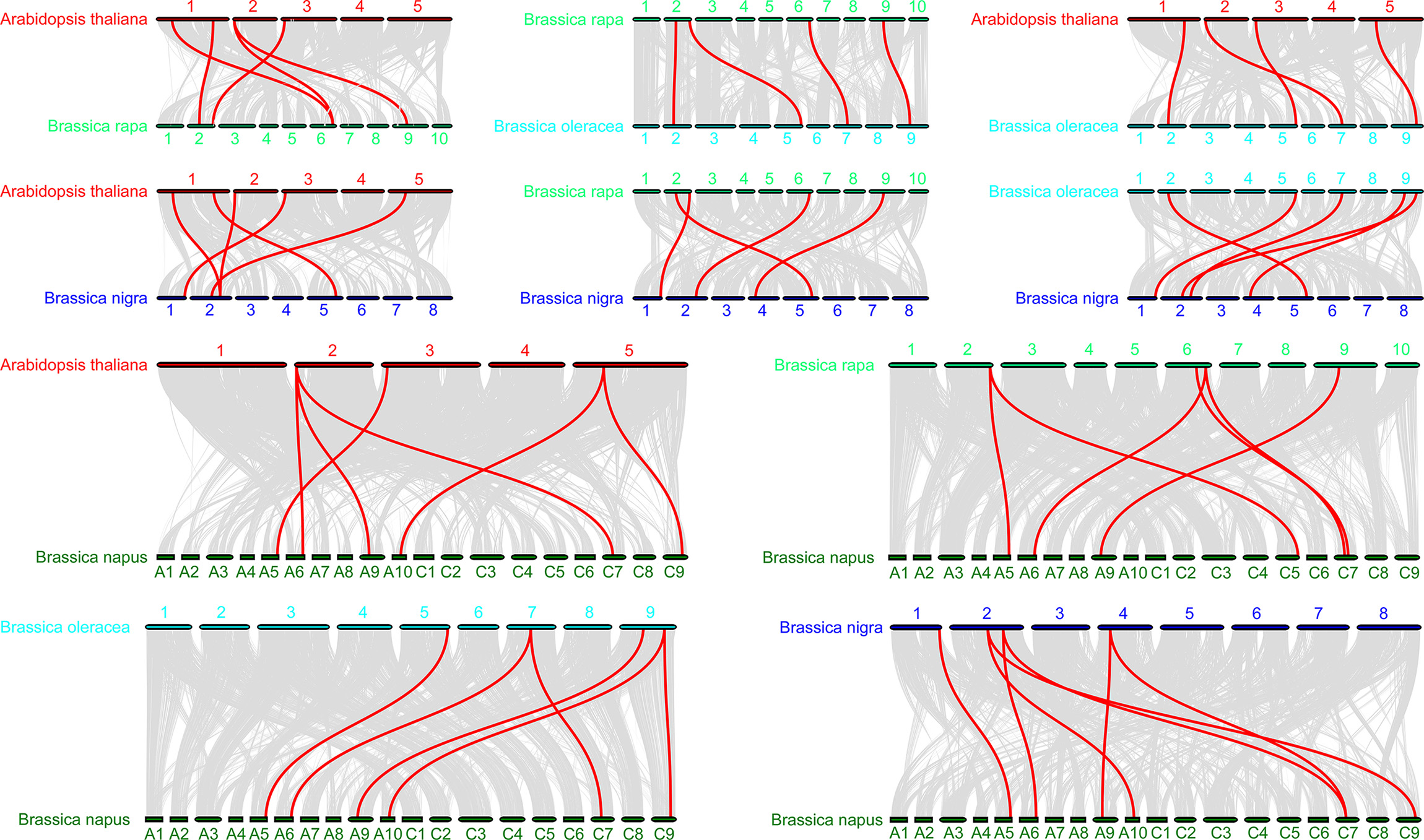 Fig. 6.
Fig. 6.Synteny analysis of DELLA genes among Brassica rapa, Brassica oleracea and Arabidopsis species. The Blue lines indicate the syntenic DELLA gene pairs; while the gray lines in the background indicate the collinear blocks within Brassica rapa, Brassica oleracea and Arabidopsis genomes.
To understand of the potential functions and regulatory mechanisms of BraDELLAs, we identified that cis-elements of DELLA genes contain transcriptional regulation elements, photosensitive regulatory elements, stress response elements, hormone response elements, plant tissue-specific elements and specific binding elements (Table 3). CAAT box and TATA, which are the key promoter elements for eukaryotic regulation of transcription initiation box, were identified in all promoters of BraDELLAs. Interestingly, all the BraDELLAs contain light response elements (TCT motif, I-box, G-box, box 4), which are also involved in the GA signals. The DELLA protein negatively regulates the GA signal pathway in light signal regulation. Generally, BraDELLA genes promoter contains four hormone response elements (GA, auxin, ETH, ABA and JA) as well as abiotic stress response elements related to drought, salt and low temperature. These specific binding sites demonstrate that BraDELLA responds to stress regulation pathways, participates in a variety of biological pathways and regulates biological processes. In our study, we discovered that the cis-elements in BraDELLAs were diverse. ABRE played an important role in stress response and P-box was crucial for gibberellin-responsive. All BraDELLAs had ABRE and P-box in their promoter regions, except for BraA06gRGA. Likewise, BraA06gRGA, BraA05gRGL2 and BraA02gRGL1 had 2-5 TGACG-motif cis-elements in their promoter region, which are related to MeJA-responsiveness. Thus, these findings suggest that the availability of dynamic cis-elements on prompter regions can diversify the function of BraDELLAs and promote defense mechanisms to induce biotic and abiotic stress tolerance.
| Cis-element | Gene name | Function | ||||
| BraA02gRGL1 | BraA05gRGL2 | BraA10gRGL3 | BraA06gRGA | BraA09gRGA | ||
| CAAT-box | 43 | 32 | 38 | 21 | 38 | cis-element common in promoter and enhancer regions |
| TATA-box | 38 | 64 | 8 | 24 | 8 | cis-element common in promoter and enhancer regions |
| TCT-motif | 0 | 2 | 0 | 2 | 0 | part of a light responsive element |
| TGACG-motif | 2 | 3 | 0 | 5 | 0 | cis-element involved in the MeJA-responsivenes |
| MYB | 4 | 0 | 0 | 4 | 0 | Drought responsive element |
| MYC | 4 | 3 | 4 | 3 | 4 | cis-element involved in ABA reaction |
| MBS | 0 | 1 | 0 | 1 | 0 | MYB binding site involved in drought-inducibility |
| TC-rich repeats | 0 | 0 | 2 | 2 | 2 | cis-element involved in defense and stress responsiveness |
| TGA-element | 1 | 0 | 3 | 1 | 3 | auxin-responsive element |
| LTR | 1 | 1 | 0 | 1 | 0 | cis-element involved in low-temperature responsiveness |
| ABRE | 3 | 2 | 2 | 0 | 2 | cis-element involved in the abscisic acid responsiveness |
| ARE | 4 | 4 | 2 | 4 | 2 | cis-element regulatory element essential for the anaerobic induction |
| CAT-box | 0 | 1 | 0 | 0 | 0 | cis-aelement regulatory element related to meristem expression |
| I-box | 2 | 1 | 2 | 1 | 2 | part of a light responsive element |
| G-Box | 1 | 3 | 1 | 0 | 1 | part of a light responsive element |
| Box 4 | 2 | 1 | 4 | 1 | 4 | part of a light responsive element |
| P-box | 1 | 1 | 1 | 0 | 1 | gibberellin-responsive element |
LTR, Long Terminal Repeat.
In our previous study, the correlation between gene matrix of different modules and different stages of Y2 and Y7 was analyzed by WGCNA. As a result, 25,406 differentially expressed genes (DEGs) between Y2 and Y7 were identified and grouped into 29 modules based on their expression patterns [45]. The expression pattern of genes in “turquoise” and “red” module does not exhibit any clear regularity while the expression of genes in “brown” module was higher in Y7 but lower in Y2 during the whole growth (Fig. 7A). According to previous results, Y2 showed the bigger size and faster growth rate than Y7, indicating that genes in “brown” module probably negatively regulated the growth and development of Chinese cabbage. In this study, based on the WGCNA analysis, we divided these 5 DELLA genes into three different modules. Among them, BraA06gRGA is in the “turquoise” module, BraA05gRGL2 was located in the “red” module, and BraA09gRGA, BraA02gRGL1, and BraA10gRGL3 were in the “brown” module. The FPKM of BraA09gRGA, BraA02gRGL1, and BraA10gRGL3 were higher in Y7 than in Y2 (Fig. 7B). Therefore, we speculated that “brown” module was the key module that regulates growth and development of Chinese cabbage, and BraA09gRGA, BraA02gRGL1, and BraA10gRGL3 were key genes.
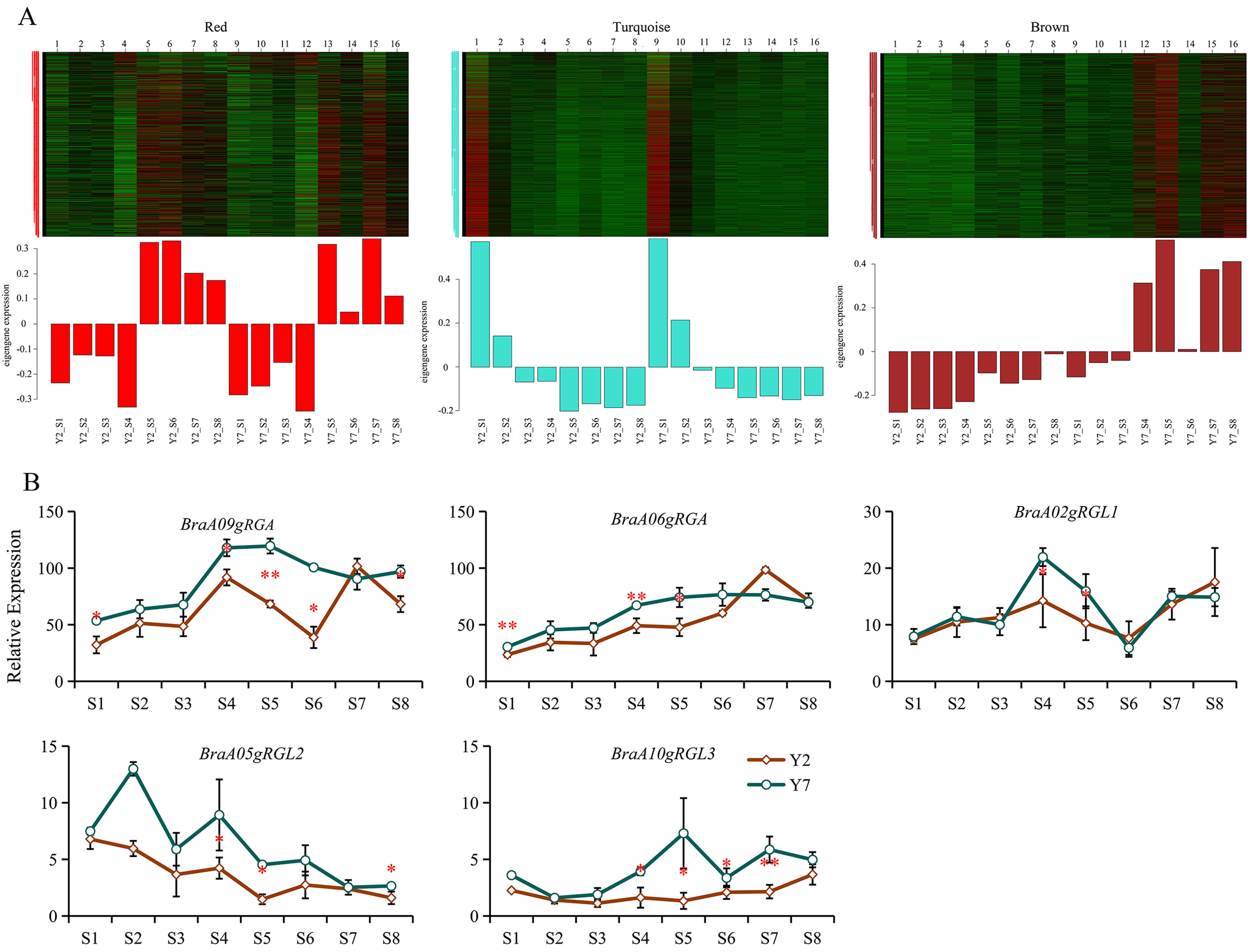 Fig. 7.
Fig. 7.Module analysis and expression pattern of BraDELLAs in
Y2 and Y7 at different stages of growth. (A) Module analysis expression pattern
of BraDELLAs in Y2 and Y7 at different stages of growth in RNA sequencing (RNA-Seq); (B)
expression pattern of BraDELLAs in Y2 and Y7 at different stages of
growth Asterisks on vertical bar shows significant difference at * p
To further understand the biological functions of the BraDELLAs in regulating the growth and development of Chinese cabbage and its response to environment stress, we selected Chinese cabbage high-generation self-crossing lines Y2 and Y7 as experimental materials, which have significant differences in size and stress resistance. As shown in the Fig. 8A,B, during the seedling stage, the cotyledons of Y2 were significantly larger than those of Y7, and the root system was thicker and longer than that of Y7. In previous studies, we also compared the size and growth rate of Y2 and Y7 plants during field growth and found that Y2 plants were larger than Y7 throughout the entire growth period, with a higher growth rate than Y7 [45]. Additionally, we compared the resistance of Y2 and Y7 to various stress conditions and found that under high temperature, low temperature, salt, and osmotic stress, the root length of Y7 was significantly longer than that of Y2, indicating that Y7 has higher stress resistance than Y2.
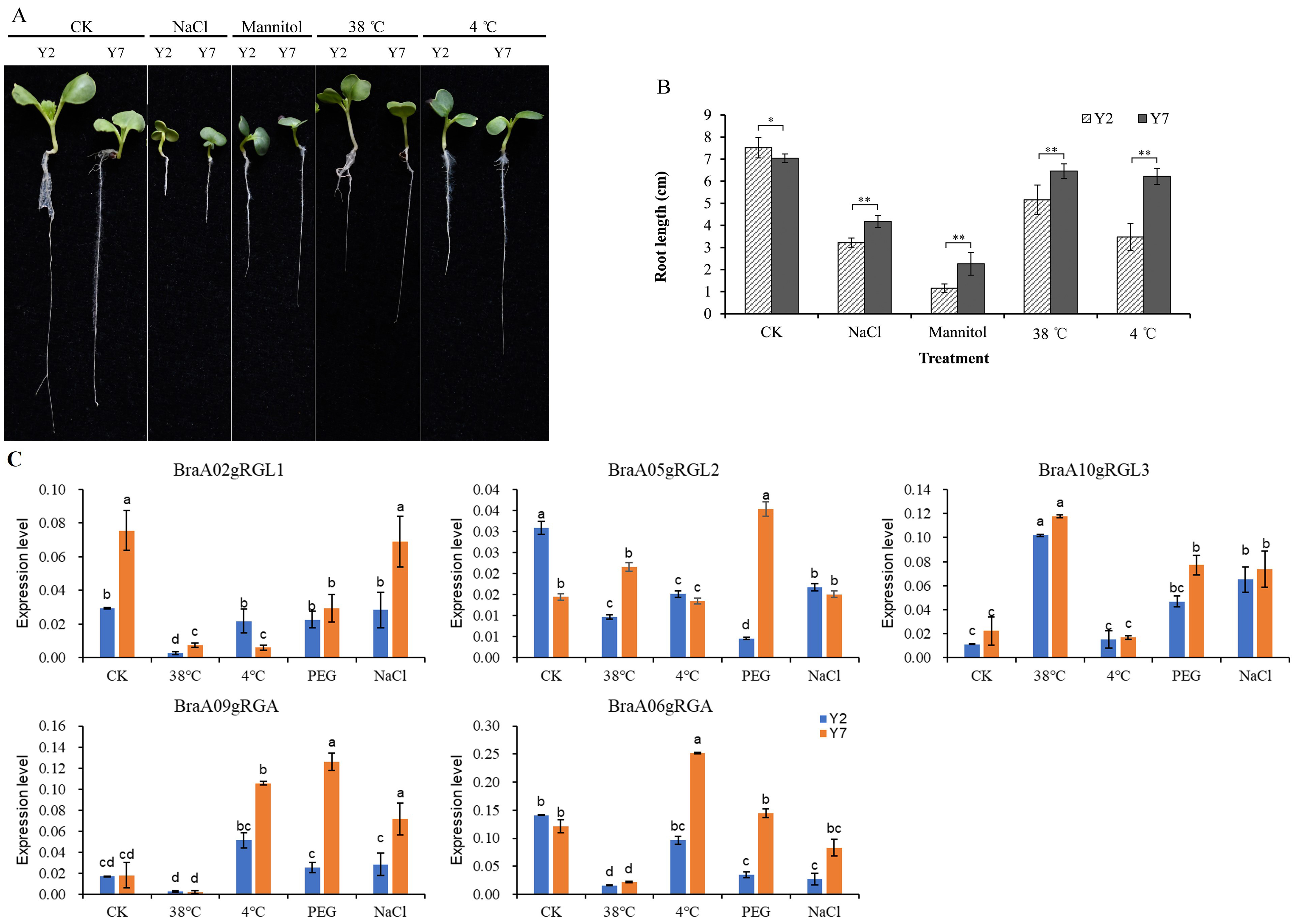 Fig. 8.
Fig. 8.Phenotypic analysis of Y2 and Y7 Chinese cabbage and
expression analysis of BraDELLAs under different abiotic stresses. (A)
Under different stress treatments, the phenotype difference between Y2 and Y7.
(B) The root length of Y2 and Y7 under different stress treatments; Asterisks on
vertical bar shows significant difference at * p
qRT-PCR results indicated that, there were significant expression differences between different members of BraDELLAs in Y2 and Y7, under stress treatments (Fig. 8C). Under cold treated, the expression level of BraA02gRGL1 both in Y2 and Y7 was down regulated, while BraA09gRGA is rapidly upregulated. Under high temperature, the expression levels of BraA06gRGA, BraA09gRGA, and BraA02gRGL1 significantly decrease in Y2 and Y7, while the expression level of BraA10gRGL3 significantly increases. Under osmotic stress (PEG), the expression level of BraA10gRGL3 significantly increased both in Y2 and Y7, while the expression level of BraA09gRGA only significantly increased in Y7. The NaCl increased the BraA10gRGL3 and BraA09gRGA expression. In addition, the expression level of BraA06gRGA in Y2 decreases under salt treatment. In general, different BraDELLAs play different role in the response to the environmental stress but were more sensitive to temperature stress. BraA10gRGL3 showed the most pronounced response to various stresses, especially in Y7. This aligns with the observation that the plant is larger in size when compared to the Y2 inbred line than the Y7 inbred line. However, its ability to respond to adverse stress is inferior to that of Y7. Therefore, BraA10gRGL3 of Y7 will also be the focus of our next research.
To further investigate the functional differences between the BraA10gRGL3-Y2 and BraA10gRGL3-Y7, we constructed plant overexpression vectors for BraA10gRGL3-Y2 and BraA10gRGL3-Y7 and transformed them into Arabidopsis. After 2–3 generations of self-pollination and screening, we obtained homozygous transgenic lines overexpressing the genes. Phenotypic observations revealed that the overexpression of BraA10gRGL3-Y2 in Arabidopsis (OE-Y2) did not show any significant differences compared to the WT (Fig. 9A). However, the leaves of the overexpressing BraA10gRGL3-Y7 Arabidopsis (OE-Y7) were smaller in size and had lower fresh weight compared to the WT (Fig. 9B), indicating that OE-Y7 can suppress plant growth, while OE-Y2 does not have a significant role in regulating plant growth.
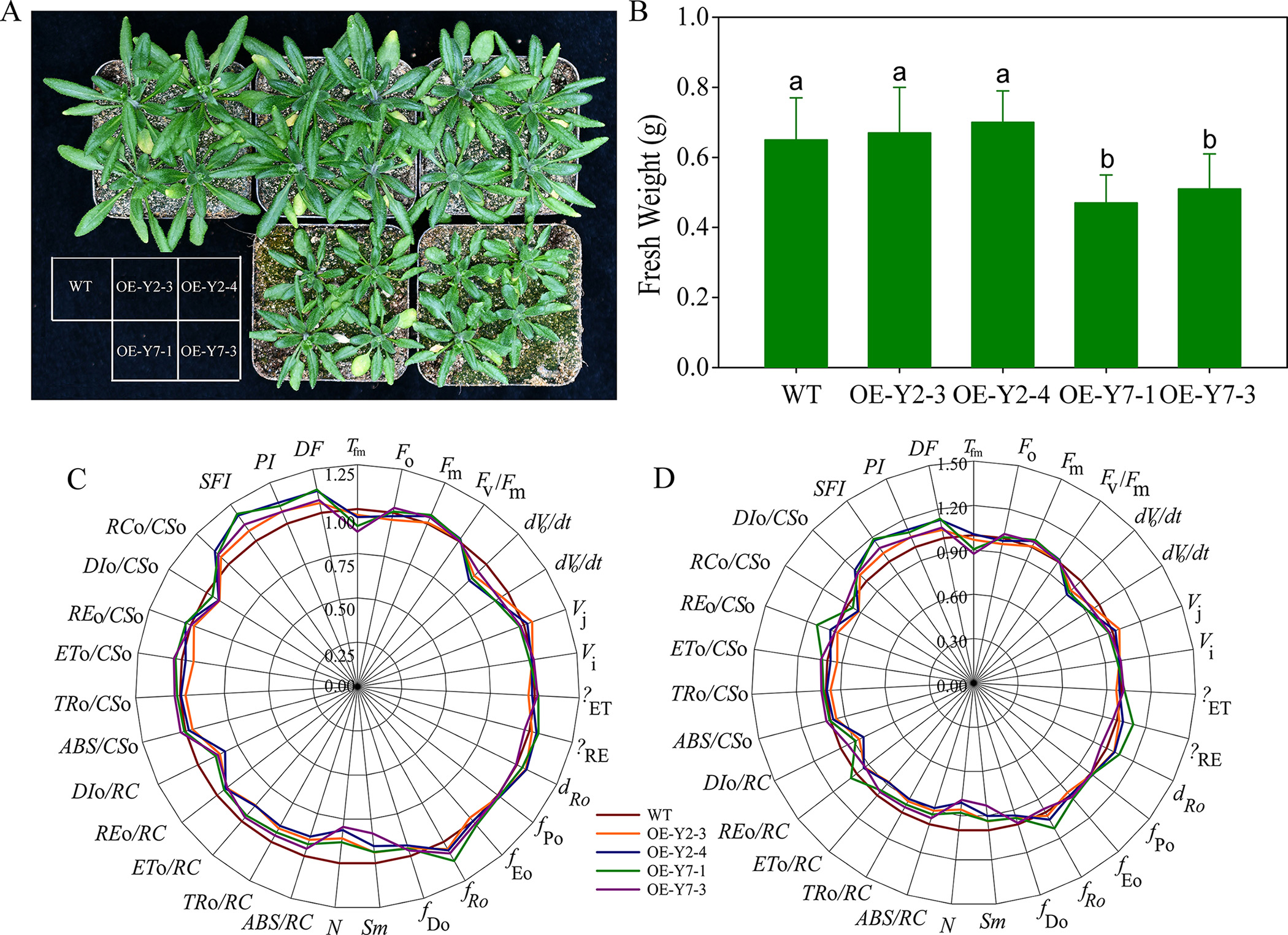 Fig. 9.
Fig. 9.Phenotypic and physiological-biochemical analysis of
Arabidopsis with the overexpression of BraA10gRGL3. (A)
Phenotypic analysis of Arabidopsis with the overexpression of
BraA10gRGL3; (B) Fresh weight of Arabidopsis with the
overexpression of BraA10gRGL3. OE-Y2-3 and OE-Y2-4 represent two lines
of BraA10gRGL3-Y2 transgenic Arabidopsis respectively; OE-Y7-1 and
OE-Y7-3 represent two lines of BraA10gRGL3-Y7 transgenic Arabidopsis
respectively.
We further compared the differences between transgenic BraA10gRGL3 Arabidopsis and WT under 38 °C and Control (room temperature treatment),
respectively. The results showed that compared to control, significant changes
occurred in the fluorescence parameters of transgenic BraA10gRGL3 Arabidopsis and WT under 38 °C stresses (Fig. 9C,D). The Tfm of OE-Y2 and
OE-Y7 was no significant changes compared to WT. The Fo (minimum
fluorescence intensity after dark adaptation) of OE-Y2 and OE-Y7 were
significantly lower than of the in WT, while the Fm (maximum
fluorescence intensity after dark adaptation) was slightly higher than of them in
WT, indicating that it has less heat dissipation. The dV/dt
Sequence alignment revealed that BraA04gGID1b1, BraA07gGID1b2, and BraA06gGID1c1 have identical sequences in both Y2 and Y7 Chinese cabbage, while BraA05gGID1a and BraA09gGID1b3 showed significant sequence differences between the two inbred lines (Supplementary Fig. 1). To investigate the interaction between BraA10gRGL3-Y2, BraA10gRGL3-Y7, and the BraGID1 family proteins, we constructed the BraA10gRGL3-Y2 and BraA10gRGL3-Y7 genes into the AD vector, and subsequently inserted the BraGID1 gene family from Chinese cabbage Y2 and Y7 into the BD vector. The yeast two-hybrid experiment showed that the AD and BD plasmids containing the target gene fragments did not exhibit self-activation. Specifically, BraA10gRGL3-Y7 interacted stably with BraA05gGID1a-Y7, BraA04gGID1b1, BraA09gGID1b3-Y2, and BraA06gGID1c, while BraA10gRGL3-Y2 did not interact with the BraGID1 family proteins (Fig. 10). We speculated that the differences in amino acid sequences between BraA10gRGL3-Y2 and BraA10gRGL3-Y7 may result in variations in their protein binding sites, thus affecting their interaction with the BraGID1 family proteins. These findings suggested that these two gene segments have distinct functions, providing a solid foundation for further investigation of key interaction sites.
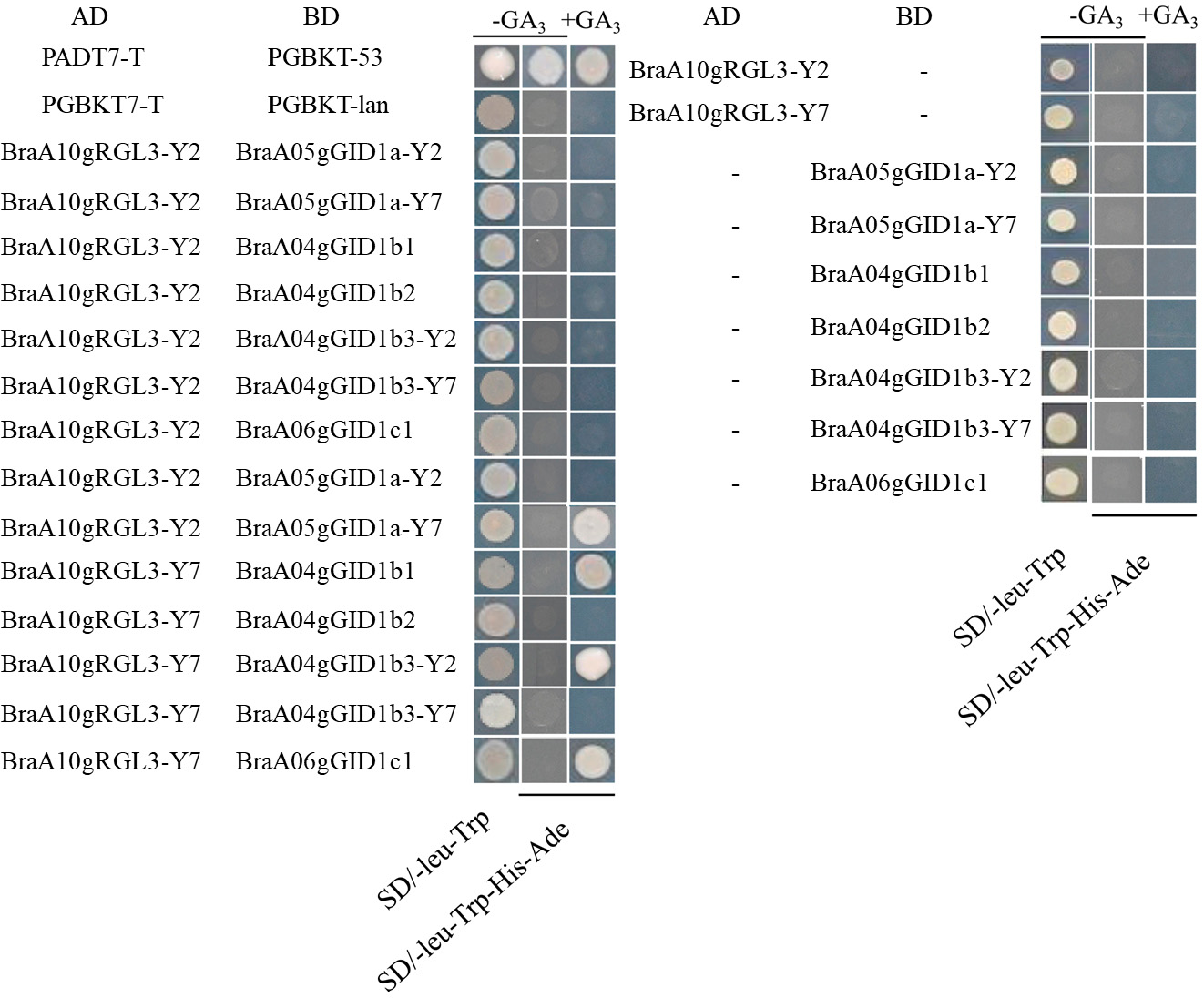 Fig. 10.
Fig. 10.Interaction of BraA10gRGL3-Y2 and BraA10gRGL3-Y7 with BraGID1 family proteins.
Chinese cabbage is one of the essential vegetables in the world. They are the most widely grown vegetables in China and northern areas and account for over one-quarter of the total annual vegetable consumption. The resistance and quality of Chinese cabbage are influenced by various environmental conditions. During environmental influences, DELLAs response to enhance plant survival and confine growth [39]. DELLA protein is an important growth regulator [46] that involved in various signaling pathway, such as environmental stress responsive signals [47, 48] as well as hormonal biosynthesis. Whether DELLAs can make contributions in plant developmental process and enhance abiotic stress tolerance in B. rapa remained unclear. If the network mechanism of DELLA regulation in Chinese cabbage can be clearly understood, it will provide a theoretical basis for improving the quality and resistance of Chinese cabbage through molecular-assisted breeding methods, and thus obtain higher economic value.
Currently, the regulatory mechanism of DELLA has been studied more thoroughly in other plants. In Arabidopsis, 5 DELLA genes have been identified, mainly involved in flower development and seed germination [16]. In rice, 1 DELLA gene has been identified, mainly involved in stem elongation. In soybean, 7 DELLA genes have been identified, mainly involved in seed formation. In strawberry, DELLA affects asexual reproduction, while in other plants, DELLA genes also participate in regulating leaf senescence, flowering, and wood formation [49, 50]. In rice and maize, 1 DELLA gene has been identified in each, mainly participating in plant dwarfing [37, 51]. In this study, we used the bioinformatics to analyze the physicochemical properties of the DELLA gene family in Chinese cabbage. 5 BraDELLAs were identified from the B. rapa genome and grouped into three subfamilies based on their homology, namely BraA02gRGL1, BraA05gRGL2, BraA10gRGL3, BraA06gRGA, and BraA09gRGA. Their pI ranges from 4.83 to 5.58, all of which are acidic proteins. Through sequence alignment and phylogenetic tree construction with rice, wheat, maize, cabbage, and rapeseed, we found that the 5 DELLA proteins in Chinese cabbage have a high homology with DELLA proteins in Arabidopsis, cabbage, and rapeseed, but a lower homology with wheat, maize, and rice. Previously, the expansion of DELLA protein family was found widely contributed to abiotic stress tolerance and development and growth of plants [42, 52]. Recent studies reported that AtRGA and AtGAI are involved in flower induction, cell division in hypocotyl and promote plant growth [12]. AtRGLs have great responsive to a lot of plant hormones [18], thus lead to activate plant defense mechanism to reduce the harmful effects of abiotic stress [53]. The phylogenetic analysis suggested that AtRGL3 is closely related to BraA10gRGL3 while AtRGL1 and BraA02gRGL1 are in the same evolutionary branch. The DELLA proteins from Arabidopsis and Brassica rapa are closely related which suggested they may have similar molecular mechanism during stress conditions. In order to investigate the role of the DELLA protein in the growth and development of Chinese cabbage, we conducted an analysis of its expression patterns by combining the transcription analysis according to our previous study [45]. We found that the expression pattern of BraA02gRGL1, BraA05gRGL2, and BraA10gRGL3 in different development stages of Y2 and Y7 were consistent to the “brown” module, indicating the important roles of them in the growth and development of Chinese cabbage. In addition, the leaf size and growth rate were bigger and higher in Y2 than them in Y7, which is opposite to the expression pattern of “brown” module. It was probably that the genes in “brown” module including BraA02gRGL1, BraA05gRGL2, and BraA10gRGL3 negatively regulated the growth and development of Chinese cabbage. Previous studies have also reached similar conclusions. When model plants such as Arabidopsis and rice overexpress the DELLA gene, they exhibit dwarfing traits. After introducing the plum DELLA gene into Arabidopsis, the leaf size reduces, flowering is delayed, and embryonic axis growth is restricted [54]. In transgenic MdRGL1a tobacco, the root system significantly decreases, and the plant becomes dwarfed [55]. This is consistent with the results obtained in this experiment, which indicate that the BraA10gRGL3-Y7 protein plays a negative regulatory role in GA signaling in Chinese cabbage, thereby inhibiting growth and development processes.
According to the phenotype analysis of Chinese cabbage under stress conditions, Y7 showed a stronger tolerance than Y2. Meantime, the expression of BraA10gRGL3 in Y7 was significantly higher than CK and Y2 after heat stress treated. This is consistent with the fact that Y7 has stronger heat resistance than Y2. In addition, the overexpression of BraA10gRGL3-Y7 gene can significantly enhance the heat tolerance of Arabidopsis, while the heat resistance of the BraA10gRGL3-Y2 gene overexpression was not obvious. Yeast two-hybrid experiments have revealed distinct interaction patterns between BraA10gRGL3-Y2 and BraA10gRGL3-Y7 with GID1 proteins. Specifically, BraA10gRGL3-Y7 showed a stable interaction with BraGID1a-Y7, BraGID1b1, and BraA09gGID1b3-Y2, whereas BraA10gRGL3-Y2 did not interact with the BraGID1 protein family. These variances in interaction implied potential disparities in their respective biological functions. In recent years, more DELLA proteins have been reported to enhance plant stress tolerance. Arabidopsis CBF3 and DELLA can interact with each other to respond to low-temperature stress [15]. DELLA-mediated degradation of PIF 4 regulates auxin biosynthesis under high-temperature conditions [56]. RGL3 responds to low-temperature stress by participating in LCBK2 signaling regulation [57]. This study found that BraA10gRGL3 can enhance plant heat resistance, confirming the view that DELLA can improve plant stress tolerance and laying the foundation for further exploration of new functions of DELLA. Therefore, our research provides clues for improving plant varieties to adapt to expected environmental conditions using molecular breeding technology. The growth and development of plants are influenced by both hormones and the external environment. The DELLA protein, an integrative factor of multiple hormone and environmental signal systems, plays a crucial role. DELLA protein genes have been cloned in numerous plants. Their expression can be regulated through hormone treatments, environmental stress, transgenic technology, and other methods. This regulation impacts seed germination, seedling growth, plant height, flowering period, fruit quality, crop yield, secondary metabolite content, resistance, and disease resilience. Studying the DELLA protein has become increasingly popular due to its significant role. Under adverse conditions, the DELLA protein inhibits plant growth and development, enhancing its resistance to adversity. This finding aligns with the results of this study. This research provides a theoretical foundation for breeding heat-resistant cabbage through molecular biology and suggests the potential for broader application of the DELLA protein in future modern agricultural production.
DELLA protein is a key inhibitory factor in GA signal transduction and a negative regulatory factor involved in plant growth and development. Currently, the DELLA gene has been cloned from dozens of plants, and it has been found to be an integrating factor for multiple hormone and environmental signal systems, regulating physiological processes such as seed germination, seedling growth, plant height, flowering time, crop yield, secondary metabolite content, stress resistance, and disease resistance. In this study, we conducted bioinformatics analysis and functional validation of BraDELLAs. In this study, five DELLA genes were identified in Brassica rapa through genome wide analysis. The comparative analysis and genetic evolution revealed BraDELLAs had similar structure and function as AtDELLAs. We conducted transcriptomics, Phenotypic analysis of Chinese cabbage under abiotic stress, and overexpression on the regulation of growth and development and abiotic stress tolerance of Chinese cabbage. The result showed that BraA10gRGL3-Y7 play an important role in the regulation of the tolerance of Chinese cabbage to the temperature stress but negatively regulated the development and growth of Chinese cabbage. This information will help to better understand the BraDELLAs biological function in stresses resistance and development and growth of Chinese cabbage and provides the theoretical basis and technical support for improving the variety of Chinese cabbage through molecular-assisted breeding methods, aiming to increase its yield and stress resistance.
We are grateful to the NCBI database (https://www.ncbi.nlm.nih.gov/cdd/) for providing the data. All of our data can be accessed through the corresponding author Fengde Wang.
Supervised and conceived this project (FW and JG); conceptualization and design of the manuscript (LW and QZ); wrote the paper and revised the paper (YY, LW and QZ); performed the formal analysis (XL, JL, and NQ); carried out the experiments of mutant construction (YY, CLiu, XF, MQ, YZ and CLi). All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Chinese cabbage of high generation inbred line “Y2” and “Y7” were used in this study. Y2 and Y7 are advanced inbred lines independently selected in our laboratory (Molecular Breeding Lab, Shandong Branch of National Vegetable Improvement Center, Institute of Vegetables, Shandong Academy of Agricultural Science, Jinan, China).
We would also like to thank all the colleagues in our group who have supported us.
This study was supported by the Nature Foundation of Shandong Province, China (ZR2022QC113); the Formation Mechanism and Regulation of Fruit and Vegetable Quality in Sa-line-alkali Land (GYJ2023004); the Taishan Scholars Program of Shandong Province, China (tsqn201909167); the National Natural Science Foundation, China (32172591); the Natural Science Foundation of Shandong Province, China (ZR2020MC144); the Modern Agricultural Industrial Technology System Funding of Shandong Province, China (SDAIT-05); and the Prospect of Shandong Seed Project, China (2022LZGC008).
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.










