1 Division of Gastroenterology & General Surgery, Department of Surgery, Chi Mei Medical Center, 710 Tainan, Taiwan
2 Department of Medical Technology, Chung Hwa University of Medical Technology, 717 Tainan, Taiwan
3 Institute of Biomedical Sciences, National Sun Yat-sen University, 804 Kaohsiung, Taiwan
4 Department of Pharmacology, School of Medicine, China Medical University, 404333 Taichung, Taiwan
5 Department of Clinical Pathology, Chi Mei Medical Center, 710 Tainan, Taiwan
6 Division of Urology, Department of Surgery, Chi Mei Medical Center, 710 Tainan, Taiwan
7 Department of Medical Science Industries, College of Health Sciences, Chang Jung Christian University, 711 Tainan, Taiwan
8 Division of Colon and Rectal Surgery, Department of Surgery, Chi Mei Medical Center, 710 Tainan, Taiwan
9 Institute of Precision Medicine, National Sun Yat-sen University, 804 Kaohsiung, Taiwan
10 Department of Medical Research, Chi Mei Medical Center, 710 Tainan, Taiwan
11 National Institute of Cancer Research, National Health Research Institutes, 704 Tainan, Taiwan
12 Trans-Omic Laboratory for Precision Medicine, Chi Mei Medical Center, 710 Tainan, Taiwan
13 Division of Hematology and Oncology, Department of Internal Medicine, Chi-Mei Medical Center, 71004 Tainan, Taiwan
14 College of Pharmacy and Science, Chia Nan University, 71710 Tainan, Taiwan
Abstract
Background: Cholangiocarcinoma (CCA) is a malignant tumor with an
increasing incidence worldwide. Although radiation therapy has improved the
therapeutic efficiency of CCA treatment, differential expression of genes among
cholangiocarcinoma subtypes has been revealed through precise sequencing.
However, no specific molecular therapeutic targets or biomarkers have been
figured out for use in precision medicine, and the exact mechanism by which
antitumorigenic effects occur is still unclear. Therefore, it is necessary to
conduct further studies on the development and mechanisms associated with CCA.
Methods: We examined the clinical data and pathological features of
patients with cholangiocarcinomas. We investigated the associations between DNA
Topoisomerase II Alpha (TOP2A) expression and patient outcomes, such as
metastasis-free survival (MFS) and disease-specific survival (DSS), as well as
clinical characteristics and pathological results. Results:
TOP2A expression was shown to be upregulated in CCA tissue sections by
immunohistochemistry staining and data mining. Moreover, we observed that the
TOP2A expression correlated with clinical features, such as the primary
tumor stage, histological variants, and patients with hepatitis. Furthermore,
high expression of TOP2A was associated with worse survival outcomes in
terms of the overall survival (p
Keywords
- cholangiocarcinoma
- DNA topoisomerase (ATP-hydrolyzing activity) activity
- topoisomerase II α (TOP2A)
- prognostic biomarker
Cholangiocarcinoma, which is a type of cancer that affects the epithelial cells, can arise from various locations of the bile duct [1]. Depending on its location, cholangiocarcinoma can be classified as intrahepatic, perihilar, or distal. Intrahepatic cholangiocarcinoma starts from the secondary bile ducts located close to the liver, while perihilar cholangiocarcinoma occurs between the cystic duct and the secondary bile ducts. Distal cholangiocarcinoma is found between the ampulla and the origin of the cystic duct [2, 3]. According to previous studies, the risk factors for cholangiocarcinoma (CCA) are sporadic and are correlated with geographic variations [4, 5]. For example, infection with hepatobiliary flukes is associated with chronic inflammation and is also correlated with the development of CCA in individuals from Southeast Asia [6]; Hepatolithiasis is also a risk factor of developing CCA (mainly intrahepatic cholangiocarcioma (iCCA)) in Asians [5]. In the West, primary sclerosing cholangitis (PSC) carries the greatest risk for patients with CCA [7, 8]. Accordingly, CCA is diagnosed within two years of developing PSC. Although risk factors for CCA development, such as smoking and alcohol, have been identified in PSC patients, direct research data are lacking [9].
The TOP2A gene is responsible for regulating the chromosome bond
pathway by encoding DNA topoisomerase, which in turn controls the topological
state of DNA transcription and replication [10]. Studies have linked the
expression of TOP2A with various levels of cancer progression and the
development of nasopharyngeal carcinoma [11], breast cancer [12], adrenal cancer
[13], and endometrial cancer [14]. Abnormal expression of TOP2A is often
linked to irregular cell proliferation, while decreased expression of the gene
can lead to changes in several molecular signaling pathways, such as the
In recent years, few studies involving genetic factors (tumor suppressors and oncogenes) and epigenetic alterations associated with CCA progression have been con-ducted [18]. However, there is no definitive result to identify the major oncogenes or suppressor genes associated with CCA development. In this study, we explore the molecular role of TOP2A in CCA and try to identify the biological functions of TOP2A in CCA progression.
The gene expression profile of cholangiocarcinoma (GSE26566) was investigated
through the Gene Expression Omnibus (GEO) database. Significant gene expression
changes were identified, particularly those in molecular pathways involving DNA
topoisomerase (ATP-hydrolyzing activity) activity in Gene Ontology (GO:2000371),
through comparative and functional analyses. The databases were analyzed, and
focus was placed on p-values of
The Chi Mei Medical Center enrolled 182 iCCA patients who underwent curative surgery between 1990 and 2010. Patients with metastatic disease or nodal metastases were excluded. This study was authorized by the Institutional Review Board (IRB) of The Chi Mei Medical Center with approval number 09912003. All participants provided their informed consent. Patients’ demographics and clinical details was collected retrospectively, including pathological characteristics, oncological survival follow-up, and cause of death. The Tumor, Node, Metastasis (TNM) system created by the eighth edition American Joint Committee on Cancer (AJCC) in 2017 was used to measure the tumor stage. Two pathologists examined the tumor samples.
The formalin-fixed tissues were embedded in paraffin and sectioned into 10
The relative expression and knockdown efficacy of the TOP2A gene in iCCA cell lines were determined using RT-PCR, as previously published [19]. Total RNA was collected from iCCA cell lines, and real-time RT-PCR was employed to determine the transcription level of TOP2A. The mRNA abundance of TOP2A (Hs01032137_mL) was assessed using predesigned TaqMan assay reagents (Cat. No. Hs01032137_mL, Applied Biosystems, Waltham, MA, USA) and the ABI StepOnePlusTM system (Applied Biosystems, Waltham, MA, USA), with POLR2A (Hs01108291_mL) used as the internal control for normalization.
The SNU1079 and SNU1196 cell lines were procured from a cell bank based in
Seoul, South Korea. Initially, these cell lines were cultured in ACL-4 medium
supplemented with 5% heat-inactivated fetal bovine serum. After establishment,
the cell cultures were maintained in RPMI 1640 medium and supplemented with 10%
heat-inactivated fetal bovine serum. These cells were cultured under controlled
conditions in a humidified incubator at 37 °C in the presence of 5%
CO
The SNU1196 and Huh28 cell lines were modified to create stable TOP2A-silenced clones using lentiviral vectors pLKO.1-shLacZ (TRCN0000072223: 5′-TGTTCGCATTATCCGAACCAT-3′) and pLKO.1-shTOP2A (#1: TRCN0000049280: 5′-GCTCCAAATCAA TATGTGATT-3′; #2: TRCN0000049278: 5′-GCCCAA GTGTTCTTTAGCTTT-3′), which were obtained from the Taiwan National RNAi Core Facility in Taipei, Taiwan. These cell lines initially had high TOP2A expression that was reduced by shRNAs against TOP2A (shTOP2A). To generate viruses for the modification, HEK293 cells were transfected with the three vectors mentioned above, using Lipofectamine 2000 from Thermo Fisher Scientific in Waltham, MA, USA, as previously described [19].
Primary antibodies against TOP2A (clone AA6, 1:500; Millipore, Beverly,
MA, USA) were utilized, and a previously reported western blotting technique was
utilized to assess TOP2A expression and the effect of TOP2Aknockdown in SNU1196 and Huh28 cell lines. Glyceraldehyde 3-phosphate
dehydrogenase (GADPH) was used as a control for protein loading (6C5, 1:10,000;
Millipore, Beverly, MA, USA) [22]. To immobilize the protein, cell lysates
containing 25
The enzyme-linked immunosorbent assay (ELISA)-based and colorimetric
bromodeoxyuridine (BrdU) assay (Roche Holding AG, Basel, Switzerland) was
utilized to quantify DNA synthesis. At 24, 48, and 72 hours, the amount of DNA
synthesis was measured in the TOP2A-knockdown or shLacA control SNU1196
and Huh28 cell lines. After three hours of BrdU incubation at 37 °C and
5% CO
The experimental protocol for cell migration and invasion was carried out in accordance with a previously documented procedure [22]. For the cell invasion experiment, the 24-well Collagen-based Cell Invasion Assay from Millipore, Beverly, MA, USA, and Falcon HTS FluoroBlok 24-well inserts from BD Biosciences, Franklin Lakes, NJ, USA, were used. The inserts were rehydrated using serum-free medium and then placed in the upper chamber, which contained a serum-free suspension with an equal number of cells. Over a 12- to 24-hour incubation period, the cells were allowed to migrate towards the lower chamber, which contained a medium with 10% fetal bovine serum. Following removal of the non-invading cells from the upper chamber, the invading cells were stained, lysed in extraction buffer, and then transferred to 96-well plates for 560 nm colorimetric readings.
In order to determine the unknown functions of TOP2A in iCCA, the transcription level of TOP2A and its coexpressed genes contained in the cholangiocarcinoma dataset (n = 51, Firehose Legacy, TCGA) were analyzed to establish correlations. Subsequently, the top 200 differentially expressed genes with positive or negative correlations with TOP2A were selected for functional annotation using the Gene Ontology (GO) classification system and rated by fold enrichment.
The statistical analyses were conducted using SPSS software (version 28, IBM Corp., Chicago, IL, USA). The Chi-square test was utilized to evaluate the association between TOP2A expression stage and clinicopathologic characteristics. Local recurrence-free survival (LRFS), metastasis-free survival (MFS), and disease-specific survival (DSS) were calculated from the beginning of therapy to the date of the event, and the latest follow-up date was noted for patients who were lost to follow-up during the study period. Kaplan-Meier analysis was employed to generate survival curves, and log-rank tests were utilized to identify prognostic differences between groups. All analyses were performed using two-sided significance tests, and a p-value less than 0.05 was considered significant.
We mined the public CCA transcriptome dataset (GSE26566) and compared the
results with DNA topoisomerase (ATP-hydrolyzing activity) activity (GO:2000371).
We discovered that five genes were significantly associated with DNA
topoisomerase (ATP-hydrolyzing activity) activity (Table 1), and TOP2Awas found to be a high-ranking differently expressed candidate gene that showed a
significant difference (p
| Probe | CCA vs Non-tumor |
CCA vs Normal intrahepatic bile duct |
Gene Symbol | Molecular function | Biological process | ||
| log ratio | p-value | log ratio | p-value | ||||
| ILMN_1686097 | 1.7717 | 1.7928 | TOP2A | DNA topoisomerase (ATP-hydrolyzing) activity, protein homodimerization activity, histone deacetylase binding, protein heterodimerization activity, ATP binding, chromatin binding, DNA-dependent ATPase activity, nucleotide binding, protein C-terminus binding, ubiquitin binding, protein kinase C binding, drug binding | DNA ligation, DNA topological change, positive regulation of apoptosis, phosphoinositide-mediated signaling, DNA repair, chromosome segregation, DNA replication, apoptotic chromosome condensation | ||
| ILMN_1777663 | 0.3019 | 0.0018* | 0.339 | 0.1852 | TOP2B | DNA topoisomerase (ATP-hydrolyzing) activity, histone deacetylase binding, protein heterodimerization activity, ATP binding, chromatin binding, nucleotide binding, protein C-terminus binding, protein kinase C binding | DNA topological change |
| ILMN_1659651 | 0.2556 | 0.6474 | TOP1MT | DNA topoisomerase (ATP-hydrolyzing) activity, DNA topoisomerase type I activity | DNA topological change | ||
| ILMN_1796508 | 0.185 | 0.0010* | 0.4342 | 0.0122* | |||
| ILMN_1735572 | 0.0875 | 0.1265 | 0.1408 | 0.3608 | TOP1 | DNA topoisomerase (ATP-hydrolyzing) activity, chromatin binding, DNA topoisomerase type I activity, protein binding | DNA topological change |
| ILMN_1687970 | –0.1256 | 0.0006* | –0.1417 | 0.1639 | SPO11 | DNA topoisomerase (ATP-hydrolyzing) activity, hydrolase activity, DNA binding, ATP binding | DNA topological change, female gamete generation, spermatogenesis, meiotic recombination, meiosis |
| ILMN_1796655 | –0.1309 | 0.0009* | –0.0693 | 0.5052 | DNA topological change, female gamete generation, spermatogenesis, meiotic recombination, meiosis | ||
| #, Comparing cholangiocarcinoma (CCA, n = 104) to surrounding liver (n = 59) and normal intrahepatic bile duct (n = 6); &, Comparing cholangiocarcinoma (CCA, n = 104) to normal intrahepatic bile duct (n = 6); * statistically significant. | |||||||
 Fig. 1.
Fig. 1.Data mining of genes expression compared between cholangiocarcinoma, surrounding liver and non-tumor biliary epithelium from the public domain (GSE26566).
Our previous results indicate that high expression of TOP2A might be correlated with CCA progression. We further analyzed associations between TOP2A expression and the clinicopathologic characteristics of CCA patients. We included data from 182 iCCA patients, 108 males and 74 females, where 107 patients were older than 65 and 75 patients were younger than 65. These cases were classified as 105 of the large duct type and 77 of the small duct type based on histological variants; 61 cases were classified as well-differentiated, 66 as moderately differentiated, and 55 as poorly differentiated according to histological grades. A total of 163 cases were classified as R0 and 19 cases were classified as R1 based on the surgical margin, and in terms of the primary tumor size, 87 cases were classified as T1, 61 as T2, and 34 as T3 (Table 2). Taken together, our results indicate that TOP2A overexpression is highly correlated with the co-occurrence of hepatitis (p-value = 0.002), the primary tumor size (p-value = 0.001), and the histological variants (p-value = 0.002) of clinicopathological parameters in cholangiocarcinoma patients with iCCA (Table 2). Moreover, we determined that TOP2A is overexpressed in cholangiocarcinoma patients (Fig. 1). We confirmed this by IHC staining of cholangiocarcinoma patient tissue sections (Fig. 2). Moreover, we calculated the immunohistochemical staining H-score which showed significantly higher in iCCA cells than hepatocytes and cholangiocytes (Fig. 3). These data suggest that TOP2A expression is significantly associated with clinicopathological variables in iCCA patients.
| Parameter | Category | Case No. | TOP2A expression | p-value | |
| Low | High | ||||
| Gender | Male | 108 | 51 | 57 | 0.365 |
| Female | 74 | 40 | 34 | ||
| Age (years) | 107 | 49 | 58 | 0.175 | |
| 75 | 42 | 33 | |||
| Hepatitis | Hepatitis B | 72 | 38 | 34 | 0.002* |
| Hepatitis C | 29 | 22 | 7 | ||
| Non-B, non-C | 81 | 31 | 50 | ||
| Intrahepatic lithiasis | Not identified | 102 | 56 | 46 | 0.135 |
| Present | 80 | 35 | 45 | ||
| Surgical margin | R0 | 163 | 84 | 79 | 0.225 |
| R1 | 19 | 7 | 12 | ||
| Primary tumor (T) | T1 | 87 | 56 | 21 | 0.001* |
| T2 | 61 | 24 | 37 | ||
| T3 | 34 | 11 | 23 | ||
| Histological variants | Large duct type | 105 | 42 | 63 | 0.002* |
| Small duct type | 77 | 49 | 28 | ||
| Histological grade | Well differentiated | 61 | 31 | 30 | 0.887 |
| Moderately differentiated | 66 | 34 | 32 | ||
| Poorly differentiated | 55 | 26 | 29 | ||
| * Statistically significant. | |||||
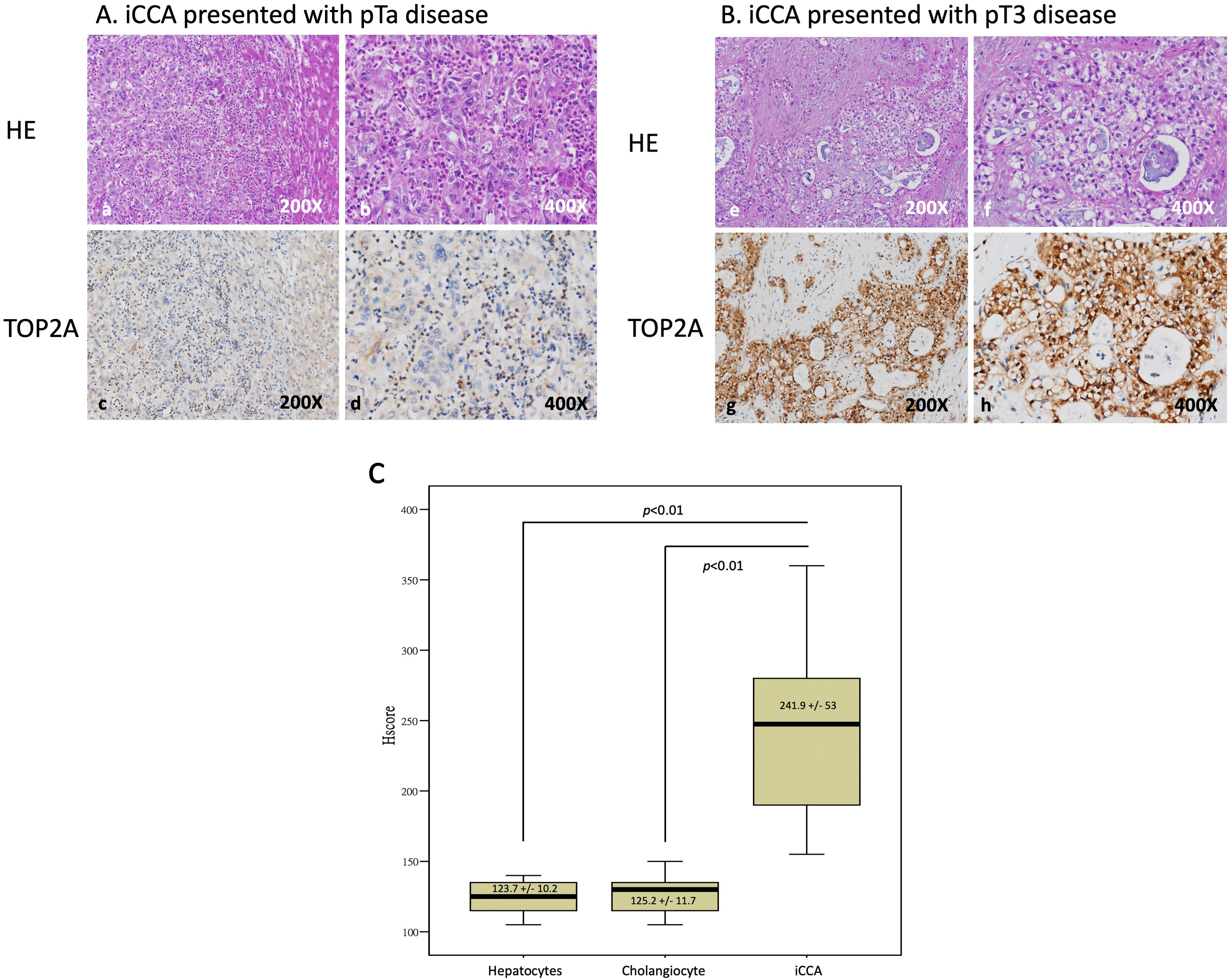 Fig. 2.
Fig. 2.Identify the TOP2A expression in iCCA tissues. (A) In low-stage tumor tissues, TOP2A positive cells are weak detected (a–d). (B) In high-stage tumor tissues, tumor cells showed positive TOP2A staining (e–h). (C) A comparison of H-score showed significantly higher TOP2A expression in iCCA cells than hepatocytes and cholangiocytes (N = 182 in each group).
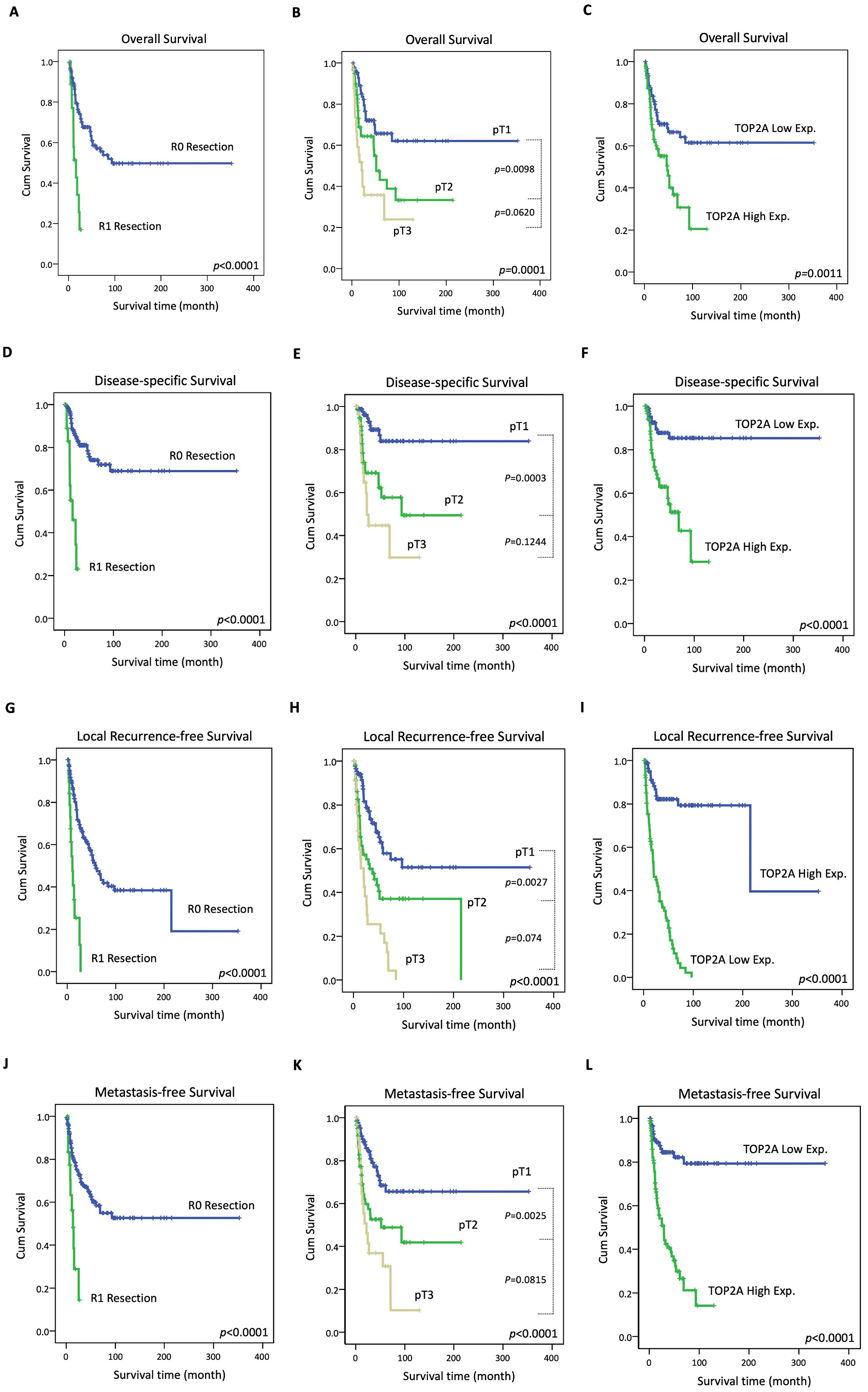 Fig. 3.
Fig. 3.Kaplan–Meier survival curves in the CCA patients according to surgical margin, primary tumor staging and TOP2A expression. R0 Resection, pT1 stage and low TOP2A expression correlated with better overall survival (A,B,C), better disease-specific survival (D,E,F), better local recurrence-free survival (G,H,I) and better metastasis-free survival (J,K,L).
To determine whether TOP2A expression is correlated with CCA patients’ survival outcomes, we analyzed the association between TOP2A expression and the overall survival rate, disease-specific survival rate, local recurrence-free survival rate, and metastasis-free survival rate in CCA patients. For overall survival, an R0 stage surgical margin, T1 stage primary tumor, and low TOP2A expression (Fig. 3A–C) were associated with better outcomes in both the univariate and multivariate analyses (Table 3). The dis-ease-specific survival endpoints analysis showed the same results whereby R0 stage, T1 stage, and low TOP2A expression patients had better outcomes (Table 3) (Fig. 3D–F) than other patients. Moreover, patients with a T1 stage primary tumor, large duct type histological variant, and histological low grade were shown to have better outcomes in the univariate analysis of local recurrence-free survival (Table 4). Importantly, an R0 stage surgical margin, T1 stage primary tumor, and low TOP2A expression were all significantly associated with better outcomes (Fig. 3G–I) in both the univariate analysis and the multivariate analysis of local recurrence-free survival and metastasis-free survival (Table 4) (Fig. 3J–L).
| Parameter | Category | Case No. | Overall survival | Disease-specific survival | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||||||
| No. of event | p-value | R.R. | 95% CI | p-value | No. of event | p-value | R.R. | 95% CI | p-value | |||
| Gender | Male | 108 | 50 | 0.0254* | 1 | - | 0.087 | 9 | 0.0072* | 1 | - | 0.044* |
| Female | 74 | 21 | 1.569 | 0.937–2.629 | - | 32 | 2.158 | 1.021–4.558 | - | |||
| Age (years) | 107 | 37 | 0.2626 | - | - | - | 28 | 0.2125 | - | - | - | |
| 75 | 34 | - | - | - | 13 | - | - | - | ||||
| Hepatitis | Hepatitis B | 72 | 32 | 0.2379 | - | - | - | 16 | 0.4561 | - | - | - |
| Hepatitis C | 29 | 8 | - | - | - | 19 | - | - | - | |||
| Non-B, non-C | 81 | 31 | - | - | - | 6 | - | - | - | |||
| Intrahepatic lithiasis | Not identified | 102 | 36 | 0.2831 | - | - | - | 19 | 0.1613 | - | - | - |
| Present | 80 | 35 | - | - | - | 22 | - | - | - | |||
| Surgical margin | R0 | 163 | 59 | 1 | - | 0.002* | 31 | 1 | - | |||
| R1 | 19 | 12 | 2.913 | 1.466–5.789 | 10 | 4.962 | 2.196–11.213 | |||||
| Primary tumor (T) | T1 | 87 | 25 | 0.0001* | 1 | - | 0.037* | 9 | 1 | - | 0.020* | |
| T2 | 61 | 27 | 1.608 | 0.920–2.812 | - | 19 | 2.769 | 1.234–6.211 | - | |||
| T3 | 34 | 19 | 2.317 | 1.205–4.455 | - | 13 | 3.281 | 1.321–8.153 | - | |||
| Histological variants | Large duct type | 105 | 43 | 0.4281 | - | - | - | 27 | 0.1984 | - | - | - |
| Small duct type | 77 | 28 | - | - | - | 14 | - | - | - | |||
| Histological grade (Differentiation) | Well | 61 | 20 | 0.1663 | - | - | - | 12 | 0.3881 | - | - | - |
| Moderately | 66 | 28 | - | - | - | 16 | - | - | - | |||
| Poorly | 55 | 23 | - | - | - | 13 | - | - | - | |||
| TOP2A Exp. | Low expression | 91 | 28 | 0.0011 | 1 | - | 0.036*- | 10 | 1 | - | 0.002*- | |
| High expression | 91 | 43 | 1.714 | 1.035–2.838 | - | 31 | 1.929 | 1.198–3.886 | - | |||
| * Statistically significant. | ||||||||||||
| Parameter | Category | Case No. | Local recurrence-free survival | Metastasis-free survival | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||||||
| No. of event | p-value | R.R. | 95% CI | p-value | No. of event | p-value | R.R. | 95% CI | p-value | |||
| Gender | Male | 108 | 54 | 0.2170 | - | - | - | 21 | 0.1008 | - | - | - |
| Female | 74 | 31 | - | - | - | 44 | - | - | - | |||
| Age (years) | 107 | 55 | 0.2993 | - | - | - | 42 | 0.2936 | - | - | - | |
| 75 | 30 | - | - | - | 23 | - | - | - | ||||
| Hepatitis | Hepatitis B | 72 | 33 | 0.7333 | - | - | - | 26 | 0.8762 | - | - | - |
| Hepatitis C | 29 | 13 | - | - | - | 11 | - | - | - | |||
| Non-B, non-C | 81 | 39 | - | - | - | 28 | - | - | - | |||
| Intrahepatic lithiasis | Not identified | 102 | 41 | 0.0551 | - | - | - | 31 | 0.1000 | - | - | - |
| Present | 80 | 44 | - | - | - | 34 | - | - | - | |||
| Surgical margin | R0 | 163 | 71 | 1 | - | 0.010* | 54 | 1 | 0.009* | |||
| R1 | 19 | 14 | 2.702 | 1/294–5.640 | 11 | 2.674 | 1.306–5.474 | |||||
| Primary tumor (T) | T1 | 87 | 28 | 1 | - | 0.051 | 21 | 1 | - | 0.046* | ||
| T2 | 61 | 32 | 1.741 | 0.961–3.154 | 26 | 1.759 | 0.978–3.164 | |||||
| T3 | 34 | 25 | 1.899 | 0.958–3.764 | 18 | 1.912 | 0.965–3.788 | |||||
| Histological variants | Large duct type | 105 | 58 | 0.0085* | 1 | - | 0.988 | 43 | 0.0759 | - | - | - |
| Small duct type | 77 | 27 | 1.108 | 0.592–1.751 | 22 | - | - | - | ||||
| Histological grade (Differentiation) | Well | 61 | 28 | 0.0299* | 1 | - | 0.829 | 22 | 0.1794 | - | - | - |
| Moderately | 66 | 27 | 1.154 | 0.630–2.114 | 22 | - | - | - | ||||
| Poorly | 55 | 30 | 1.208 | 0.638–2.283 | 21 | - | - | - | ||||
| TOP2A Exp. | Low expression | 91 | 15 | 1 | - | 14 | 1 | - | ||||
| High expression | 91 | 70 | 4.658 | 2.481–8.745 | 51 | 4.695 | 2.514–8.768 | |||||
| * Statistically significant. | ||||||||||||
Taken together, our results indicate that TOP2A is highly expressed in
CCA patients, and high TOP2A expression is correlated with poor
prognosis and notably worse overall survival (p = 0.0011, Fig. 3C),
disease-specific survival (p
To determine the unknown functions of TOP2A in
IHCC, a set of the top 200 differentially expressed genes with positive
(Supplementary Table 1) or negative correlations (Supplementary
Table 2) with TOP2A were downloaded from the cholangiocarcinoma dataset
(n = 51, Firehose Legacy, TCGA). These genes were then utilized for a functional
annotation analysis using the Gene Ontology (GO) classification system. The
biological processes most positively correlated with TOP2A were positive
regulation of DNA endoreduplication (GO: 0032877, fold enrichment:
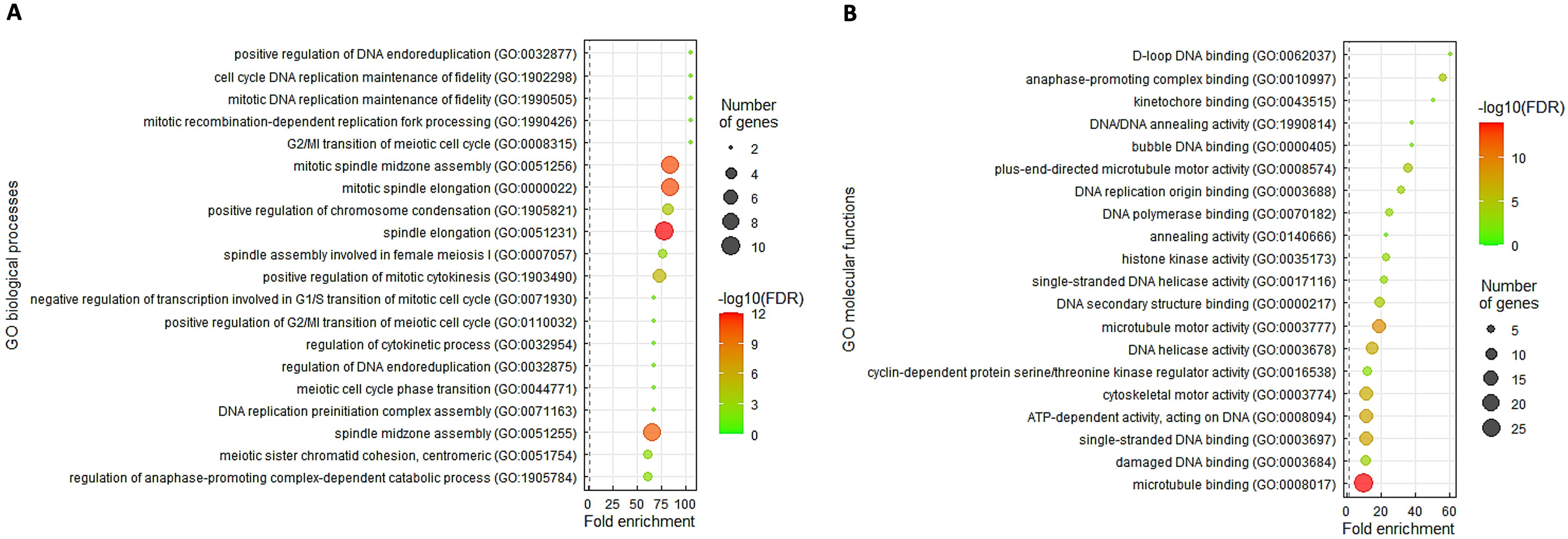 Fig. 4.
Fig. 4.The characteristic GO terms enriched in TOP2A upregulation. The top 200 differentially expressed genes with positive relationship to TOP2A were annotated utilizing the GO classification system depending on (A) biological processes and (B) molecular functions and were rated by fold enrichment.
In order to assess the impact of TOP2A, we initially measured the levels of endogenous TOP2A expression in four different cholangiocarcinoma cell lines. We found that SNU1196 and Huh28 cells had the highest amounts of TOP2A transcripts and protein expression (Fig. 5A). We then used short hairpin RNA (shRNA) to successfully knock down TOP2A in both SNU1196 (Fig. 5B, left) and Huh28 (Fig. 5B, right) cell lines. TOP2A-silenced SNU1196 (Fig. 5E, left) and Huh28 (Fig. 5E, right) cells had considerably reduced proliferation (viability). We also looked at TOP2A’s role in cholangiocarcinoma cell migration and invasion. TOP2A knockdown significantly reduced SNU1196 (Fig. 5C, left and Fig. 5D, left) and Huh28 (Fig. 5C, right and Fig. 5D, right) cell migratory and invasive abilities.
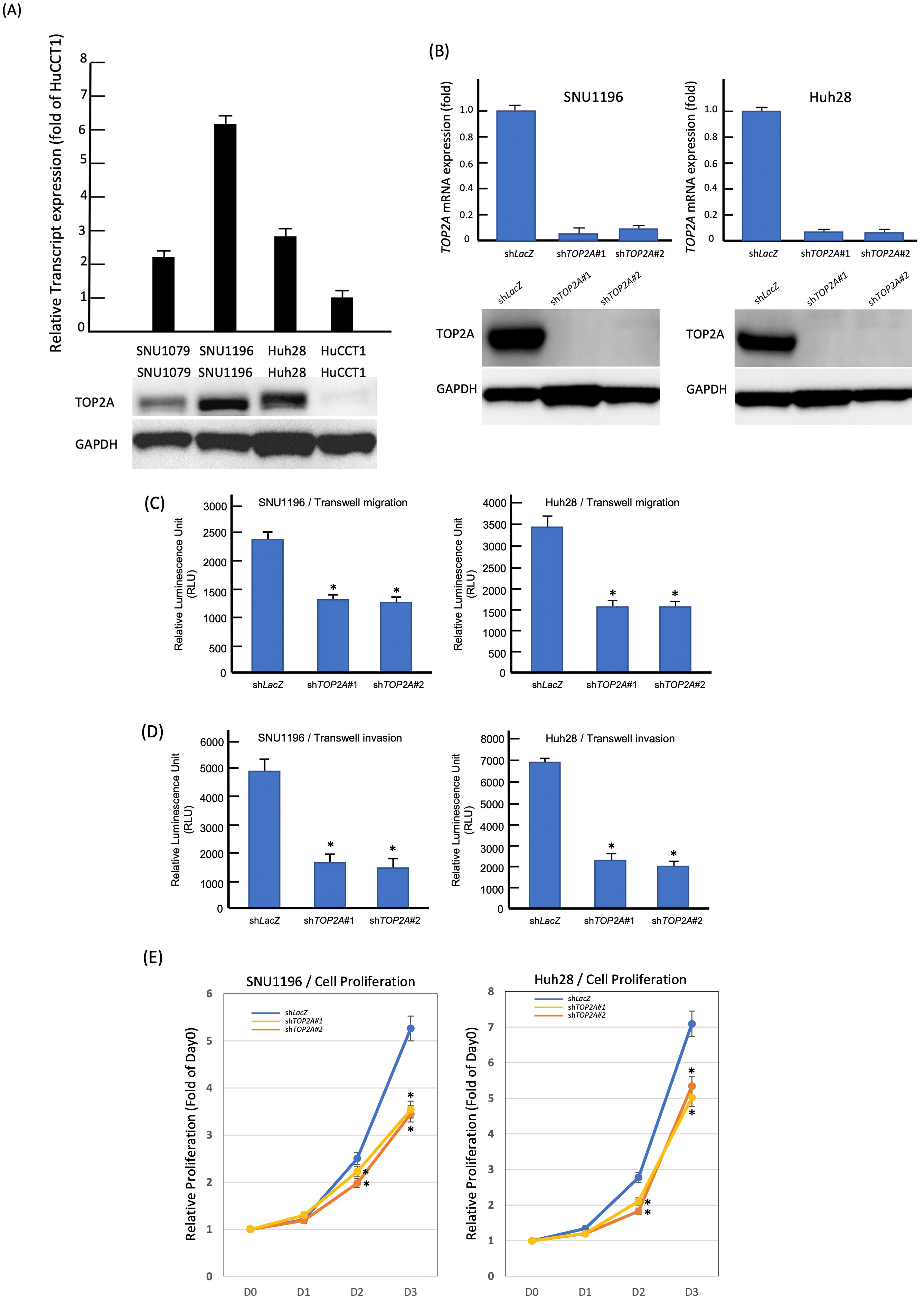 Fig. 5.
Fig. 5.TOP2A expression promotes the growth of CCA cells
in vitro. (A) SNU1196 and Huh28 cells had the highest amounts of
TOP2A mRNA and protein expression among four CCA cell lines. (B) The two
cell lines with high endogenous TOP2A expression were stably silenced by
a lentiviral vector bearing one of the two clones of TOP2A shRNA with
different sequences for both SNU1196 (left panel) and Huh28 (right panel) cells.
(C) Cell migration ability was significantly reduced in TOP2A- knockdown
SNU1196 and Huh28 cell lines. (D) Cell invasion ability was significantly reduced
in TOP2A- knockdown SNU1196 and Huh28 cell lines. (E) Using an
ELISA-based colorimetric assay to assess the rate of BrdU uptake, cell
proliferation was significantly reduced in TOP2A-knockdown SNU1196 and
Huh28 cell lines (* p
Cholangiocarcinoma is a heterogeneous group of tumors that initiate from a
number of cells from the biliary tree [23]. According to previous research, the
risk and the molecular mechanisms associated with CCA pathogenesis involve
inflammation and cholestasis or CCA development [24]. Accordingly, it has been
observed that CCA patients have increased total serum bilirubin, alkaline
phosphatase, and gamma-glutamyl transpeptidase levels or obstructive jaundice
[25, 26]. Although no specific biomarkers for CCA have been identified, some
references indicate that Carbohydrate antigen 19-9 (CA 19-9) could be used as be
a marker for the detection of CCA, since CA 19-9 levels
TOP2A is a member of the TOP2 family that plays a vital role in promoting transcriptional initiation in DNA replication, chromosome condensation, and mitosis [37]. Furthermore, TOP2A has been reported to play a crucial role in cancer progression. For example, TOP2A expression is usually increased or down-regulated depending on the presence of Erb-B2 Receptor Tyrosine Kinase 2 (ERBB2). Based on this characteristic, these two genes are often co-tested before treating patients with Herceptin as an anti-cancer drug [38, 39]. Other studies have demonstrated that TOP2A is upregulated in lung cancer patients by IHC staining, and it has also been discovered that TOP2A expression is correlated with poor overall survival in lung cancer patients [40]. Moreover, high expression of TOP2A and EZH2 is positively correlated with aggressive disease progression in prostate cancer patients [41]. Recently, TOP2A was considered as a therapeutic target of anti-cancer drugs. For example, TOP2A may serve as a predictor of responses to anthracycline therapy in breast cancer patients, and etoposide is used for the treatment of different cancers (lung cancer, ovarian, lymphoma, and acute myeloid leukemia) [42, 43, 44]. Furthermore, several researchers have clarified that topoisomerase II inhibitors affect cancer cell proliferation by inducing apoptosis, altering metabolism, and regulating the JAK2-STAT1-CXCL1 molecular pathway [45, 46]. In addition, topoisomerase inhibitors directly affect nucleic acid metabolism, which hints at their potential lethality [47]. Nevertheless, the functions and regulation of TOP2A in CCA patients were still unknown prior to this study.
In order to gain insight into the impact of TOP2A on CCA progression and prognosis, we conducted an analysis of TOP2A expression in patient tissues using an online database. The results revealed that TOP2A expression was markedly elevated in CCA patients, particularly in those with advanced primary T stage and those with poor overall survival rates and R1 stage surgical margins.
DNA double-strand breaks (DSBs) can be repaired by homologous recombination (HR) or non-homologous end joining (NHEJ) in response to DNA damage [48]. Homologous recombination is a DNA repair process that involves RAD51 recombinase and its regulator BRCA2 [49]. Since topoisomerase II may also generate transient DSBs in DNA [50], it is not surprisingly that the BRCA2, RAD51, and RAD51AP1 genes were found to be significantly positively correlated with TOP2A (Supplementary Table 1 and Fig. 4). BRCA2 is generally regarded as a tumor suppressor, and its mutation may contribute to an increased risk for the development of various cancers, especially breast and ovarian cancers [51]. However, the recovery of BRCA2 function owing to secondary BRCA2 mutation has been considered a mechanism associated with acquired resistance to cisplatin, suggesting that this genetic reversion is beneficial for cell survival [52]. In addition, high RAD51 expression has also been indicated to increase drug resistance and genome instability in tumor cells [53]. Moreover, as an RAD51 activator, RAD51AP1 upregulation has been correlated with inferior survival in hepatocellular carcinoma patients [54]. Collectively, the involvement of TOP2A, BRCA2, RAD51, and RAD51AP1 in IHCC development deserves further investigation.
In summary, the expression of TOP2A has been recognized to be upregulated in CCA patients, and this was confirmed by immunohistochemistry staining of CCA tissue sections. The expression of TOP2A was significantly correlated with the primary tumor stage (according to AJCC stages) and histological variant (large duct type). Moreover, high expression of TOP2A is predictive of worse overall survival, disease-specific survival, and metastasis-free survival rates. In addition, increased expression of TOP2A may contribute to tumor progression in CCA patients. This information indicates that TOP2A can be considered for use in future prospective prognostic analyses. Moreover, we need more evidence to clarify the molecular mechanisms and explore the biological functions of TOP2A to determine its potential as a therapeutic target for CCA in clinical trials.
The transcriptome dataset (GSE26566) used in this study is publicly available in the Gene Expression Omnibus (GEO) database, which is maintained by the National Center for Biotechnology Information in Bethesda, MD, USA.
Conceptualization—KHO and YHK; methodology—KHO, HYL, DPS, TJC, SKHH, YFT, CLC, YLS, TCC, and CFL; investigation—KHO, HYL, DPS, TJC, SKHH, YFT, CLC, YLS, TCC, and CFL; formal analysis—KHO, HYL, DPS, TJC, SKHH, YFT, CLC, YLS, TCC, and CFL; resources—CLC, YLS, TCC, and CFL; validation—KHO, HYL, DPS, TJC, SKHH, and YFT; visualization—KHO, HYL, DPS, TJC, SKHH, and YFT; writing - original draft—KHO and YHK; writing - review & editing—KHO and YHK; funding acquisition—YHK; supervision—KHO and YHK. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
All participants in this study provided informed consent before their samples were deposited in the biobank. The Ethics Committee and Institutional Review Board of Chi Mei Medical Center (IRB09912003) approved the study’s use of deidentified tumor samples from the biobank, indicating that the research was conducted in compliance with ethical guidelines outlined in the Declaration of Helsinki and regulations set forth by the government.
Not applicable.
This research received no external funding.
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.





