1 Montreal Centre for Experimental Therapeutics in Cancer Segal Cancer Center, Lady Davis Institute for Medical Research, Jewish General Hospital, McGill University, Montréal, QC H3T 1E2, Canada
Abstract
Background: Poly(ADP-ribose) polymerases 1 and 2 (PARP1, 2), and 3 mediate protein modifications that facilitate the recruitment of DNA repair factors to single and double strand breaks. PARP3 is unique in that it is also required for efficient mitotic progression and stabilization of the mitotic spindle. Eribulin, an anti-microtubule agent used clinically to treat breast cancer, exerts its cytotoxicity by altering microtubule dynamics resulting in cell cycle arrest and apoptosis. Herein, we hypothesize that the pan PARP inhibitor olaparib has the potential to enhance the cytotoxicity of eribulin by halting mitosis through inhibition of PARP3. Methods: The effect of olaparib on eribulin cytotoxicity was assessed using the Sulforhodamine (SRB) assay, with two triple negative breast cancer cell lines and an estrogen receptor positive (ER+)/human epidermal growth factor receptor 2 negative (HER2-) breast cancer cell line. Alteration by the treatments on PARP3 activity and microtubule dynamics were assessed utilizing a chemiluminescent enzymatic assay and immunofluorescence, respectively. The effect of the treatments on cell cycle progression and apoptosis induction were assessed by flow cytometry using propidium iodide and Annexin V staining, respectively. Results: Our results demonstrate that non-cytotoxic concentrations of olaparib sensitize breast cancer cells regardless of ER status. Mechanistically, our results indicate that olaparib potentiates eribulin-induced cell cycle arrest at the G2/M boundary, PARP3 inhibition and microtubule destabilizing resulting in mitotic catastrophe and apoptosis. Conclusions: In breast cancer (regardless of ER status) settings, treatment outcomes could be improved by the incorporation of olaparib in eribulin treatment regimens.
Keywords
- eribulin
- olaparib
- PARP3 inhibitor
- drug combination
- breast cancer cell lines
- microtubule dynamics
The poly (ADP-ribose) polymerase (PARP) superfamily includes 17 members involved in a variety of cellular processes such as transcription, chromatin status regulation, telomere function, and DNA repair [1]. PARPs play an important role in DNA repair because they facilitate the recruiting of DNA repair components to single strand breaks (SSBs) and double strand breaks (DSBs). The observation that deficiencies in PARP and DNA DSB repair processes are synthetically lethal prompted the clinical development of PARP inhibitors to target BRCA1/2 driven cancers. These efforts led to the development of olaparib. Olaparib is an FDA approved pan PARP inhibitor that shows promising effects in breast cancer gene (BRCA) 1/2-deficient breast and ovarian cancer cells plus in BRCA-mutated patients bearing these tumors [2, 3, 4, 5, 6, 7, 8]. BRCA1 has a role in the repair of DNA damage, especially cytotoxic double-stranded DNA breaks (DSBs), which are important types of DNA lesions. Homologous Recombinational Repair (HRR) is a critically important mechanism leading to DNA damage correction. BRCA1 is central to several macromolecular complexes and as one of the major tumor suppressor proteins drives HRR and cell cycle progression [9]. HRR proficiency is one of the major determinants of cellular sensitivity to PARP inhibitors because restoring homologous recombination repair limits sensitivity to PARP inhibitors [9, 10, 11]. As well, PARP inhibition by olaparib was shown to sensitize tumor cells to DNA-damaging drugs and radiation [8, 12, 13]. PARP1 is the best characterized family member whose catalytic poly (ADP-ribosyl) ation activity is one of the earliest responses to SSBs and DSBs [14, 15]. As well as other PARP family members, PARP3 interacts with both classical and alternative nonhomologous end-joining (NHEJ) proteins herein facilitating DSB repair. PARP3 participating in DSB repair pathway(s) by interact with the classical nonhomologous endjoining (C-NHEJ) proteins, DNA-PK, KU70, KU80, and DNA ligase IV [16]. In contrast, only PARP3 is required for efficient mitotic progression as it stabilizes the mitotic spindle and promotes telomere integrity [16, 17, 18]. Although, PARP3 is a core component of the centrosome [19], it appears to regulate cell cycle progression without interfering with centrosome duplication [20]. Even though the mechanistic role of PARP3 in mitotic spindle stabilization is not completely understood, it is plausible that PARP3 inhibitors affect mitotic progression by delaying both DSB repair and spindle assembly [19, 21].
Our working hypothesis is that because of its role in mitotic progression, reduction of PARP3 activity has the potential to enhance the efficacy of anticancer agents targeting microtubule dynamics. We previously demonstrated that in a triple negative breast cancer (TNBC) cell lines, a nontoxic dose of a pan PARP inhibitor (olaparib) or a selective PARP3 inhibitor (ME-0328) leads to enhanced antitumor activity of a vinca alkaloid (vinorelbine) but not a taxane (paclitaxel) [22]. These results suggest that inhibition of PARP3 sensitizes breast cancer cells to anticancer agents that cause dissolution of the mitotic spindle such as vinorelbine but not to taxanes which stabilize the mitotic spindle. In the current work, we examined the effect of olaparib on eribulin cytotoxicity in human breast cancer cell lines [23, 24]. Eribulin mesylate is an FDA-approved non-taxane microtubule inhibitor. Eribulin is utilized to treat patients with metastatic breast cancer, especially those bearing the HER2 negative genotype [25, 26, 27, 28]. Eribulin inhibition of microtubule dynamics occurs through a novel mechanism of action. In contrast to taxanes that shorten microtubules, eribulin binds to a unique part of tubulin site, leading to suppression of microtubule polymerization with no effect on microtubule depolymerization. Furthermore, eribulin (but not taxanes) can promote the tubulin degradation to nonfunctional aggregates [23, 26]. Consequently, eribulin generates fine structural changes of the mitotic spindle which are sufficient to cause cell cycle arrest at the G2-M boundary [25, 29] leading to an irreversible block in mitosis and apoptosis [23, 26, 27]. The effect of olaparib on eribulin cytotoxicity was evaluated in breast cancer cell lines.
MDA-231 and MDA-436 are triple negative breast cancer cell lines and MCF-7 is an ER+/HER2- breast cancer cell line. They were obtained from the American Type Culture Collection (ATCC) and maintained following the manufacturer instructions. The cell lines used have been tested and are negative for mycoplasma. Eribulin mesylate 0.5 mg/mL was obtained from Eisai LTD (Mississauga, ON, Canada) and olaparib was provided by Selleckchem Company (Houston, Texas, USA).
The SRB cytotoxicity assay in breast cancer cell lines was assessed 5 days after
treatment using the SRB colorimetric assay as described by us [30, 31]. The
IC
Apoptotic cell death levels were assessed by monitoring drug-treated cultures for Annexin V content using flow cytometry as described by us [34]. After 24 hours, the breast cancer cells treated with the stated drug combinations and concentrations were assayed by using the PE Annexin V Apoptosis Detection Kit (BD Pharmingen, Waltham, MA, USA) as previously described [22].
Breast cancer cell lines were grown in T25 flasks including Roswell Park Memorial Institute culture media (RPMI) and 10% Fetal Bovine Serum (FBS), and then treated with olaparib and eribulin alone and drug combinations for 24 hours as described [22]. Cell cycle analysis was performed using a BD Fortessa flow cytometer (BD Bioscences, San Jose, CA, USA) flow cytometry [30, 32]. A minimum of 20,000 events was recorded for each sample.
Breast cancer cell lines (5
To assess alterations in microtubule networks that associated with the
microtubule depolymerization effects the breast cancer cells were seeded, treated
and immunostained for tubulin in multichamber slides 24 hours after treatment as
indicated [35, 36, 37]. Briefly, after treatment, the cells were fixed and
permeabilized using 4% formaldehyde for 15 min at room temperature, followed by
washing with PBS/BSA for 5 min three times. Then the cells were incubated in
PBS/BSA (5%)/Triton X100 (0.1%) for 30 minutes at room temperature. Next, the
cells were incubated overnight at 4 °C with
Cells showing abnormal
The experiments were performed in triplicate. Analysis of variance and two-sided
or paired t-test were performed using Sigma Plot Version 13.0 Systat.
Software, Inc., San Jose, California, CA, USA. Differences were considered significant
with p values
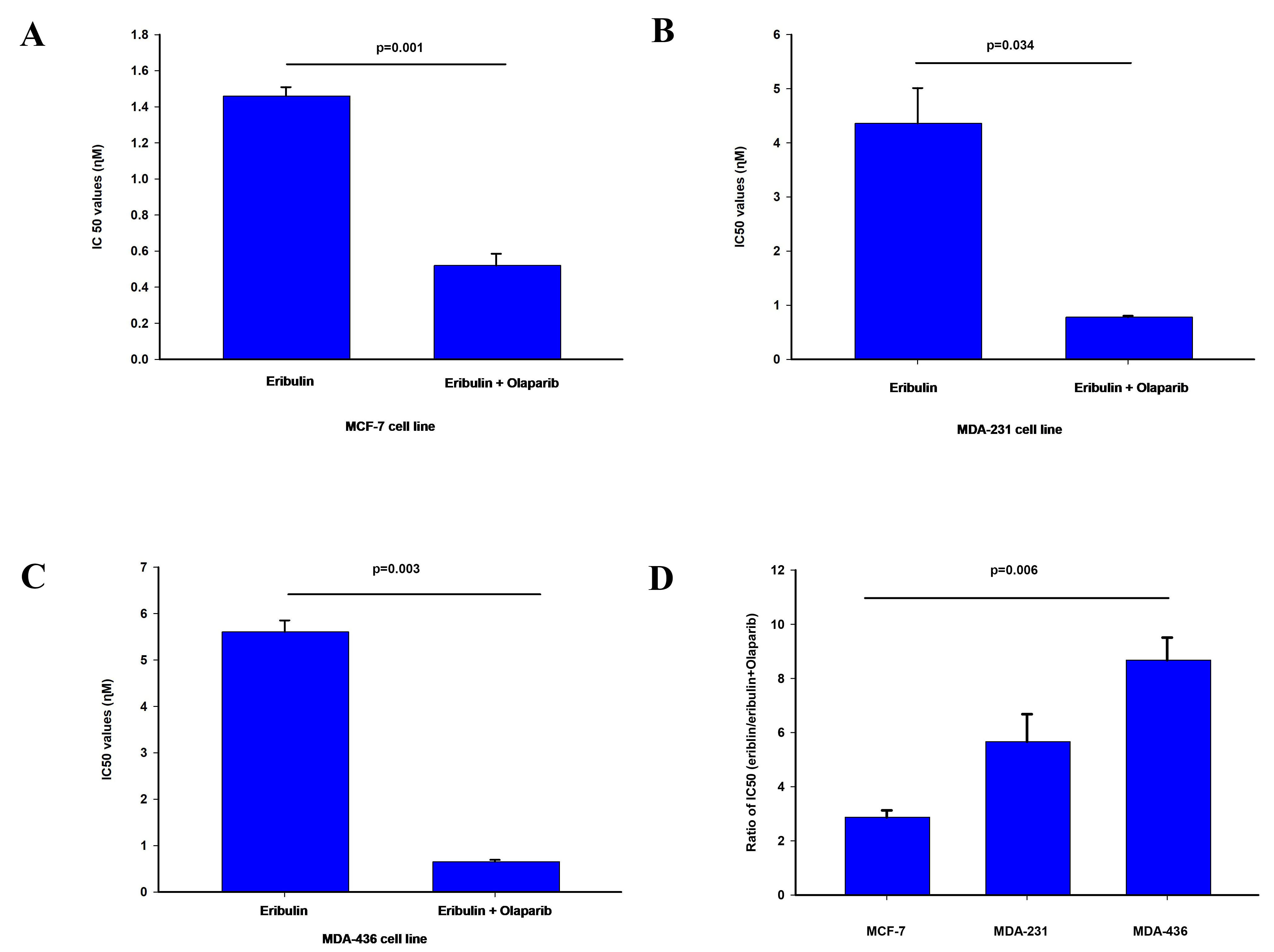
The cytotoxicity of olaparib and eribulin were determined using the SRB assay to
obtain the IC
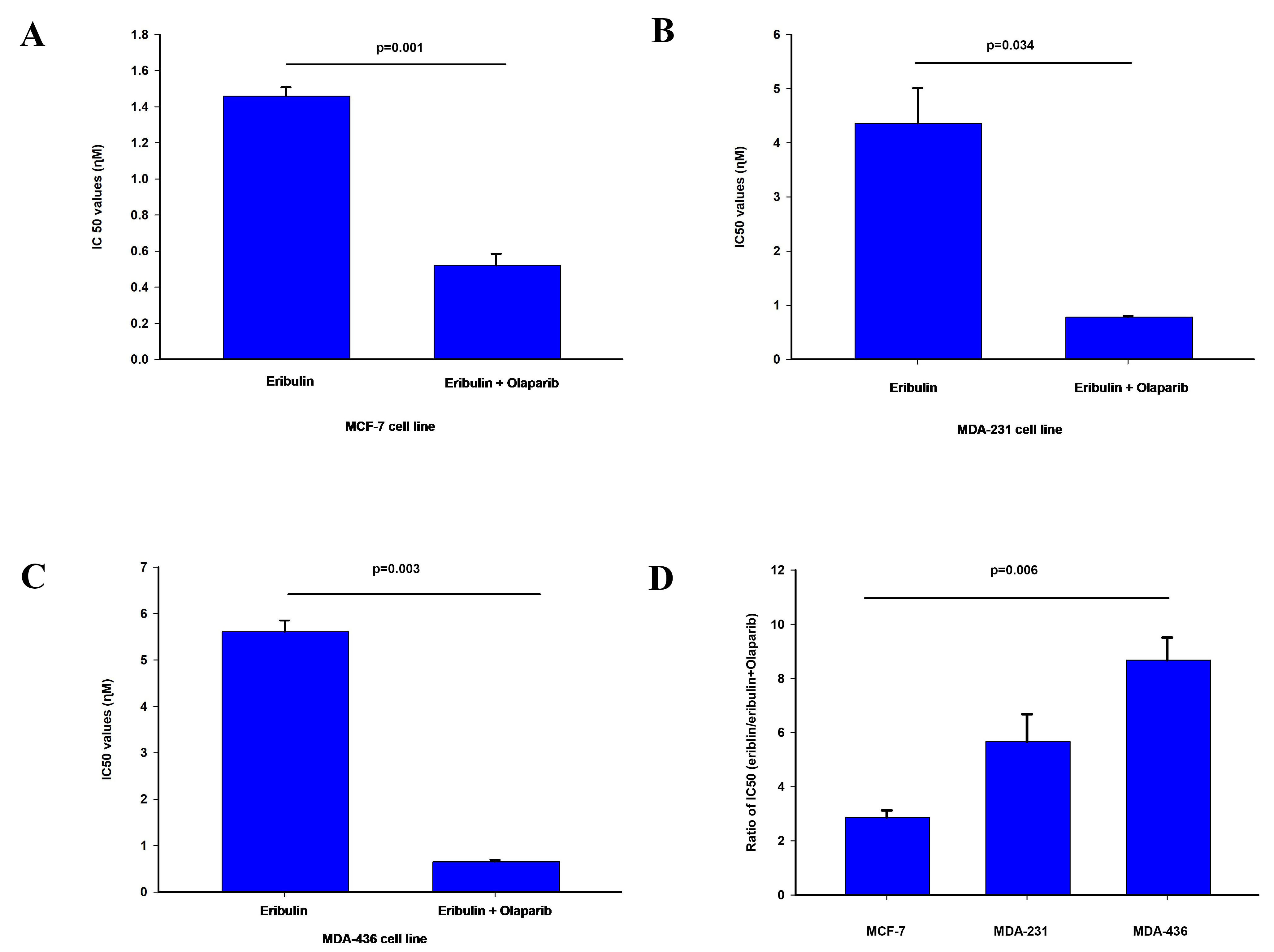 Fig. 1.
Fig. 1.Effect of olaparib on eribulin cytotoxicity. The
IC
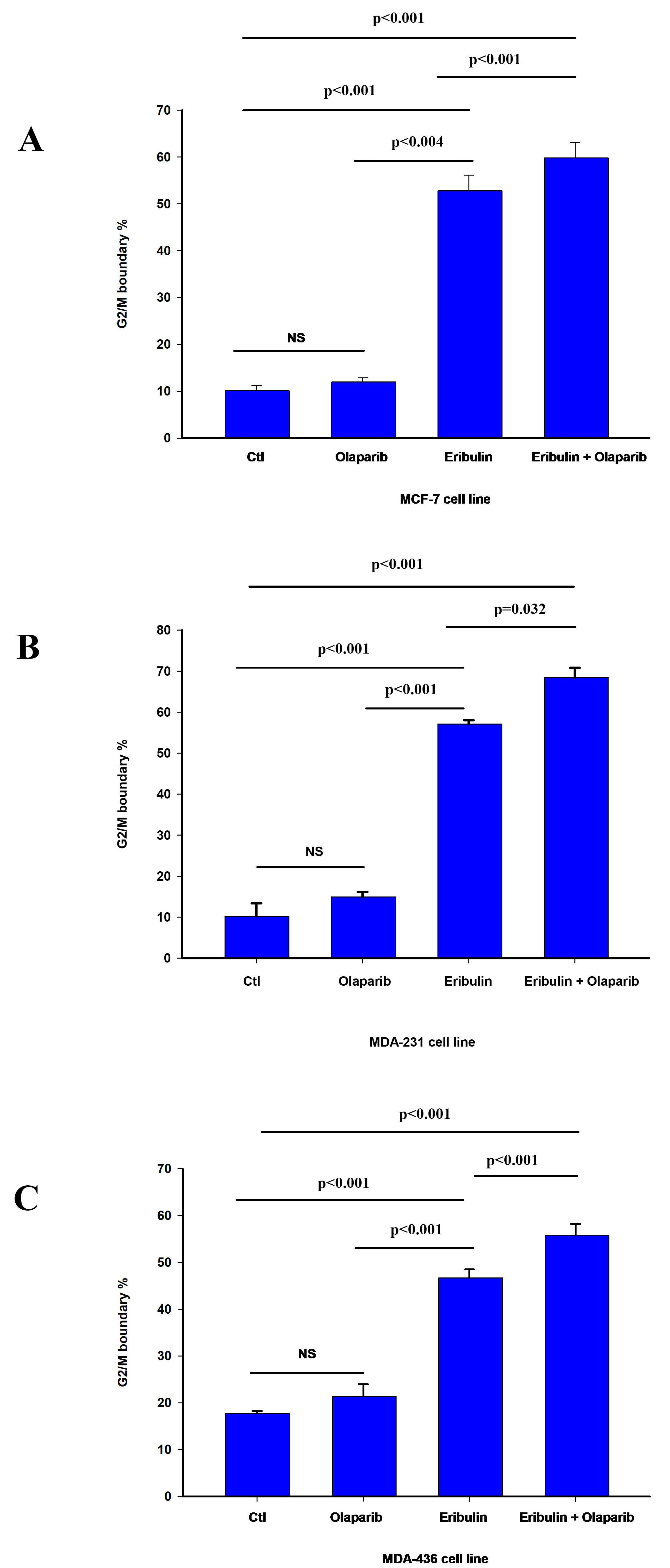
We next assessed the contribution of programmed cell death and cell cycle arrest
to the sensitization effect of olaparib on eribulin cytotoxicity. Treatment with
eribulin IC
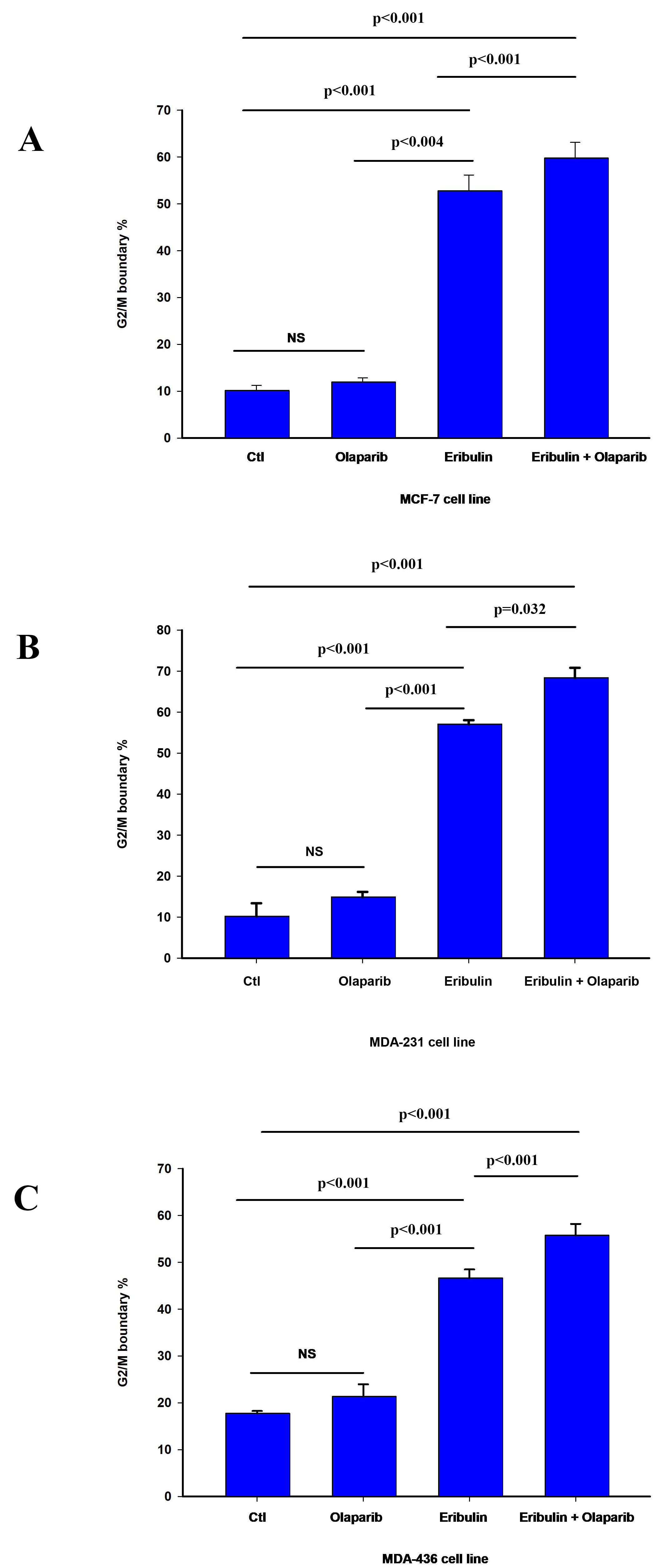 Fig. 2.
Fig. 2.Effect of olaparib on eribulin-induced G2/M cell cycle arrest.
Cell cycle analysis was performed by flow cytometry as described in material and
methods. The bar graphs represent the percentage of cells at the G2/M cell cycle
boundary in MCF-7 (A), MDA-231 (B), and MDA-436 (C) cell line 24 h after
treatment as indicated. The data is presented as the mean value
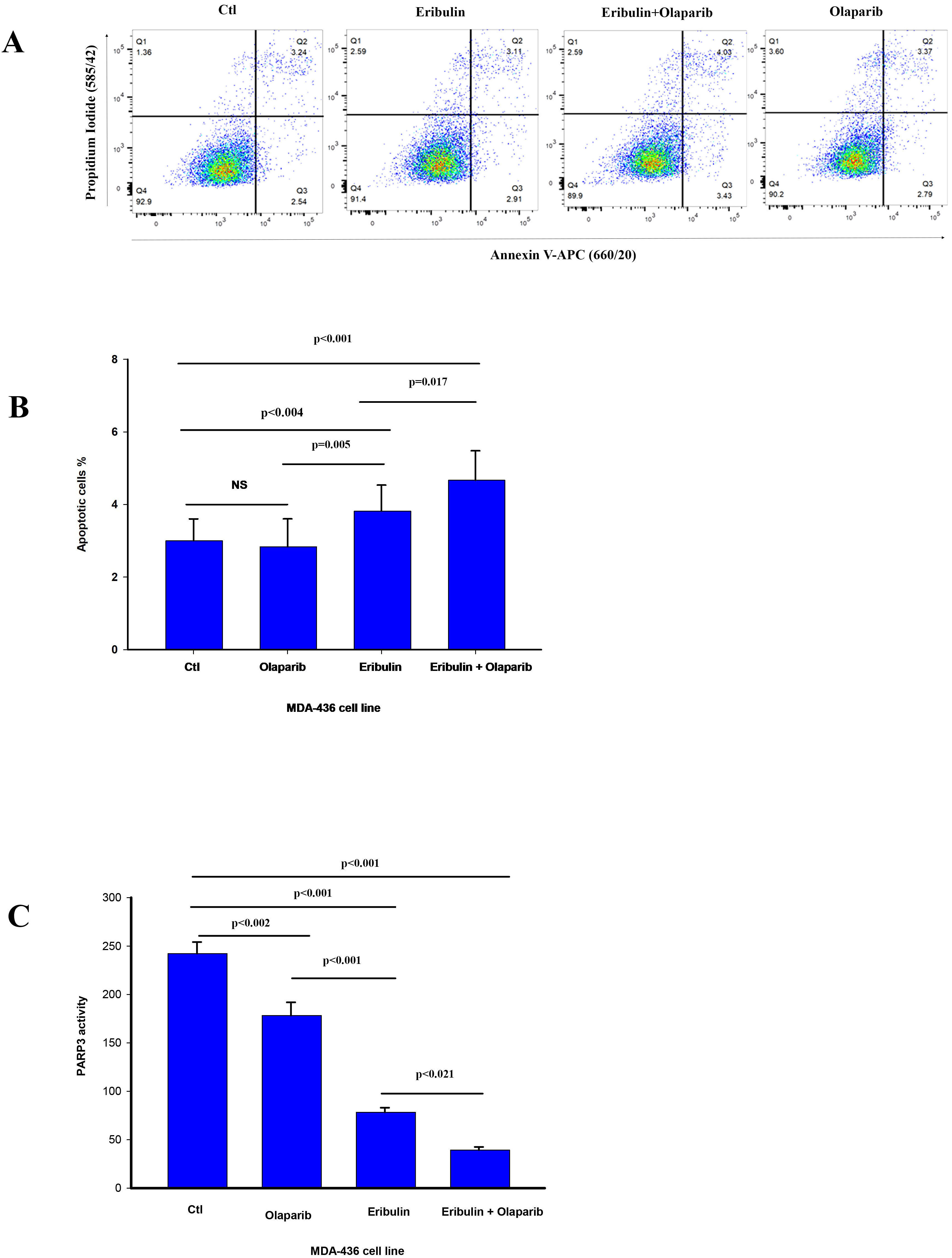
Because PARP3 activity is required for efficient mitotic progression [16, 17, 18], we
assessed for changes in PARP3 activity in MDA-436 cells by the drug treatments.
The Eribulin IC
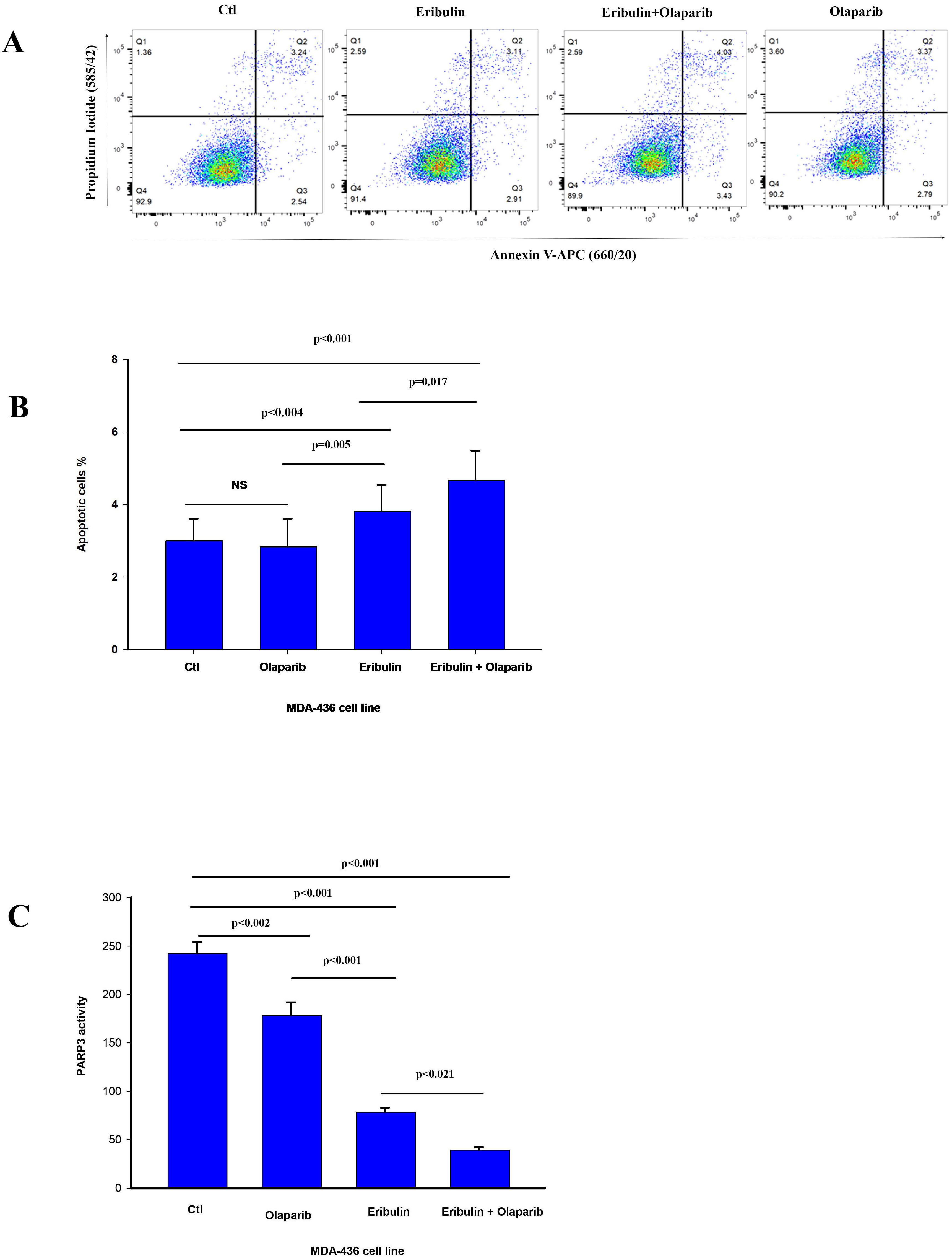 Fig. 3.
Fig. 3.Olaparib enhances eribulin induced apoptosis and reduces PARP3
activity. (A) The percentage cells undergoing apoptosis was assessed by flow
cytometry analysis of Annexin V staining 24 h after treatment with vehicle (CTL)
and eribulin IC
In summary, our results show that eribulin induces cell cycle arrest at the G2/M boundary. Furthermore, while our results suggest that olaparib enhanced eribulin-induced cell cycle arrest in all cells, there is a heterogeneous response to the combination of both drugs on apoptosis. This agrees with the reported effect of eribulin on cell cycle progression and heterogenous induction of programmed cell death [39, 40].
Importantly, 2
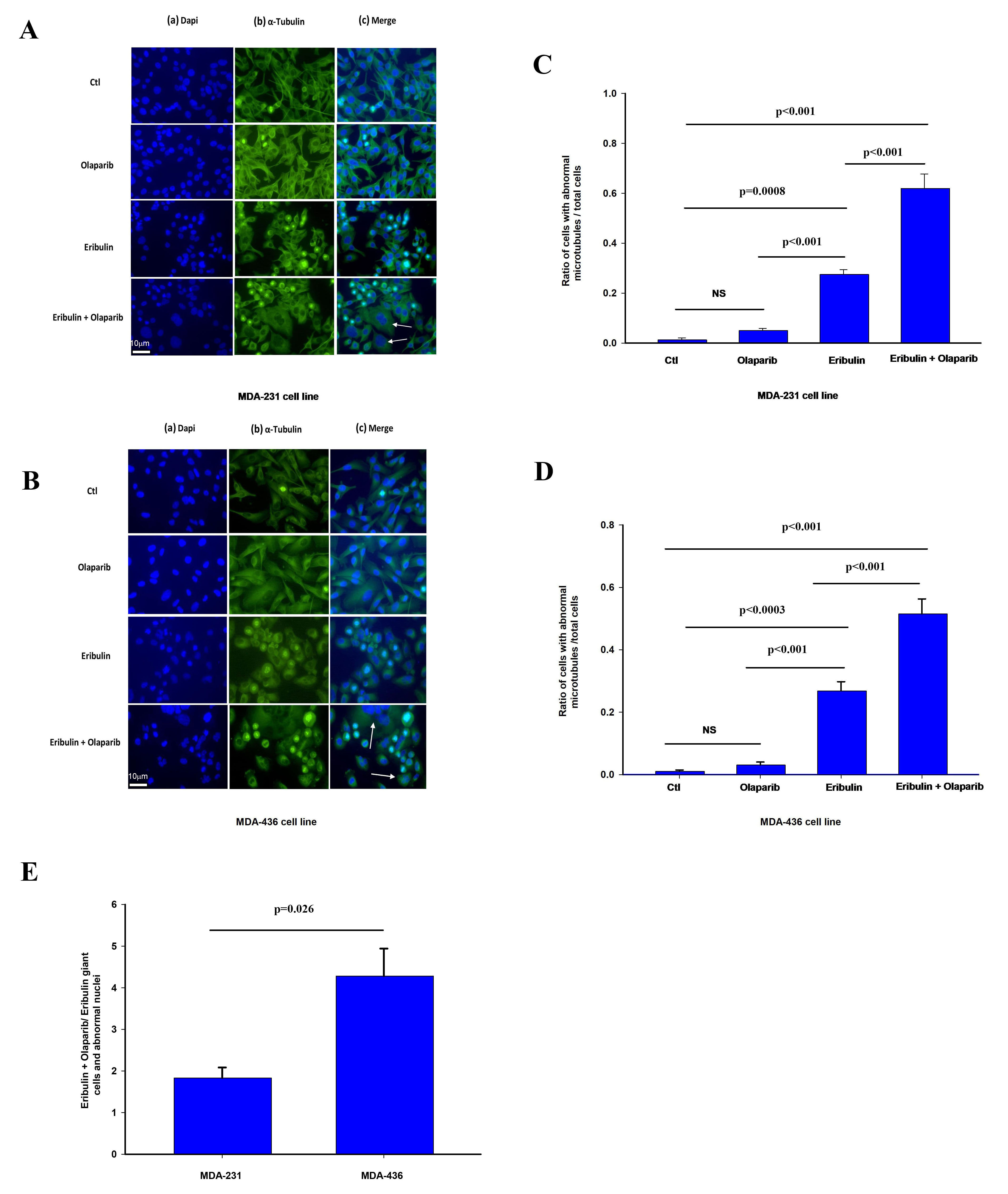
To test the impact of PARP3 activity on microtubule dynamics and markers of
mitotic arrest in cells treated with eribulin, we assessed the effect of eribulin
and olaparib alone or in combination on tubulin staining and nuclear morphology
that have been associated with altered microtubule dynamics by eribulin and other
microtubule targeting agents [35, 36, 37, 41]. MDA-231 and MDA-436 24 hours after
treatment with vehicle (CTL), eribulin IC
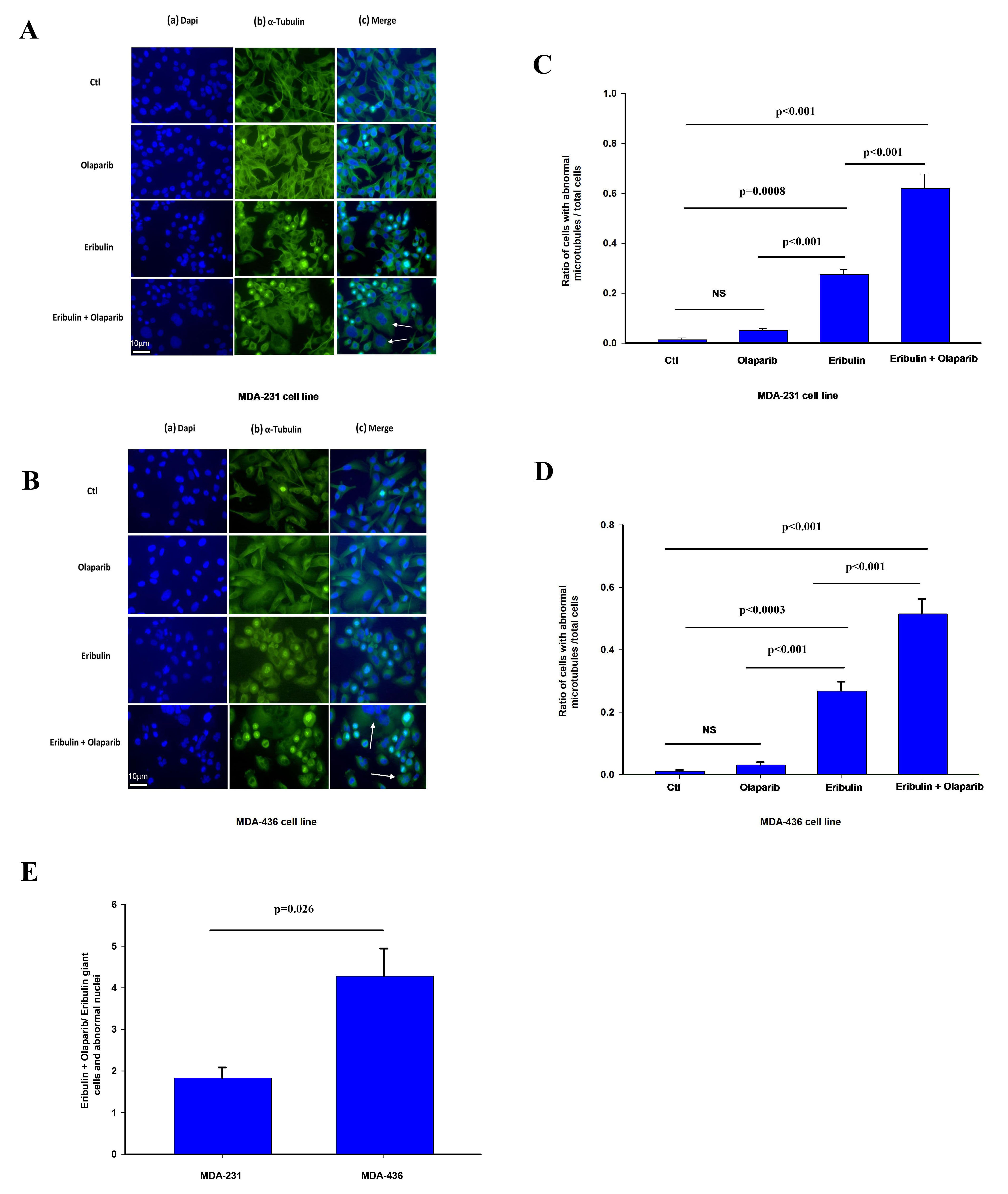 Fig. 4.
Fig. 4.Effect of olaparib and eribulin on
The effect of olaparib on eribulin microtubule depolarization was significantly more pronounced in MDA-436 cells with respect to MDA-231 cells (Fig. 4E). We observe a similar effect by these treatments in the frequency of cells with abnormally looking nucleus in shape and size such as giant cells shown in Fig. 4 (Supplementary Fig. 3A,B).
Overall, our findings suggest that eribulin and olaparib when combined result in greater disruption of microtubule dynamic and altered nuclear morphology than when used alone, effects that have previously been associated with increased chromosome misalignment and splayed microtubule spindles leading to mitotic arrest [23, 24, 25, 29].
Targeting PARP by small-molecule inhibitors is a promising anticancer approach to the treatment of metastatic breast cancer [2]. The most studied PARP inhibitors which are currently used in clinical trials are PARP1 and PARP2 inhibitors [15, 42, 43]. Inhibitors of PARP3, which have a unique function on DSBs repair pathway and mitotic spindle dynamics have been developed recently [19, 21]. Olaparib, although first described as PARP1 and PARP2 inhibitors, has recently demonstrated a critical role in PARP3 inhibition [2, 8]. Olaparib, which has been approved by the US Food and Drug Administration (FDA), providing significant advances in the treatment of patients with HER2 negative BRCA mutated metastatic breast cancer [4, 5, 8, 13]. Despite this, resistance of cancer cells to PARP inhibitors has been reported with olaparib like many other anticancer drugs [44]. Moreover, patients with BRCA-mutated tumors represent a small minority of patients.
Eribulin is an FDA-approved non-taxane microtubule inhibitor. Eribulin is
utilized to treat patients with metastatic breast cancer, especially those
bearing the HER2 negative genotype [25, 26, 27, 28]. The antiproliferative mechanisms of
eribulin are characterized by suppression of the centromere and changes in
spindle microtubule dynamics during mitotic arrest [45]. Eribulin, like other
vinca alkaloids, binds to the
Considering the important role of PARP3 in the mitotic pathway and centrosome
function, we tested the effect of olaparib as a PARP3 inhibitor on eribulin
cytotoxicity. Our results shed light on a new method of using PARP3 inhibitors
with anti-microtubule agents. Based on our results, the drug combination of
eribulin with a PARP3 inhibitor, olaparib demonstrated a significant reduction of
IC
Based on our results, eribulin or olaparib alone inhibited PARP3 activity, but the eribulin and olaparib drug combination reduced PARP3 activity significantly more than eribulin alone. In a similar fashion, the same significant reduction of PARP3 activity has been shown by us, using the olaparib and vinorelbine drug combination against TNBC [22].
During mitosis, microtubules undergo rapid polymerization and depolymerization to allow chromosome movement [52]. Effective bipolar spindle organization in the mitotic phase is highly regulated by centrosome duplication during each cell cycle of dividing cells [53]. Anti-microtubule targeting agents like eribulin bind tubulin and prevent its incorporation into growing microtubules leading to microtubule disassembly and increased splayed and defective tubulin via alteration of microtubule network and suppression of mitotic spindle dynamics [24, 25, 45]. As expected, eribulin experimentally arrests cells during mitotic metaphase leading to mitotic catastrophe and apoptosis [24, 25, 52, 54].
Eribulin induced microtubule instability and defective mitotic spindle dynamics in treated breast cancer cells, resulting in mitotic arrest and subsequent abnormal mitotic divisions with a more compact, irregular spindle fiber appearance and increased chromosome misalignment [23, 26, 27, 45]. Based on our results, these types of mitotic catastrophe effects are enhanced by the combination of eribulin with the PARP inhibitor, olaparib. Inhibition of PARP3 with olaparib potentiates eribulin-induced mitotic catastrophe due to microtubule network alterations, abnormal mitosis, and cell cycle arrest.
Interestingly, inhibition of PARP3 sensitizes breast cancer cells to vinorelbine and eribulin in spite of their different mechanisms of interacting with the tubulin structure but not to paclitaxel which interacts with tubulin in a different fashion then the above two anticancer agents.
In conclusion, the results of our current study should lead the way to new approaches for targeting HER2 negative metastatic breast cancer. However, combination of olaparib with vinorelbine or eribulin in clinical trials should be done utilizing the known pharmacokinetics and toxicities of these drugs; i.e., specifically utilizing limited exposure time (5–7 days) of olaparib with every 2 week schedules of anticancer drugs to allow for the use of granulocyte stimulating agents to overcome increased myelotoxicity and maximize potentially synergistic antitumor activity. A previous phase II trial of olaparib given daily continuously and eribulin days 1 and 8 resulted in significant myelosuppression [55].
Not applicable.
BSA, LP and RA designed the research study. BSA performed the experimental work and data acquisition. BSA and RA analyzed and interpreted the results. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Not applicable.
The authors acknowledge the Cancer Research Society.
Cancer Research Society (Montreal, Grant number 01713).
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.