1 Institute of Cell Genetics, Medical University Innsbruck, A6020 Innsbruck, Austria
Academic Editor: Amedeo Amedei
Abstract
Following gene expansion during evolution, today’s phylogenetic tree of the NR2F family of nuclear orphan receptors in mammals is represented by three different isoforms: NR2F1, NR2F2, and NR2F6. Structural analysis of the NR2F family members has revealed that NR2F1 and NR2F2 are closely related and grouped together apart from NR2F6, which is more divergent in its biochemical characteristics. In this review, we highlight current knowledge on the cellular functions of NR2F family members. NR2F family members have been reported to be causally involved in carcinogenesis. Mechanistically, NR2F proteins are localized in the nucleus, where they bind to target DNA enhancer sequences and have been implicated in the regulation of de novo gene transcription, though this is not sufficiently understood. Based on apparently divergent and non-uniform expression patterns of the NR2F isoforms in different tissues and cell types, non-redundant functions of the individual family members appear to exist. Notably, NR2F2 appears to be more closely related functionally to NR2F6 than NR2F1. Along these lines, NR2F2 and NR2F6 have been reported to be involved in cellular neoplasia. Furthermore, enhanced expression of NR2F isoforms has been established as prognostic biomarkers in various cancer entities. Therefore, it is tempting to speculate that NR2F isoforms represent innovative targets for therapeutic intervention in defined types of cancer. Thus, NR2F family nuclear receptors can be viewed as gatekeepers balancing cell type-specific regulation of proliferation and the suppression of terminal differentiation in health and disease.
Keywords
- nuclear receptors
- NR2F family
- NR2F isoform-specific functions
- development
- cell differentiation
- metabolism
- carcinogenesis
Nuclear receptors (NRs) are ligand-activated transcription factors involved in the regulation of a wide array of physiological and developmental processes. The NR superfamily consists of 48 transcription factors in humans, including the receptors for steroid hormones, thyroid hormones, cholesterol metabolites, and lipophilic vitamins. However, approximately half of the NRs are categorized as orphan receptors because their ligands are not known [1]. As the name suggests, NRs are located in the nucleus, where they function as transcriptional regulators with direct DNA binding activity, regulating gene expression. Due to their excellent druggability, NRs are promising candidates as novel therapeutic targets for a multitude of diseases, including cancer [2]. NRs exhibit a characteristic modular structure that is composed of five to six homologous domains designated A to F (from the N-terminal to the C-terminal end). This nomenclature is based on the region of conserved sequence and function. The defining features of NRs are the DNA binding domain (DBD; region C) and ligand binding domain (LBD; region E), which are both highly conserved. They are the two most important regions and can function independently from each other. The variable N-terminal A/B region, as well as the D region, are less conserved. The C-terminal F region does not exist in all receptors, and its function is not well understood [3]. The first NR was described in 1985 [4], and the NR superfamily is classified according to their evolutionary distance into six subfamilies, numbered from 1 to 6. These six subfamilies are further divided into several groups, starting with the letter A and further specified by an additional number [3, 5].
The NR2F family, also known as the nuclear orphan receptors of the chicken ovalbumin upstream promoter transcription factor (COUP-TF) family, consists of three members: NR2F1 (synonyms: COUP-TFI, EAR-3), NR2F2 (synonyms: COUP-TFII, ARP-1), and NR2F6 (synonyms: COUP-TFIII, EAR-2) [5, 6, 7, 8]. Among the NR2F family members, NR2F1 and NR2F2 have the highest homology, especially in the functionally important DBD and LBD, with a homology of 98 percent and 96 percent, respectively [7]. The third NR2F family member, NR2F6, is more divergent but still functionally closely related [9]. The NR2F family members homo- or heterodimerize with retinoid X receptor (RXR/NR2B1) and other NRs, which results in binding to a variety of response elements containing imperfect AGGTCA direct or inverted repeats with various spacing on the cognate DNA sequence. All of the members of the NR2F family are orphan receptors because endogenous ligands have not yet been identified [10].
NR2F1 is regarded as one of the main transcriptional regulators governing cortical arealization, cell-type specification, and maturation. Multi-faceted functions of NR2F1 in the development of different mouse brain structures, including the neocortex, hippocampus, and ganglionic eminences, have been observed [11]. Other functions of NR2F1 include regulating migration [12, 13], controlling temporal identity specification of neuronal progenitor cells [14, 15], and constituting area-specific identity in progenitors and neurons [16, 17, 18]. In humans, NR2F1 haploinsufficiency has been associated with the rare disease Bosch-Boonstra-Schaaf optic atrophy syndrome (BBSOAS), a complex neurodevelopmental disease affecting intellectual ability and resulting in optic atrophy [19, 20]. The Nr2f1 heterozygous knockout mouse model exhibits some of the neurological symptoms of BBSOAS and, mechanistically, impaired hippocampal synaptic plasticity has been observed. This suggests that a deficit or alteration in hippocampal synaptic plasticity contributes to the intellectual disability symptoms present in BBSOAS [21].
In vertebrates, NR2F1 and NR2F2 are expressed during early development. In general, NR2F1 is predominantly expressed in the developing peripheral and central nervous system, whereas NR2F2 is present in the mesenchymal area of the internal organs. In mice, homozygous deletions of NR2F1 result in perinatal lethality due to defects in the central nervous system. In contrast, mice with homozygous deletions of NR2F2 die due to growth retardation in the vasculature of the head, spine, and heart [22]. Evidently, NR2F2 is involved in tissue homeostasis and maintenance, functioning as a major regulator of cell differentiation and angiogenesis [23, 24, 25]. Moreover, studies with genetically modified mouse models have shown major regulatory functions of NR2F2 in the development of several tissues and organs, including the kidney, stomach, and diaphragm [23, 26, 27].
Mice deficient in NR2F2 exhibit defects in angiogenesis, particularly in the
appearance of venous and lymphatic vessels, which suggests the involvement of
NR2F2 in angiogenesis [25] (Table 1, Ref. [25, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40]). The major drivers of this process are the
Notch signaling pathway and vascular endothelial growth factor (VEGF), alongside
the BMP/TGF-
| Family member | Area | Effect | Ref. |
| NR2F2 | Lymphatic vessel formation in constitutive knockout mice | Venous and lymphatic vessel formation defects | [25] |
| NR2F2 | Notch pathway inhibitor | FoxC1 and NR-1 binding upstream of Notch | [28] |
| Hey2 binding downstream of Notch | [28] | ||
| NR2F2 | Repressor of artery-specific genes | Reduced NR2F2 expression associated with higher artery marker expression | [29, 30] |
| Promotes cell proliferation and sprouting in endothelial cells | [29, 30] | ||
| NR2F2 | Maintenance of venous identity | High NR2F2 expression in endothelial progenitor cells (EPCs) in vein patch | [31, 32] |
| Hypoxia-induced NR2F2 downregulation | [32] | ||
| Regulatory function on VEGF-C and G, VEGF-R3, and NP-2 expression | [33, 34] | ||
| Humans: NR2F2-mediated activation of cell cycle genes, activation of vein-specific enhancers, repression of artery enhancers | [35] | ||
| NR2F2 | Lymphatic endothelial cell fate regulator | NR2F2 and Sox18 induce Prox1 expression | [36] |
| NR2F2-Prox1 heterodimers allow for lymph marker expression | [36, 37, 38, 39, 40] |
In the vein patch, endothelial progenitor cells (EPCs) express high levels of
vein markers, such as NR2F2, whereas arterial markers Dll-4 and Hey2 are
expressed at lower levels [31, 32]. The oxygen sensor HIF
In addition to the involvement of NR2F2 in Notch signaling, NR2F2 also induces vein and lymph node identity by regulating the expression of VEGF-C and -D, as well as VEGF-R3 and NP-2 [33, 34]. The expression of NR2F2 in vein cells depends on the transcription factor brahma-related gene 1 (BRG1) [45]. Interestingly, BRG1 expression has also been reported to be necessary for the expression of Notch ligands [46, 47].
In a study that focused on identifying regulatory elements in the human genome responsible for controlling artery and vein gene expression, several thousand artery- and vein-specific regulatory elements were identified. This genomic characterization of endothelial enhancers exhibited overrepresentation of NR2F2 sites in vein-specific enhancers, suggesting a direct role in promoting vein identity and a multifunctional role of NR2F2 in the regulation of arteriovenous gene expression. In particular, NR2F2 has been shown to regulate three distinct aspects of arteriovenous identity. First, in accordance with previous studies, they observed that NR2F2 directly activates enhancer elements flanking cell cycle genes to drive their expression. Second, NR2F2 appears to be necessary for direct activation of vein-specific enhancers and their associated genes. Third, NR2F2 directly represses artery enhancers in venous cells, such as Hey2, preventing their activation. Apparently, NR2F2 functions in multiple roles to maintain venous identity [35].
In lymphatic vessels, sustained expression of the transcription factor Prospero homeobox protein 1 (Prox1) is required to maintain the lymphatic identity in adults. Mice deficient in Prox1 do not develop lymphatic structures [48]. The transcription factor SRY-related gene (Sox) 18 is expressed before Prox1 and required for Prox1 expression during development. However, Sox18 alone is not enough for the induction of Prox1 expression. Presumably, some arterial-specific gene hinders Prox1 induction. Alternatively, some vein-specific factors could cooperate with Sox18 to induce Prox1. One of those factors might be NR2F2 since it was observed that NR2F2 acts jointly with Sox18 to induce Prox1 expression in embryonic veins [36]. NR2F2 binds directly to a conserved site within the upstream regulatory region of the Prox1 promoter and forms heterodimers with Prox1 [36, 37]. Though NR2F2 homodimers inhibit the Notch pathway, NR2F2 heterodimers with Prox1 are not able to accomplish this and, thus, Notch effectors are partially expressed in lymphatic vessels. Furthermore, heterodimers allow for the expression of lymph markers, such as VEGF-R3, which binds to VEGF-C and induces the expression of cyclin E1 [37, 38, 39]. Further evidence of the importance of NR2F2 in lymphatic specification is the observation that NR2F2 and Prox1 are suppressed by Hey1 and Hey2. Moreover, the expression of Notch effectors changes lymph cells into arterial-like cells. This suggests that lymph cells are extremely plastic and external stimuli could give rise to all types of endothelial cells. This is reinforced by the observation that all three master regulators of endothelial specification, namely Notch, NR2F2, and Prox1, are co-expressed in lymph cells, indicating a cross-control mechanism between these cell fate regulators. Consequently, a slight change in the expression of these regulators may lead to reprogramming of the lymphatic endothelial cell fate [40].
The role of the NR2F family in cancer progression appears to be diverse (Table 2, Ref. [49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70]). Depending on the cell type and biological processes examined, the NR2F family members have been proposed to act negatively or positively on cancer progression [49]. In some tumors, NR2F1 is re-expressed in the process of dedifferentiation and associated with the tumor phenotype [50], with higher expression compared to normal tissue samples [49, 50, 51]. In contrast, some cancer types have been reported to have lower NR2F1 expression compared to normal tissue [52, 53]. High NR2F1 expression is linked to increased cell proliferation and migration in breast cancer [50]. In contrast, high NR2F1 expression appears to function as a cell cycle break in other tumor types, such as prostate cancer or head and neck squamous cell carcinoma (HNSCC), which causes long-term quiescence in dormant cancerous cells [54]. One study reported the discovery of an NR2F1-specific agonist that activates dormancy programs in malignant cells, suppressing metastasis by inducing cancer cell dormancy [71]. Thus, even in tumor tissues, NR2F1 apparently has a multitude of functions; the expression levels and molecular roles in cancer development and progression differ based on the context and tissue type and whether cell proliferation or migration is affected [11].
| Family member | Area | Effect | Ref. |
| NR2F1 | Expression in tumor cells | Associated with tumoral phenotype | [49, 50, 51] |
| NR2F1 expression lower than in normal cells | [52, 53, 54] | ||
| NR2F2 | Angiogenesis regulator within the tumor microenvironment | Conditional ablation of NR2F2 compromises neoangiogenesis and suppresses tumor growth | [55] |
| NR2F2 | Facilitator of prostate tumorigenesis | NR2F2-mediated inhibition of TGF- |
[56] |
| Correlation of NR2F2 expression with disease progression | [56] | ||
| NR2F2 | Involvement in estrogen receptor expression | NR2F2 expression associated with loss of estrogen receptor expression | [57] |
| NR2F6 | Regulator in leukemia | Expression significantly induced in leukemia | [58] |
| Tumor suppressor in mixed-lineage leukemia acute lymphocytic leukemia 1-fused gene from chromosome 4 (MLL-AF4) acute leukemia | [59] | ||
| Patients with missense NR2F6 mutations in mast cell leukemia with associated hematological neoplasm (MCL-AHN) leukemia | [60] | ||
| NR2F6 | Prognostic marker | Muscle-invasive bladder cancer | [61] |
| Head and neck squamous cell carcinoma | [62] | ||
| Early-stage cervical cancer | [63] | ||
| Ovarian cancer | [65, 66] | ||
| NR2F6 | Colorectal cancer | Expression of NR2F6 significantly induced in colorectal cancer | [64] |
| NR2F6 | Involvement in liver cancer | Expression is implicated in progression of hepatocellular carcinoma | [67] |
| Expression contributes to rapid progression of hepatoblastoma | [68] | ||
| NR2F6 | Non-small cell lung cancer | Inhibition of NR2F6 expression results in suppression of proliferation, migration, and invasion of cancer cells | [69] |
| Single nucleotide polymorphism in nonsmall cell lung cancer (NSCLC) patients associated with better overall survival | [70] |
As mentioned above, NR2F2 plays an important role in angiogenesis. This is also relevant in the context of tumorigenesis and tumor progression because tumor growth depends on nutrients and oxygen supply via the vasculature through angiogenesis [72, 55]. One study described NR2F2 as a major regulator of angiogenesis within the tumor microenvironment. Conditional ablation of NR2F2 in adults massively compromised neoangiogenesis and suppressed tumor growth in xenograft mouse models. The same study observed that the absence of NR2F2 in a spontaneous mammary-gland tumor model resulted in impaired tumor growth and tumor metastasis. Overall, it appears that NR2F2 is an important regulator of the pathological neovascular response [55].
Mutations in phosphate and tensin homologue (PTEN) have frequently been observed
in human prostate cancer [73, 74]. PTEN loss can lead to upregulation of
TGF-
In mammary cancer cell lines, NR2F2 expression is associated with loss of estrogen receptor expression [57]. Estrogen receptor expression serves as a major indicator of the hormone-dependent cancer differentiation state. Tumors without estrogen receptor expression, so-called ER-negative tumors, are histologically less differentiated and have superior metastatic potential [76]. Interestingly, one study observed that estrogen receptor-positive breast cancer cells have increased NR2F2 expression, whereas estrogen receptor-negative cells expressed a low amount of NR2F2 [77].
The third family member, NR2F6, has been reported by various studies to be
involved in carcinogenesis and progression. For example, NR2F6 expression is
significantly induced in leukemia [58]. Mechanistically, the same group reported
that NR2F6 inhibits hematopoietic cell differentiation and induces myeloid
dysplasia [78]. One study focusing on t (4;11) MLL-AF4 acute leukemia, a specific
form of acute lymphoblastic leukemia characterized by the MLL-AF4 fusion gene
associated with poor prognosis, identified NR2F6 as a novel tumor suppressor of
MLL-AF4
NR2F6 has been proposed as a prognostic biomarker of muscle-invasive bladder cancer (MIBC) and HNSCC [61, 62]. NR2F6 expression has also been reported to correlate with pelvic lymph node metastasis and overall poorer prognosis in early-stage cervical cancer [63]. NR2F6 expression is significantly induced in colorectal cancer [64]. NR2F6 upregulation has also been detected in ovarian cancer and is associated with significantly worse overall survival [65]. In breast cancer, one study reported the involvement of NR2F6 in regulating the docetaxel chemosensitivity [66]. Furthermore, overexpression of NR2F6 was described to promote the chemoresistance of epithelial ovarian cancer by activating the Notch3 signaling pathway. The same study proposed NR2F6 as a biomarker for identifying patients who are likely to respond to therapy with gamma-secretase inhibitors, which inhibit Notch signaling [79].
In hepatocellular carcinoma (HCC), NR2F6 expression is implicated in disease progression. Knocking out NR2F6 inhibits the growth, migration, and invasion of HCC cells [67]. Corresponding to these observations, NR2F6 upregulation has been reported to be higher in hepatoblastoma than non-cancer livers, and NR2F6 expression was suggested to contribute to the rapid progression of residual liver tumor in hepatoblastoma [68].
NR2F6 also plays a role in the progression of non-small cell lung cancer (NSCLC). Mechanistically, the microRNA miR-142-3p directly inhibits NR2F6 expression, resulting in suppression of proliferation, migration, and invasion of cancer cells [69]. In accordance with this observation, one study reported that an analysis of single nucleotide polymorphisms at miRNA target sites in 782 early-stage NSCLC patients revealed that patients carrying the NR2F6 rs2288539 TT genotype have significantly better overall survival than patients with the NR2F6 rs2288539 CC or CT genotypes [70].
Evidently, NR2F family members can act as promoters or inhibitors of phenotypic modifications during cancer development and progression depending on the cellular context, expression levels, or possibly other transcription factors and signaling pathways [49].
The expression of NR2F2 in metabolic tissues was described for the first time
about 20 years ago [80]. Since then, numerous studies have elucidated the role of
NR2F2 in various metabolic systems, such as adipogenesis, lipid metabolism,
insulin secretion, and hepatic gluconeogenesis (Table 3, Ref. [25, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93]) [81]. NR2F2 heterozygous
mice are characterized by less white adipose tissue than wild-type mice fed a
high-fat diet and maintain a lean body mass phenotype. These heterozygous mice do
not develop obesity. Mechanistically, they have reduced expression of NR2F2 in
3T3L1 cells, leading to increased Wnt signaling, which is known to be a
repressive factor in adipogenesis [82]. Furthermore, Wnt/
| Family member | Area | Effect | Ref. |
| NR2F2 | Adipose tissue regulation in NR2F2 heterozygous mice | Less white adipose tissue, no obesity when fed high-fat diet | [82] |
| NR2F2 | Wnt/ |
Reduction of NR2F2 expression increases Wnt signaling | [82] |
| Activation of NR2F2 expression due to Wnt/ |
[83] | ||
| Potentially important |
[84] | ||
| NR2F2 | Adipocyte differentiation | Necessary for adipocyte differentiation | [83] |
| NR2F2 | Insulin secretion | Tissue-specific knockdown of NR2F2 in |
[85] |
| Reciprocal regulation of insulin and NR2F2 due to forkhead box protein O1 (FOXO1) | [85] | ||
| Humans: single nucleotide polymorphism observed with lower blood insulin concentrations | [86] | ||
| NR2F2 | Liver function | Expression in liver inhibited by glucose and insulin | [80, 85] |
| Induces upregulation of gluconeogenic enzymes and |
[87] | ||
| Interaction with glucocorticoid receptor | [81] | ||
| NR2F2 | Mitochondrial dysfunction | Overexpression causes defects in electron transport chain activity and higher reactive oxygen species (ROS) production | [88] |
| NR2F2 | Skeletal muscle and heart muscle | Levels in skeletal muscle regulate Glut4 expression | [89] |
| Important for myogenesis in skeletal muscle | [90] | ||
| Overexpression reduces cardiac performance and results in cardiomyopathy and heart failure | [25, 88] | ||
| NR2F2 | Fibrosis | Increased NR2F2 expression in myofibroblasts | [92] |
| NR2F6 | Adipocyte differentiation | Downregulation of NR2F6 inhibits adipogenesis | [91] |
| NR2F6 | Liver function | NR2F6 upregulation in patients with non-alcoholic fatty liver disease | [92] |
| Beef cattle: key regulator of hepatic inflammatory response | [93] |
NR2F2 appears to be important for
Analogous to the pancreas, expression of NR2F2 in the liver is inhibited by
glucose and insulin in vitro and in vivo [80, 85]. NR2F2
expression in mice is upregulated under fasting conditions and downregulated upon
re-feeding. During fasting, NR2F2 has been suggested to be involved in hepatic
glucose production via upregulation of gluconeogenic enzymes and
NR2F2 is expressed in multiple tissues and organs; therefore, its role in
metabolism may not be restricted to the liver and pancreas [94]. Skeletal muscle
is one of the most metabolically active tissues, and it is necessary to regulate
the high energy demand via metabolic processes [95]. SiRNA-mediated knockdown of
NR2F2 expression in mouse C2C12 myoblast cells reduces important genes implicated
in the fatty acid
However, the involvement of NR2F2 is not limited to skeletal muscle cells; it is also observed in heart muscle cells. One study reported that overexpression of NR2F2 in mice markedly reduces cardiac performance [25]. Although evidence supports NR2F2 being crucial for the correct function and development of the cardiovascular system, its overexpression appears to be detrimental and may increase the risk of heart diseases. In mice, overexpression of NR2F2 in the myocardium results in dilated cardiomyopathy and heart failure [88].
A connection between metabolic disturbance and the development of fibrosis has been described in recent studies. One study reported increased NR2F2 expression in myofibroblasts from human fibrotic kidneys, lungs, kidney organoids, and mouse kidneys after injury. Genetic attenuation of NR2F2 in mice mitigates injury-induced kidney fibrosis. Mechanistically, suppression of fatty acid oxidation and enhancement of glycolysis pathways were the result of NR2F2 overexpression in fibroblasts [97].
NR2F6 has been implicated in metabolic regulation and has been reported to play a role in obesity control, as NR2F6 has been observed to be required for adipocyte differentiation and that downregulation of NR2F6 inhibits adipogenesis [91]. One study investigated the function of NR2F6 in the context of hepatic triglyceride homeostasis and reported NR2F6 as an important regulator and a causal factor in the development of non-alcoholic fatty liver disease. Mechanistically, the fatty acid translocase CD36, as a transcriptional target of NR2F6, has been implicated in this process. Furthermore, NR2F6 is upregulated in the livers of obese mice and patients with non-alcoholic fatty liver disease [92].
Taken together, these findings clearly demonstrate a crucial role of NR2F family members, particularly NR2F2, in multiple different aspects of metabolic regulation in mice and humans. A similar finding was observed in one study in which the feed efficiency in beef cattle was studied. They identified NR2F6 as a key regulator of the hepatic inflammatory response, one of the main processes associated with feed efficiency [93].
Regarding the functional importance of the NR2F family, the focus thus far has been mainly on NR2F1 and NR2F2. However, knowledge regarding the role of NR2F1 and NR2F2 can serve as an important indicator of the functionality of the third NR2F family member, NR2F6, which has not been studied as intensively as the other family members. Based on our published [98, 99, 100, 101, 102] and unpublished data, NR2F6 appears to be important in cancer immunology and T-cell functionality and needs to be investigated in more detail.
The functional importance of NR2F6 has been investigated in various immune cells. In mouse macrophages, NR2F6 acts as a transcriptional repressor of cytokines, whereas in human macrophages it acts as a transcriptional activator of chemokines [103]. Formerly introduced solely as a Th17 transcriptional repressor, the role attributed to NR2F6 has gradually broadened over the years and been extended to most known immune cell types (Table 4, Ref. [98, 100, 103, 104, 105, 106]). The influence of NR2F6 is particularly prominent among T-cell subsets, as NR2F6 regulates the differentiation and function of multiple subsets of CD4 and CD8 T cells, lending it a crucial position in adaptive immunity and anti-tumoral responsiveness. Examination of NR2F6 function in the germinal center response has revealed that Nr2f6 deficiency increases the accumulation of germinal center B cells, plasma cells, and follicular T helper cells in mice that were immunized. Mechanistically, NR2F6 governs the expression of IL-21 through direct binding at several defined sites within the Il21 locus [104].
| Cell type | Effect | Ref. |
| Macrophages | Mice: transcriptional repressor of cytokines | [103] |
| Humans: transcriptional activator of chemokines | [103] | |
| B cells | Nr2f6 deficiency leads to accumulation of germinal center B cells secondary to follicular T helper cells | [104] |
| Plasma cells | Nr2f6 deficiency leads to accumulation of plasma cells | [104] |
| Follicular T helper cells | Nr2f6 deficiency leads to accumulation of follicular T helper cells | [104] |
| Autoimmunity via T-cell functions | Loss of Nr2f6 exacerbates experimental autoimmune encephalomyelitis (EAE) due to negative regulation of Il17a expression by a direct NFAT and/or retinoic acid receptor-related orphan receptor C (RORC) antagonism | [105, 106] |
| Cancer surveillance via T-cell functions | Nr2f6-deficient mice exhibit tumor growth inhibition benefit due to increased CD4 and CD8 T-cell infiltration leading to overall survival benefit | [98, 100] |
| In approximately 50% of human NSCLC biopsies, NR2F6 upregulation in tumor-infiltrating T lymphocytes (TILs) presumably contributes to the observed T-cell exhaustion | [100] |
Investigating NR2F6 functionality in T cells, our team and others have shown that NR2F6 plays an important role in autoimmunity and immune functions during cancer surveillance [98, 105, 107]. Our team observed that loss of Nr2f6 exacerbates experimental autoimmune encephalomyelitis (EAE). Mechanistically, NR2F6 appears to act as a negative regulator of Il17 transcription via direct DNA binding to the Il17 promoter region. This abrogates NFAT/AP-1 transcription factor binding to the Il17 gene locus, leading to robust NR2F6-mediated transrepression, thereby acting as a safe guard against EAE disease progression [105, 106].
On the other hand, NR2F6 appears to be an essential signaling intermediate
governing the amplitude of host-protective cancer immunity. The observation that
immune cells play a role in preventing cancer cell progression was first
described by Burnet and Thomas more than 60 years ago [108]. In accordance with
this observation, an increased number of tumor-infiltrating T lymphocytes (TILs)
is associated with better survival prognosis [109]. In several induced and
spontaneous mouse tumor models, we observed that Nr2f6-deficient mice exhibit a
reduction in cancer growth due to increased CD4 and CD8 T-cell tumor infiltration
and augmented effector T-cell functions, such as IL-2 and IFN
Mechanistically, during T-cell activation, regulatory phosphorylation of the
NR2F6 DBD serves as an important feedback mechanism. High-affinity antigen
receptor signaling (i.e., PKC
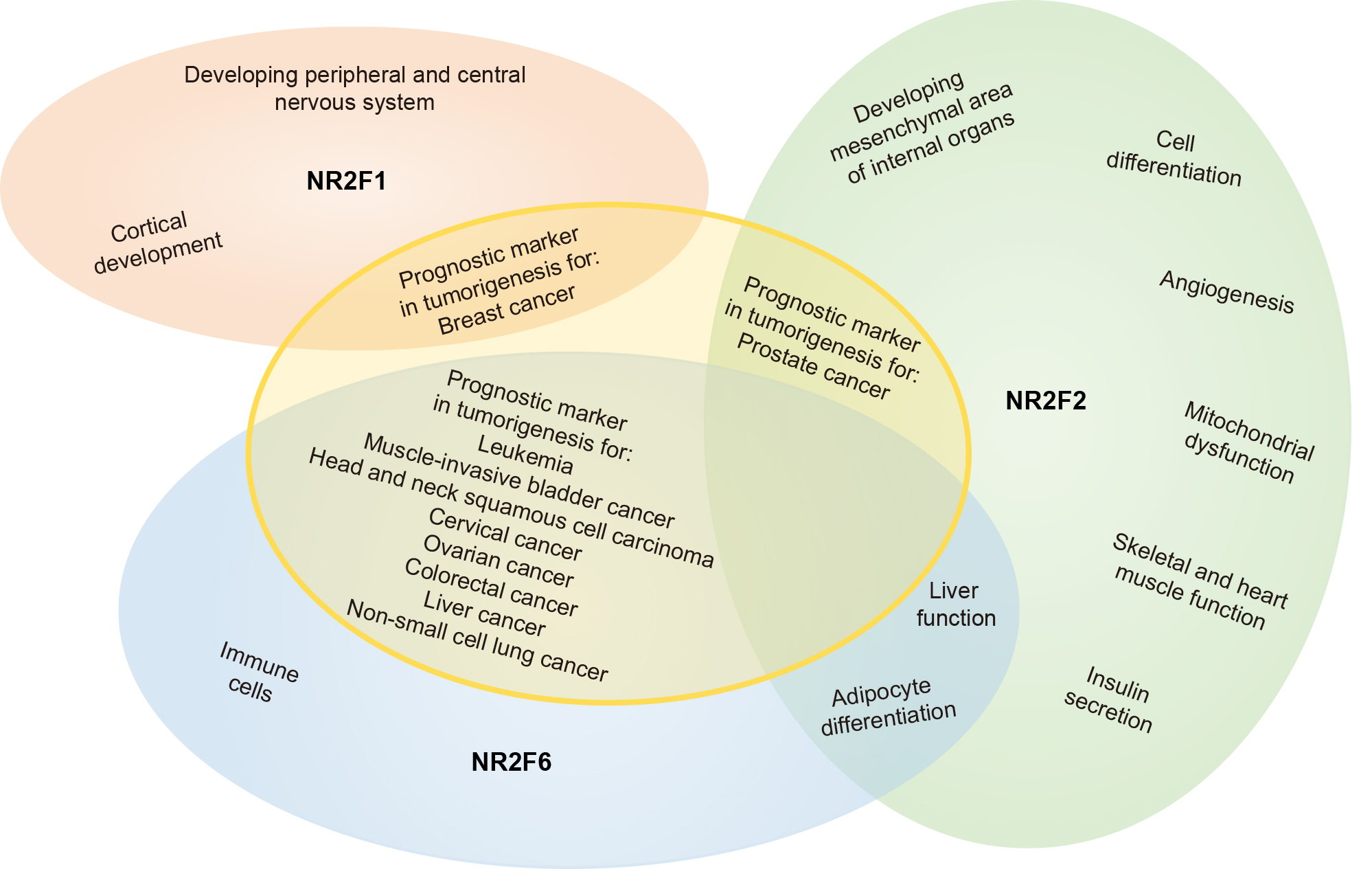
A schematic overview of NR2F family member functions, as discussed in this review and summarized per topic in Tables 1,2,3,4, is illustrated in Fig. 1.
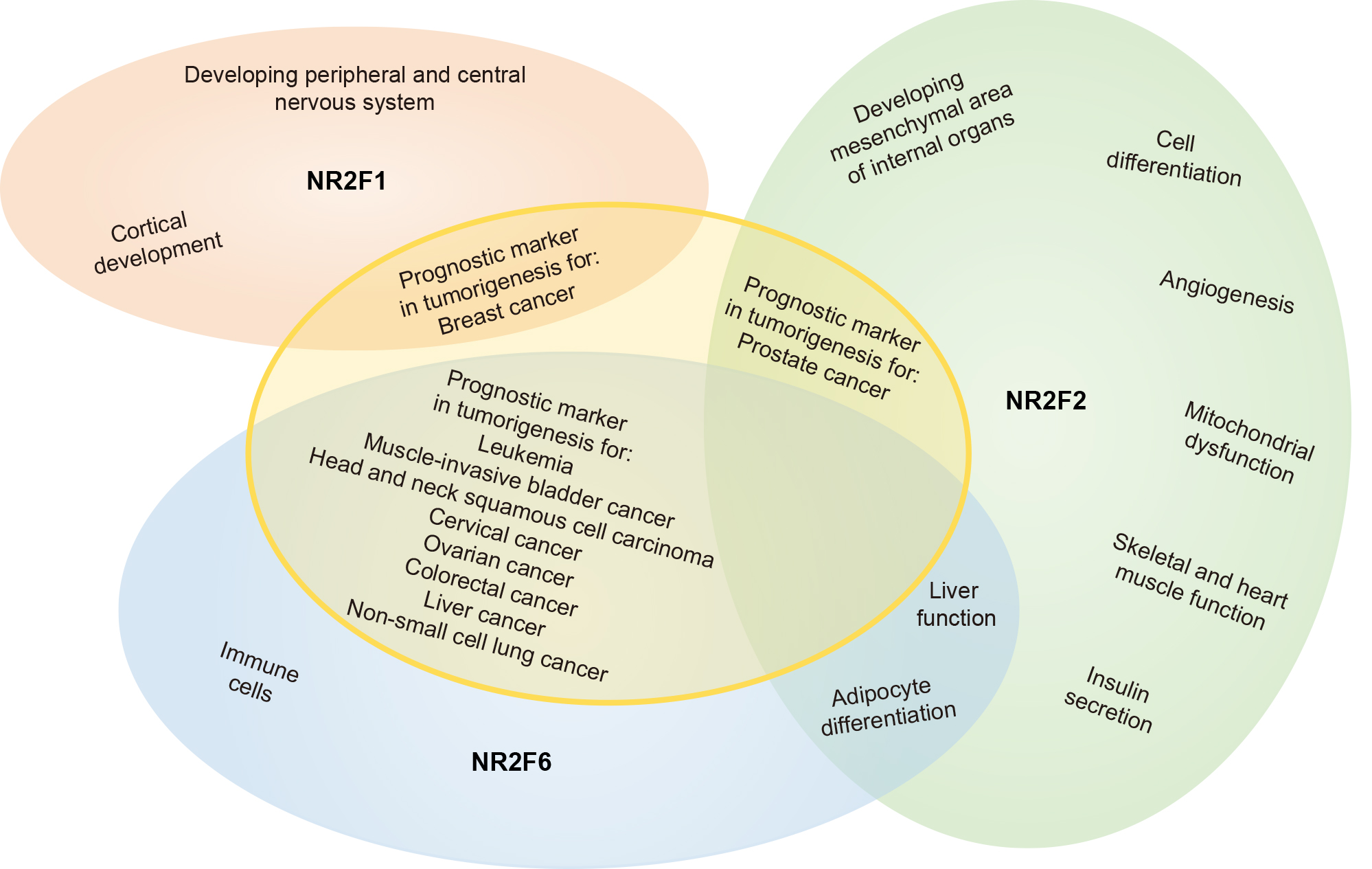 Fig. 1.
Fig. 1.Schematic overview of NR2F nuclear orphan receptor family functionality. A summary of the functions of NR2F family members in a broad range of areas. Though NR2F1 and NR2F2 are more structurally related to each other compared to NR2F6, it appears that NR2F2 and NR2F6 are more closely related functionally. An extensive summary of the functions of NR2F1, NR2F2, and NR2F6 can be found in Tables 1,2,3,4.
Profound biochemical advances have been made in understanding how NR2F family members regulate gene expression. NR2F family members are central for cell growth and developmental processes (Fig. 1). Currently under investigation at the molecular level, one major challenge is the functional connections between NR2F homo- or hetero-dimeric complexes bound to a particular promoter, their tissue-specific co-activators and co-repressors, and the specific properties of each complex with respect to gene regulation.
Yet, the NR2F family has substantial translational importance, and only the complementary information listed below will give us a better understanding of the functions of NR2F isoforms in key mechanisms of cell-type speciation. The critical details that need to be considered are (i) genome-scale validation of NR2F isoform-specific versus NR2F family-overlapping DNA-binding specificites that control gene transcription, (ii) the signaling pathways responsible for induction of NR2F gene expression as feed-forward and/or feedback processes, (iii) epigenetic regulation by DNA methylation present in NR2F family gene promoters, and (iv) the identification of signaling pathways that respond to specific cellular stimuli for post-translational regulation, such as phosphorylation and sumoylation on NR2F protein functions.
Due to their excellent druggability, NRs are promising candidates as novel therapeutic targets for cancer therapy, and numerous clinical or preclinical trials are currently underway to assess the therapeutic efficacy of estrogen receptor and androgen receptor inhibitors [2]. NR2F6 has no known ligands thus far, but recent publications describe both closely related NR2F family members NR2F1 [71] and NR2F2 [110] as druggable targets, suggesting the possibility of finding a small-molecule antagonist that targets the immune checkpoint NR2F6 due to the shared LBD between all three NR2F family members [3]. A detailed review discussing NR2F6 as a next-generation target for cancer immunotherapy was published recently [102].
Future studies promise insights into the specific roles of NR2F isoforms in defined tissues and will ultimately not only shed light on normal NR2F protein functions, but probably also lead to unique insights into human disorders, such as cancer and autoimmunity, paving the way for novel treatment options.
AP-1, activator protein 1; BBSOAS, Bosch-Boonstra-Schaaf optic atrophy syndrome;
BRG-1, Brahma-related gene 1; COUP-TF, chicken ovalbumin upstream promoter
transcription factor; DBD, DNA binding domain; Dll-4, delta-like protein 4; EAE,
experimental autoimmune encephalomyelitis; EPC, endothelial progenitor cell;
FOXO1, Forkhead box protein O1; Glut4, glucose transporter type 4; HCC,
hepatocellular carcinoma; Hey2, Hes-related family BHLH transcription factor with
YRPW motif 2; HIF
Conceptualization, writing and editing by TS and GB. All authors have read and agreed to the published version of the manuscript.
Not applicable.
We thank Victoria Klepsch, Nikolaus Thuille, Natascha Hermann-Kleiter, Thomas Gruber, and Kerstin Bellaire-Siegmund, all from our institute, for critical input.
This work was funded by FWF Austrian Science Fund P31383-B30 and ERC_ADG #786462 – HOPE.
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.