1 Department of Dermatology, Xiangya Hospital, Central South University, 410008 Changsha, Hunan, China
2 Department of Pharmacy, Xiangya Hospital, Central South University, 410008 Changsha, Hunan, China
3 Department of Clinical Pharmacology, Xiangya Hospital, Central South University, 410008 Changsha, Hunan, China
4 National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, 410008 Changsha, Hunan, China
Academic Editor: Giuseppe Ingravallo
Abstract
Background: Psoriasis vulgaris is an immune-mediated inflammatory skin disease. Although the pathogenesis of psoriasis is unclear, genetic susceptibility, such as HLA-C*06:02, is believed to be a major risk factor. However, there is a paucity of knowledge regarding the relationship between genetics and the response to systemic treatment of psoriasis. We hypothesized that genetic variations in human leukocyte antigen (HLA) genes may act as predictors of acitretin treatment in psoriasis. The aim of our study was to explore the presence of HLA gene variants in patients with moderate-to-severe psoriasis receiving acitretin treatment. Methods: A total of 100 Han Chinese patients with psoriasis completed the study. 24 patients including 16 responders and 8 non-responders underwent deep sequencing by MHC targeted region capture and 76 samples were genotyped by Sanger sequencing (SBT) based HLA typing for validation. Results: Regressions with adjustment for age, sex, body mass index (BMI), and baseline psoriasis area and severity index (PASI) revealed that two HLA alleles (HLA-DQA1*:02:01, DQB*:02:02) were associated with the response to acitretin. The DQA1*0201-positive patients showed a better response to acitretin compared to the DQA1*0201-negative patients (relative risk (RR) = 10.34, 95% confidence interval (CI): 2.62–40.77, p = 0.001), and the DQB1*0202-positive patients manifested a better response to acitretin when compared to the DQB1*0202-negative patients (RR = 21.01, 95% CI: 2.53–174.27, p = 0.005). Conclusions: Our observations support the potential role of HLA-DQA1*:02:01 and DQB*:02:02 as pharmacogenetic markers of the acitretin response in patients with psoriasis.
Keywords
- psoriasis
- acitretin
- human leukocyte antigen
- pharmacogenetics
- response
Psoriasis vulgaris is a chronic, immune-mediated inflammatory disease that affects approximately 2–3% of the general population [1, 2, 3]. It is characterized by increased proliferation of keratinocytes and abnormal activation of T-cells [4, 5, 6]. However, the pathogenesis of psoriasis remains poorly understood and may include genetic and immunological factors and abnormal metabolism [3, 7, 8]. The severity of psoriasis vulgaris is defined using the psoriasis area and severity index (PASI) or body surface area (BSA), which can be categorized into three stages: mild, moderate, and severe. For moderate and severe psoriasis vulgaris, systemic therapies or phototherapies such as methotrexate, cyclosporin, retinoids, and etanercept are used either as single agents or in combination [9, 10].
Acitretin is a second-generation retinoid that is usually used as a first-line
medication for treating moderate-severe psoriasis in China. Mechanistically,
acitretin inhibits the proliferation and differentiation of keratinocytes in the
skin and induces the differentiation of regulatory T cells [11]. However, only
50–60% of patients respond well to acitretin, and the factors involved in drug
response prediction are unclear [12]. Our previous research showed that
the rs1802073G
In the past 50 years, studies have found that genes within the human major histocompatibility complex (MHC) region at 6p21.3 are associated with the pathogenesis of psoriasis [15, 16, 17, 18]. In the past, Zhang et al. [15] found multiple new susceptibility loci for psoriasis in the HLA genes, such as HLA-C, HLA-B, and HLA-DPB1, through deep sequencing of the MHC region in controls and psoriasis cases. The underlying mechanism could be explained by a malfunction in HLA gene alleles that regulate the expression of T cell function-related key proteins, which could disrupt the balance between T-helper cells, mainly Th1/Th17, and cause the progression of psoriasis [19, 20]. The induction of regulatory T cell differentiation is a major pharmacological mechanism of acitretin. Therefore, it is rational to hypothesize that HLA gene alleles could influence acitretin efficacy by altering T cell function in patients with psoriasis.
In this study, MiSeq high-throughput sequencing was used to analyze HLA typing in 100 patients with psoriasis who were treated with acitretin for at least 8 weeks. We also sought to identify the relationship between several HLA alleles and acitretin efficacy.
A group of 100 Han Chinese patients diagnosed with moderate-to-severe psoriasis
vulgariswas recruited through the Department of Dermatology, Xiang Ya Hospital,
Central South University. The psoriasis patients who enrolled in this research
with the inclusion criteria as follows: (i) patients fulfilled the diagnostic
criteria for psoriasis vulgaris, and the PASI score
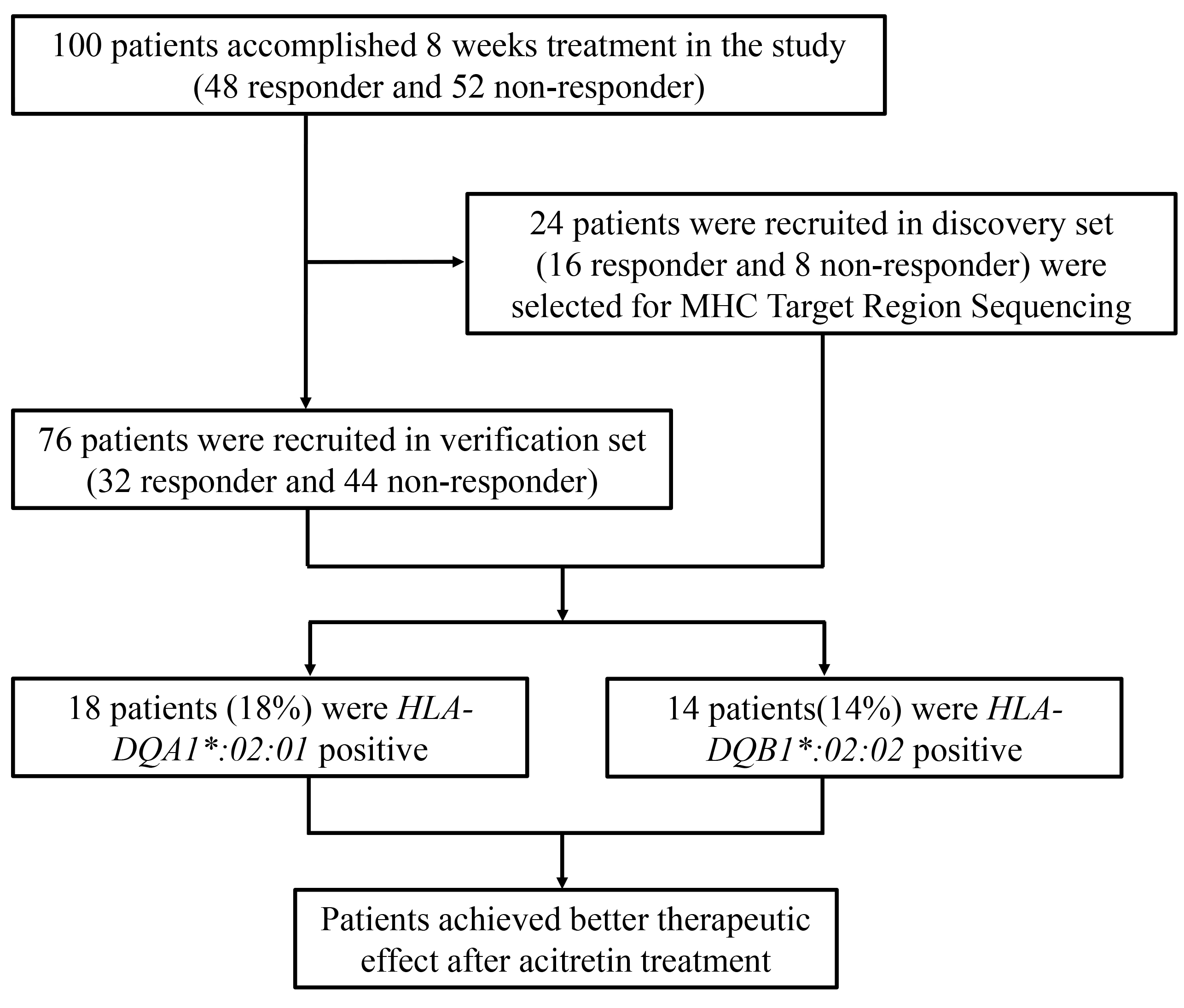 Fig. 1.
Fig. 1.Flow diagram: acitretin was prescribed and provided for all subjects at the time of enrolment. All subjects were followed for at least 8 weeks with an onsite interview at 4 weeks interval; 24 subjects were recruited in discovery phases, including 16 responders (achieved 90% PASI improvement) and 8 non-responders (failed to achieve 10% PASI improvement).
Twenty-four samples (16 responders and 8 non-responders) in the discovery phase
were sequenced using the MHC targeted region capture method described previously
[15, 21]. DNA was extracted by a commercial DNA extraction kit (QIAamp, QIAGEN
GmbH) from the PBMCs (Peripheral Blood Monouclear Cells), the shotgun libraries
were constructed using 3
The sequences of sequenced samples were mapped to the NCBI human genome
reference assembly (Hg18) using BWA48. The average depth of the sequencing was
80x for the MHC region. BAM files were realigned around known indels using GATK v
1.6 (Genome Analysis Toolkit 49). All aligned reads were subjected to
CountCovariates (GATK) on the basis of known single-nucleotide variants (SNVs)
(dbSNP135), and the base quality was then recalibrated. A high quality
single-nucleotide variant (SNV) genotype quality of
SBT-based HLA typing was used to validate the 76 samples. For samples identified as homozygous, PCR-sequence-specific primer (SSP) was used for further validation. The PCR reaction conditions were described in 2.2.
All analyses were performed using the SPSS 23.0 statistical package (IBM SPSS,
Chicago, IL, USA). Fisher’s exact test was used when data were sparse. Comparisons of
continuous variables between the different genotype groups were performed using
independent-sample t-tests. The chi-square test was used to examine the
association between HLA types and acitretin response, when one or two of
the cells have expected count less than 5, an alternative approach with 2
In total, 100 patients (28 women and 72 men) completed the study and their
samples were sequenced for the presence of DQA1 and DQB1 alleles. Interestingly, the ages of non-responders were significantly older than
those of the responders (41.6
| Characteristics | Discovery set (n = 24) | p | Verification set (n = 76) | p | Total (n = 100) | p | |||
| Responder | Non-responder | Responder | Non-responder | Responder | Non-responder | ||||
| (n = 16) | (n = 8) | (n = 32) | (n = 44) | (n = 48) | (n = 52) | ||||
| Age (mean |
34.7 |
44.6 |
0.088 | 36.5 |
41.1 |
0.102 | 35.9 |
41.6 |
0.020 |
| PASI at baseline (mean |
13.8 |
14.1 |
0.881 | 13.4 |
13.0 |
0.792 | 13.5 |
13.1 |
0.773 |
| BMI (mean |
23.3 |
25.7 |
0.187 | 22.1 |
22.9 |
0.294 | 22.5 |
23.4 |
0.235 |
| Male, n (%) | 12 (75) | 3 (37.5) | 0.180 | 27 (84.4) | 30 (68.2) | 0.107 | 39 (81.2) | 33 (63.5) | 0.048 |
| Female, n (%) | 4 (25) | 5 (62.5) | 5 (15.6) | 14 (31.8) | 9 (18.8) | 19 (36.5) | |||
Twenty four patient samples were selected for MHC target region sequencing analysis. After identification of all the HLA alleles, chi-square test analyses were used to identify the allele distribution between the responder and non-responder groups of acitretin. We obtained 11 positive HLA alleles in this study. As shown in Table 2, we found that HLA-A*:30:01, B*:13:02, C*:01:02, DQB1*:02:02, DQB1*:03:03, DQB1*:05:01, G*:01:05N, DMB*:01:01, DMB*:01:03, MICA*:008:01, and MICA*:010:01 were significantly associated with acitretin response.
| HLA alleles | Responder (n = 16) | Non-responder (n = 8) | p | ||
| 32 alleles (%) | 16 alleles (%) | ||||
| positive | negative | positive | negative | ||
| A*:30:01 | 9 (28.1) | 23 (71.9) | 0 (0) | 16 (100) | 0.05 |
| B*:13:02 | 10 (31.3) | 22 (68.7) | 0 (0) | 16 (100) | 0.033 |
| C*:01:02 | 3 (9.4) | 29 (90.6) | 6 (37.5) | 10 (62.5) | 0.05 |
| DQB1*:02:02 | 9 (28.1) | 23 (71.9) | 0 (0) | 16 (100) | 0.05 |
| DQB1*:03:03 | 3 (9.4) | 29 (90.6) | 6 (37.5) | 10 (62.5) | 0.05 |
| DQB1*:05:01 | 1 (3.1) | 31 (96.9) | 5 (31.2) | 11 (68.8) | 0.012 |
| G*:01:05N | 9 (28.1) | 23 (71.9) | 0 (0) | 16 (100) | 0.05 |
| DMB*:01:01 | 23 (71.9) | 9 (28.1) | 5 (31.2) | 11 (68.8) | 0.007 |
| DMB*:01:03 | 9 (28.1) | 23 (71.9) | 10 (62.5) | 6 (37.5) | 0.022 |
| MICA*:008:01 | 17 (53.1) | 15 (46.9) | 3 (18.8) | 13 (81.2) | 0.023 |
| MICA*:010:01 | 3 (9.4) | 29 (90.6) | 6 (37.5) | 10 (62.5) | 0.05 |
We analyzed the relationship between HLA-DQA1 and DQB1 gene polymorphisms and the response to acitretin in the verification set and found an increased RR to acitretin in patients who were DQA1*0201-positive (POS) and DQB1*0202 POS. The combination of data from the discovery and verification sets supported this finding. The adjusted RRs were consistent with the crude estimates.
DQA1*0201 was significantly more frequent in responders than in non-responders (18/30 vs 3/49, p = 0.0001), and DQA1*0201 POS was associated with a better response to acitretin compared to DQA1*0201-negative (NEG) (RR = 10.34, 95% confidence interval (CI): 2.62–40.77, p = 0.001).
DQB1*0202 was significantly more frequent in responders than in non-responders (14/34 vs 1/51, p = 0.00014), and DQB1*0202 POS was associated with a better response to acitretin compared to DQB1*0202 NEG (RR = 21.01, 95% CI: 2.53–174.27, p = 0.005) (Table 3).
| Phase | HLA alleles | Responder (n, %) | Non-responder (n, %) | p |
p |
Adjusted RR | ||
| positive | negative | positive | negative | |||||
| Verification set | DQA1*:02:01 | 9 (29.0) | 22 (71.0) | 2 (4.4) | 43 (95.6) | 0.008 | 0.015 | 8.41 (1.52–46.60) |
| DQB1*:02:02 | 6 (19.4) | 25 (80.6) | 1 (2.2) | 44 (97.8) | 0.016 | 0.03 | 12.87 (1.27–130.10) | |
| Discovery + Verification Set | DQA1*:02:01 | 18 (37.5) | 30 (62.5) | 3 (5.8) | 49 (94.2) | 0.0001 | 0.001 | 10.34 (2.62–40.77) |
| DQB1*:02:02 | 14 (29.2) | 34 (70.8) | 1 (1.9) | 51 (98.1) | 0.00014 | 0.005 | 21.01 (2.53–174.27) | |
As presented in Table 4, we observed an obvious increase in the response rate to acitretin in patients with DQA1*0201 and DQB1*0202. At week 8, PASI75 was achieved in 93.3% (14 of 15) of patients with DQA1*0201–DQB1*0202, compared to 38.0% (30 of 79) of patients without DQA1*0201–DQB1*0202 (RR = 24.69, p = 0.003, 95% CI: 2.92–208.91). Additionally, 85.7% (18 of 21) of patients with DQA1*0201 reached PASI75, compared to patients without DQA1*0201– DQB1*0202 (RR = 10.34, p = 0.001, 95% CI: 2.62–40.77).
| HLA alleles | Responder (n, %) | Non-responder (n, %) | p | p |
Adjusted RR |
| DQA1*:02:01POS DQB1*:02:02POS | 14 (93.3) | 1 (6.7) | 0.00008 | 0.003 | 24.69 (2.92–208.91) |
| VS | 30 (38.0) | 49 (62.0) | |||
| DQA1*:02:01NEG DQB1*:02:02NEG | |||||
| DQA1*:02:01POS |
18 (85.7) | 3 (14.3) | 0.0001 | 0.001 | 10.34 (2.62–40.77) |
| VS | 30 (38.0) | 49 (62.0) | |||
| DQA1*:02:01NEG DQB1*:02:02NEG | |||||
According to a previous report, HLA-DQA1*0201 and DQB1*0202 are susceptible genes for psoriasis, and 21% of patients with psoriasis were DQA1*0201 POS [22]. Although the frequency of DQA1*0201 positivity in patients with psoriasis in our study was slightly lower (18%) than that reported in Zhang’s study (24%) [15], the allele frequency of both variants was much higher in patients with psoriasis than in healthy controls (8%) [22].
In the present study, we found an association between the presence of DQA1*02:01 and DQB1*02:02 alleles and a good clinical response to acitretin in the Han Chinese population. Patients carrying the DQA1*02:01 or DQB1*02:02 loci had a better response than non-carriers to acitretin treatment, which suggests that the DQA1*02:01 or DQB1*02:02 loci may act as genetic predictors of the response to acitretin, although we cannot exclude the possibility of linkage disequilibrium between DQA1*02:01 and DQB1*02:02 and other genetic features of psoriasis. Additionally, compared to the patients who were DQA1*0201 NEG–DQB1*0202 NEG, the patients with DQA1*:02:01 POS had a good response to acitretin (RR = 10.34, p = 0.001), and patients with DQA1*0201 POS–DQB1*0202 POS were associated with a better response (RR = 24.69, p = 0.003). Therefore, DQB1*0202 is an important allele for the response to acitretin in psoriasis (Table 4).
Acitretin belongs to the family of retinoid and is also known as a member of the
retinoic acid receptors (RARs) agonist family. RARs are highly expressed in the
keratinocytes, and the expression levels of RAR
Acitretin has anti-inflammatory effects. It decreases the populations of
T-cells, Th1 cells, and Th17 cells in psoriasis lesions, which are essential for
the genesis of psoriatic plaques, further leading to the inhibition of
intra-epidermal inflammation [11, 28]. DQA1 and DQB1 encode the
DQA1 and DQB1 belong to HLA Class II alpha and beta
paralogues respectively, which consist of an alpha (DQA) and a beta chain (DQB).
The two chains anchored in the membrane form a functional protein complex, known
as the antigen-binding DQ
In summary, patients with moderate-to-severe psoriasis require an individualized approach during treatment with acitretin, and an understanding of the clinical relevance of HLA gene polymorphisms in relation to acitretin treatment for psoriasis is needed. Our data suggest that DQA1*02:01 and DQB1*02:02 alleles as the pharmacogenetic markers of the response to acitretin may provide a useful reference in psoriasis for targeted therapies.
XZ performed the experiments, analyzed data and wrote the manuscript; WZ and YK collected biological sample; XZ and YH analyzed the data; WZ and WC designed the experiments, carried out and summarized data analysis, contributed to the text of the manuscript.
The study was conducted in accordance with the Declaration of Helsinki, and approved by Ethics Committee of Xiangya Hospital (protocol code: 201512526).
We sincerely thank the participating psoriasis patients for their generous support of this study.
The study was supported by the National Natural Science Foundation of China (NSFC) (No. 81903222, 81974479), the Hunan Provincial Natural Science Foundation of China (No. 2021JJ70152), the Project of Intelligent Management Software for Multimodal Medical Big Data for New Generation Information Technology, Ministry of Industry and Information Technology of People’s Republic of China (TC210804V).
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.

