1 Department of Ophthalmology, Henan Provincial People’s Hospital, 450003 Zhengzhou, Henan, China
2 Henan Eye Hospital, Henan Eye Institute, Henan Key Laboratory of Ophthalmology and Visual Science, 450003 Zhengzhou, Henan, China
3 Department of Ophthalmology, People’s Hospital of Zhengzhou University, 450003 Zhengzhou, Henan, China
4 Department of Ophthalmology, People’s Hospital of Henan University, 450003 Zhengzhou, Henan, China
5 Department of Medicine/Hematology, Keck School of Medicine, University of Southern California, Los Angeles, CA 90033, USA
6 Department of Ophthalmology, Tianjin Medical University General Hospital, 300052 Tianjin, China
7 Laboratory of Molecular Ophthalmology, Tianjin Medical University, 300052 Tianjin, China
8 Department of Pharmacology and Tianjin Key Laboratory of Inflammation Biology, School of Basic Medical Sciences, Tianjin Medical University, 300052 Tianjin, China
9 Department of Pathology and Ophthalmology, USC Roski Eye Institute, Keck School of Medicine, University of Southern California, Los Angeles, CA 90033, USA
†These authors contributed equally.
Academic Editor: Graham Pawelec
Abstract
N6-methyladenosine (m6A) methylation/modification plays a critical role in various biological processes through post-transcriptional ribonucleic acid (RNA) modification, which involves RNA processing, nuclear export, translation and decay. Functionally, m6A modification may be involved in ocular cell growth and differentiation, stem cell identity, development, haemostasis and innate versus adaptive immunity. Aberrations in m6A methylation may mediate numerous pathological conditions in the eye, including microorganism infection, inflammation, autoimmune disease, senescence, degeneration, epithelial–mesenchymal transition, fibrosis, angiogenesis, tumorigenesis and complex eye diseases. In this review, we have discussed the relevance of m6A modification to precision medicine, stem cell directional differentiation, biomarkers of eye diseases and m6A methylation activators and inhibitors. In addition, we summarised the challenges and future research directions in the field related to visual function and eye diseases.
Keywords
- eye
- m6A
- pathogenesis
- vision function
The principle of molecular biology follows the traditional theory that genetic information flows from deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) to protein. However, it is difficult to explain the ever-changing cellular phenomenon using only the central dogma. In the last decade, it has been found that epigenetic factors control the expression and function of genes and hence determine the state of cells. Epigenetics has been a prime research focus in many disciplines, including medicine, zoology, botany and reproductive development. Several aspects regarding the pathogenesis, diagnosis and treatment of diseases have been discovered owing to the remarkable achievements and breakthroughs in the field of epigenetics. In addition to DNA methylation, histone modification and non-coding RNAs, one of the novel advances in epigenetic studies is RNA methylation, also known as RNA epigenetics or epitranscriptomics [1, 2]. RNA modifications are the focus of many epigenetic studies in the field of biomedical research and are key to the epigenetic regulation of gene expression. The collective novel evidence implies that post-transcriptional modifications of RNA are crucial for the destiny and function of messenger RNAs (mRNAs). The discovery of RNA modification transcends the view that mRNA is only a message carrier. Therefore, the traditional concept of the functions of mRNA and other non-coding RNAs has changed. Moreover, researchers working on the eye and vision should pay close attention to new studies to keep up with the rapid development of research in the field.
Several RNAs undergo varied modifications, including N6-methyladenosine (m6A),
N6,2’-O-dimethyladenosine (m6Am), N1-methyladenosine (m1A), 5-methylcytosine
(m5C), pseudouridine (
 Fig. 1.
Fig. 1.Ribose phosphate backbone
of the RNA strand and principal RNA methylation modifications. (A) Primary
structure of the RNA strand. Adenine (A), cytosine (C), guanine (G) and uracil
(U) are connected by 3
N6,2’-O-dimethyladenosine (m6Am) is a reversible modification that is widespread in different RNA molecules. It can be detected in 30% of mRNAs at the 5’ end [6]. Liu et al. [7] reported that m6Am levels inversely correlated with the corresponding protein expression; however, m6Am mapping via transcriptome-wide analysis showed that m6Am levels were relatively low in human tissues. A remarkable finding of the study was the recognition of phosphorylated CTD-interacting factor 1 (PCIF1) as a mammalian mRNA m6Am methyltransferase and the role of fat mass and obesity-associated (FTO) gene in removing m6Am deposition from mRNA molecules [8, 9]. Aberrant m6Am modification may be associated with the genesis of certain cancers [10] and stress response. Engel M et al. [11] discovered that m6A/m is altered after stress stimulation and glucocorticoid treatment, and knockdown of either methyltransferase-like 3 (METTL3) or FTO interferes with m6Am gene expression. In addition, glucocorticoid-mediated depressive diseases are closely associated with aberrant m6Am modification, suggesting that the alteration of m6Am modification may contribute to stress-related psychiatric disorders. However, several aspects of m6Am modification, its biological functions and its role in the pathogenesis of human diseases warrant further investigation.
N1-methyladenosine (m1A) is one of the common RNA modifications, which is mediated by methyltransferases (TRMT10C, TRMT61B, TRMT6 and TRMT61A), demethylases (ALKBH1 and ALKBH3) and binding proteins (YTHDF1, YTHDF2, YTHDF3 and YTHDC1) [12]. Dominissini D [3] and Li X [13] mapped the distribution of m1A and demonstrated that m1A modification was enriched near the start codon, thereby highlighting its role in regulating mRNA stability and translation in addition to regulating the functions of transfer RNA (tRNA) and ribosomal RNA (rRNA). Although the physiological function and role of m1A in human diseases remain unclear, it has been reported that m1A regulates the response to physiological stress and may lead to three types of metabolic features, namely, metabolism-excluded, metabolism-high and metabolism-intermediate phenotypes [14]. Recent studies have revealed that m1A modification is related to tumorigenesis and cancer recurrence [15, 16] and regulates metabolic heterogeneity in hepatocellular carcinoma [14]. As a new type of RNA methylation, m1A modification, its function and its relevance to human diseases should be investigated further.
5-methylcytosine (m5C) methylation is one of the major post-transcriptional RNA modifications. It occurs in almost all types of RNA and is regulated by methyltransferases (NSUN, DNMT and TRDMT family members), demethylases (TET family and ALKBH1) and binding proteins (ALYREF and YBX1). It plays an active role in regulating nuclear mRNA export, RNA alternative splicing, protein translation and RNA–protein interactions; increasing RNA stability and maintaining the normal RNA structure [17]. Amort et al. [18] reported the distribution of RNA m5C in different tissues and cells. They selected mouse embryonic stem cells (ESCs) and brain tissue as experimental samples and showed that the distribution frequency of total RNA 5mC was higher in ESCs than in the brain tissue. In addition, analysis of the association between m5C locus and different genomic regions (such as coding DNA sequence [CDS], 5’ untranslated region [5’-UTR] and 3’-UTR) indicated that m5C sites in total RNA were more likely to be distributed at transcription initiation sites. The distribution of m5C in the 3’-UTR region of brain tissue and ESCs varied greatly, suggesting that RNA m5C modification in the 3’-UTR region is closely related to cell function and types. Furthermore, Wang et al. [19] mapped m5C modification and found that m5C is located at the translation initiation site of mRNA. The analysis and comparison of different tissues of humans and mice showed that the distribution characteristics of m5C in mRNA were very conserved in mammals, and genes modified in different tissues were specific. In addition, the study revealed that dynamic m5C-modified genes were significantly enriched in sperm development-related functions during penile development in mice, suggesting that m5C modifications are involved in the regulation of reproductive development. m5C is also involved in the regulation of cell differentiation, proliferation, migration and cell cycle. Moreover, abnormal levels of m5C are closely related to nervous system defects, cardiovascular system diseases, angiogenesis and tumorigenesis [20].
Pseudouridine (
2’-O-Me modification is characterised by methylation at the 2’ position under the control of RNA methylase. It is widely distributed in mRNA, tRNA, rRNA, miRNA and other molecules. A study by Dai et al. [24] revealed the distribution characteristics of 2’-O-Me in human cells for the first time. Approximately 70.3% of 2’-O-Me sites in protein-coding transcripts are located in CDS. The signature sequence of motifs with 2’-O-Me is AGAU, with a high distribution of A or G bases. The 2’-O-Me site is also enriched in protein-coding codons for three amino acids. Studies have shown that 2’-O-Me affects the binding of mRNA to protein, regulates rRNA translation efficiency and participates in biological processes such as tRNA recognition. Recently, Bennasser et al. [25] (University of Montpellier, France) discovered a 2’-O-Me site on HIV and reported that its activity of infecting host cells was regulated by the methyltransferase FTSJ3 (2’-O-RNA methyltransferase). FTSJ3 is also involved in innate immune regulation of HIV.
As a new focus area of epitranscriptomic research, N7-methylguanosine (m7G) has been extensively investigated by many researchers. Previous studies have shown that m7G RNA methylation modifications exist in various molecules, including mRNA 5’ cap structure, mRNA interior, pri-miRNA, tRNA and rRNA. m7G modification can regulate mRNA transcription, biosynthesis and biological functions of miRNA, tRNA stability, nuclear processing and 18S rRNA maturation. m7G RNA modification is catalysed by the METTL1/WD repeat domain 4 (WDR4) complex [26]. Furthermore, METTL1 knockout decreases m7G tRNA modification and leads to impaired ESC self-renewal and differentiation, and its mutations are associated with developmental disorders, suggesting that m7G modification in tRNA has more important physiological functions in mammals [27]. Malbec et al. [28] found that the distribution of m7G in eukaryotic mRNA is species-conserved. They performed m7G-seq on mouse ESCs (mESCs) and brain tissue and found that the distribution of m7G genomic signatures in mice was highly consistent with that in humans. In a study, m7G was found to be significantly enriched in the 5’-UTR of human and mouse mRNAs, especially in the AG-rich region near the translation initiation site. However, after heat shock treatment, the distribution of m7G dynamically changed and was significantly enriched in the gene region (CDS) and 3’-UTR [28]. Furthermore, m7G formed under stress conditions can promote protein translation. METTL1 (m7G methylase) may be involved in regulating m7G formation in eukaryotic mRNA under stress conditions [28]. One of the specific examples of stress response regulated by m7G was reported by Wang et al. [29] who used m7G-MeRIP-seq and lncRNA-seq technology to analyse the m7G modification profile in hypoxic pulmonary hypertension. The results showed that the m7G modification pattern was significantly different between the disease and control groups. In the hypoxic pulmonary hypertension group, m7G modification affected RNA expression, and m7G lncRNAs were significantly upregulated compared with non-m7G lncRNAs. Although the clinical significance requires further validation, the study helped to reveal the potential role of m7G modification in the pathogenesis of hypoxic pulmonary hypertension [29]. Studies have also investigated the role of m7G in tumorigenesis. The development and malignancy of head and neck squamous cell carcinoma are highly related to the tRNA m7G methyltransferase METTL1 through the regulation of entire mRNA translation, PI3K/AKT/mTOR signalling pathway and immune response [26].
N4-acetylcytidine (ac4C) is a highly conserved RNA modification of tRNA and rRNA and has been recently studied in eukaryotic mRNA. However, the distribution and biologic functions of ac4C remain to be further elucidated. As early as 2018, Arango et al. [30] found that N-acetyltransferase 10 (NAT10) catalysed ac4C modification in mRNA and revealed the discrete acetylated regions enriched in coding sequences. In addition, ablation of NAT10 reduced ac4C detection at mapped mRNA sites and was found to be associated with target mRNA downregulation, indicating an important role of ac4C in regulating mRNA translation. In 2020, a research team from the National Institutes of Health conducted a study that provided a technical and theoretical basis for elucidating the role of ac4c modification in biology and disease using ac4C-seq, a chemogenomic approach for transcriptome-wide quantification of ac4C at single-nucleotide resolution [31]. ac4C is markedly induced in response to an increase in temperature, and acetyltransferase-deficient archaeal strains exhibit temperature-dependent growth defects.
ac4C is related to gene regulation, protein translation and stress response. The content of ac4C in the human body significantly changes under diseased conditions; therefore, ac4C may also regulate inflammatory responses, metabolic diseases and autoimmune diseases. The importance of the role of ac4C in the pathogenesis and treatment of cancer has also been reported [32]. In addition, the abnormality of ac4C modification and its catalyze enzyme NAT10 in the condition of HIV-1 infection is validated [33].
Uridine (m5U) modification was demonstrated in rRNA and tRNA a decade ago [34] but was discovered in eukaryotic mRNA in 2020 by Cheng et al. [35], who showed that m5U in mRNA can be found in various mammalian cells and tissues. However, the distribution, function and relevance of m5U modification in human diseases remain to be demonstrated.
m6A methylation is the most common and abundant RNA modification and accounts
for
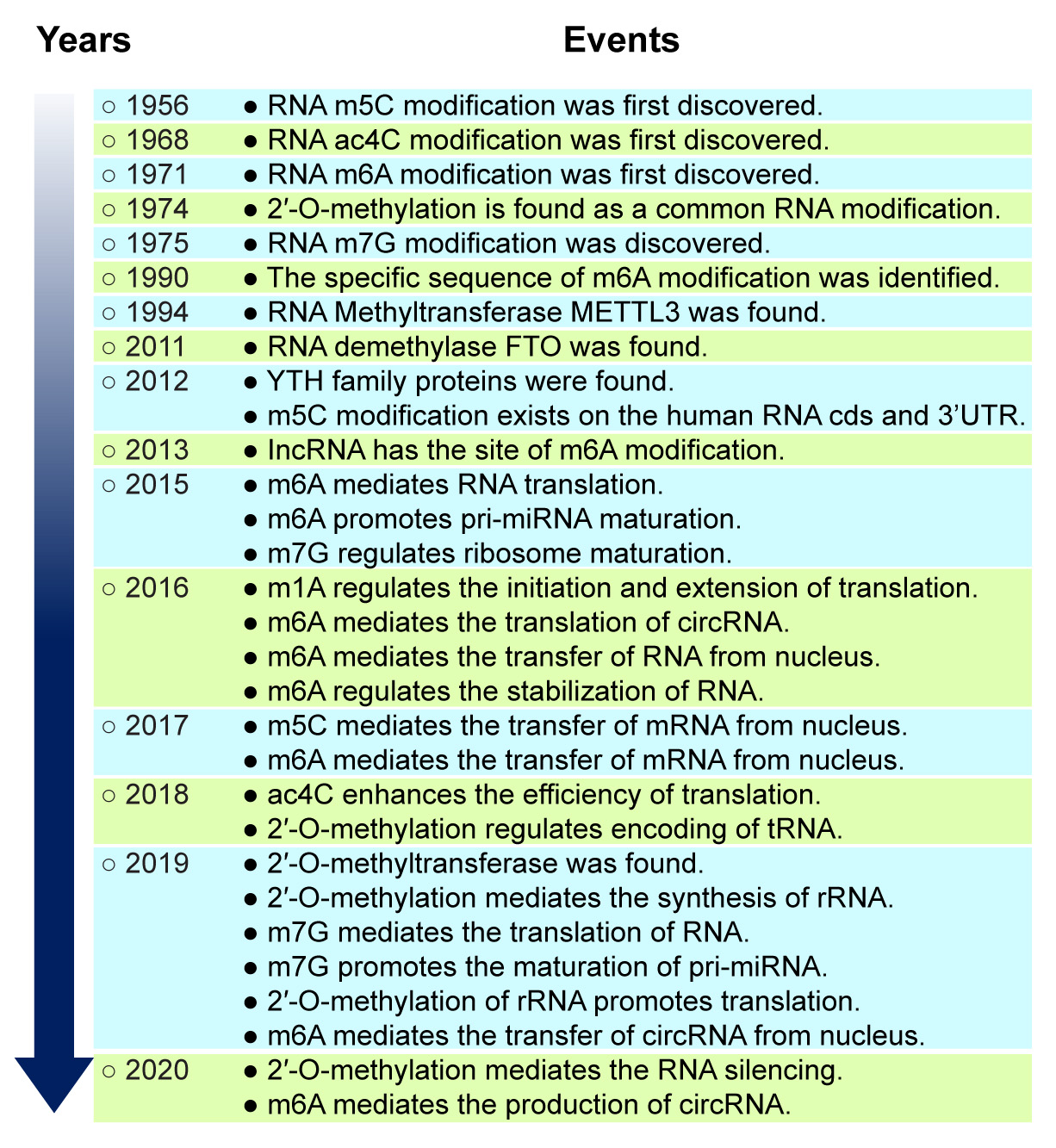 Fig. 2.
Fig. 2.Historic events of the study of RNA methylation. The discovery of m5C RNA methylation initiated the study of RNA methylation, and that of the RNA methylase METTL3 began the golden period of RNA methylation research. RNA methylation became a prime focus of epigenetic research after 2011.
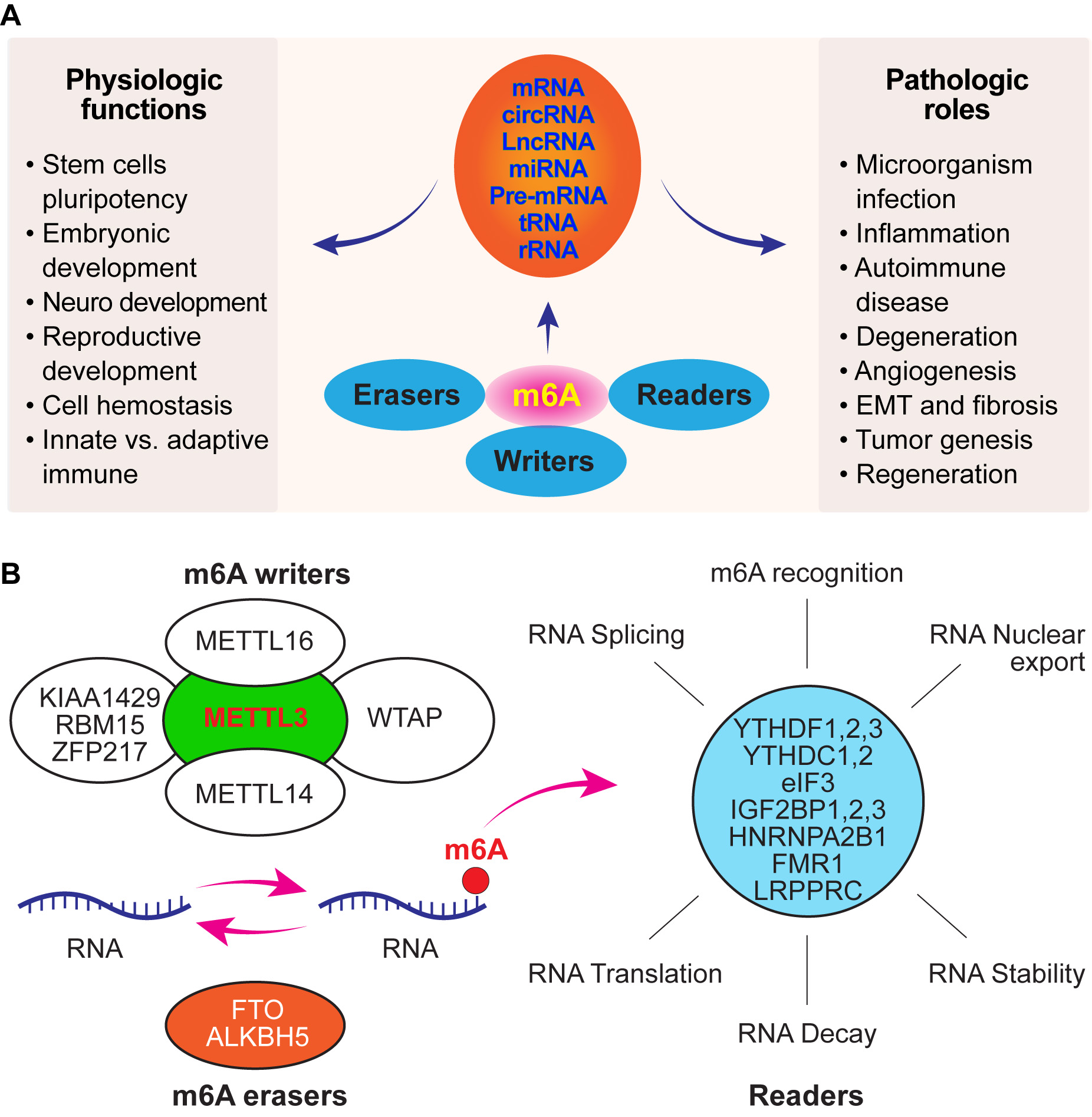 Fig. 3.
Fig. 3.The role of m6A modification in maintaining the physiological function of cells and its contribution to pathological conditions and molecular composition of m6A RNA methylation. (A) m6A modification can occur in a majority of RNAs, from mRNAs to non-coding RNAs. Therefore, aberrant expression of m6A modification-related factors may result in numerous diseases. (B) m6A methylation is a reversible process controlled by m6A methylases and demethylases. m6A reader proteins bind to m6A-containing transcripts.
In terms of its distribution in genes, m6A is located in highly conserved genomic regions. Specifically, the priority enrichment area of m6A is close to the 3’-UTR and stop codon within the longer internal exons, and m6A is found at the 3’-UTR in a majority of mRNAs. Therefore, m6A controls the translational affinity of RNA-binding proteins or unique m6A-derived transcriptomes. It can affect metabolic processes of mRNA, such as splicing, nuclear export, stabilisation and translation, and RNA–protein interactions, thereby regulating gene expression (Fig. 3B). However, it remains unclear whether the genetic code is influenced by m6A modification. A recent study (2020) [40] provided new ideas for basic biological research, which subverts the inherent unidirectional flow of genetic instructions from DNA to RNA to protein. The study reported that knocking out the m6A methylation-related enzyme METTL3 or YTHDC1 in mouse ESCs increased the openness of chromatin through chromosome-associated regulatory RNA (carRNA) in an m6A-dependent manner and activated transcription. Because RNA structural changes caused by the introduction of m6A may also alter the interaction between RNAs, proteins and chromatin, this new finding may extensively influence our comprehension of human diseases and their treatment [40], suggesting that m6A methylation regulates cellular functions and systemic diseases and is essential for vision function and eye diseases. Therefore, this review highlights the role of m6A modification in the pathogenesis of ocular diseases and vision function.
Previous studies have demonstrated the expression of enzymes required for m6A modification in ocular tissues [41, 42, 43, 44, 45]. m6A modification and its biological functions require specific methyltransferases (writers), demethylases (erasers) and proteins that recognise methylated sites (readers) [46] (Fig. 3B). m6A modification plays a key role in regulating cell division, growth and differentiation; stem cell identity [47]; embryonic and reproductive development; immunity; cell metabolism and homeostasis. Abundant expression of m6A in brain cells is related to neurodevelopment, learning and memory, synaptic plasticity, stress response and heat shock stress response [48] (Fig. 3A). In addition, the importance of m6A RNA modification in the circadian clock and retina has been addressed by Zelinger and Swaroop [49]. Furthermore, m6A mRNA methylation affects testosterone synthesis by regulating LC autophagy by affecting the stability of calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2) transcription and the translation efficiency of protein phosphatase 1A (PPM1A) [50], which provides novel insights into the functional mechanism of m6A RNA methylation.
The regulatory mechanisms of post-transcriptional gene expression in eukaryotes warrant further investigation. Aberrant expression of enzymes involved in m6A modification may mediate numerous ocular and systemic pathological conditions, including abnormal retinal development, age-related eye diseases, ocular angiogenesis, ocular surface infection [43, 51, 52, 53], learning and memory decline [54], tumorigenesis [42, 55, 56, 57, 58], degeneration [59, 60, 61, 62] and fibrosis [63, 64, 65] (Fig. 3A). Therefore, the concept of the epitranscriptomic era is well established now [66].
The essential factors that control m6A modification and its biological functions in cells, including ocular cells, are methyltransferases (writers, including METTL3, METTL14, METTL16, WTAP, KIAA1429, RBM15 and ZFP217), RNA demethylases (erasers, including FTO and ALKBH5) and specific proteins that recognise methylated sites (readers, including YTHDF1–3, YTHDC1–2, eIF3, IGF2BP1–3, HNRNPA2B1, FMR1 and LRPPRC) [67, 68, 69, 70] (Fig. 3B). METTL3 is a major methylase responsible for adding m6A to RNA molecules. FTO is the first known ‘eraser’ protein that removes methylase and regulates cellular homeostasis. ALKBH5 serves as a demethylase that is also related to DNA damage repair. Reader proteins can recognise m6A sites. YTHDF1 and 3 can promote the translation of m6A-modified mRNA, whereas YTHDF2 can promote RNA degradation. The writer and eraser proteins of mRNA determine the m6A modification level of the target mRNA, and the reader protein determines the translation efficiency of m6A-modified mRNA or affects mRNA stability [71]. The factors related to m6A modification mentioned above may be critical for regulating ocular cell function and behaviour and are discussed below.
The expression of m6A writers (METTL1, METTL3, METTL14 and WTAP), erasers (FTO and ALKBH5) and readers (YTHDF1–3, YTHDC1 and 2, eIF3 and IGF2BP1–3) in ocular tissues has been reported in our previous study and other studies as well [11, 12, 13, 15, 39]. The expression of m6A modification factors (METTL3, METTL14, WTAP, FTO, ALKBH5, YTHDF1, YTHDF3 and YTHDC1) can be found in numerous ocular tissues (cornea, lens, retina, retinal pigmental epithelium [RPE], extraocular muscles and uveal melanoma cells) [41, 42, 43, 45, 51, 72, 73, 74, 75, 76]. m6A methylation usually varies among different tissues [77, 78], indicating that m6A methylation is not only variable but also dynamic, and data obtained from one specific tissue or cell cannot represent the status of m6A methylation in other tissues or cells. For example, the outcome of m6A methylation will be different in the same tissue if the experimental time point is different.
Because of limited studies on the role of m6A in regulating vision function under normal conditions, we systemically discussed some major aspects related to its role physiologically and its implication in vision function.
m6A is known to control mammalian neurogenesis and development [79]. Knockdown of METTL3 inhibits the duplication of neural stem cells and morphological maturation of neurons, and depletion of m6A in the mRNA of histone methyltransferase EZH2 downregulates EZH2 expression, leading to defects in neuronal development [80]. The eye is derived from the neuroepithelium, ectoderm and mesenchyme and is considered a part of the brain. Therefore, it would be interesting to discover whether the above mentioned mechanism underlies the development of ocular tissues as well (eyeball, appendices of the eye and visual pathway).
The retina is the most important part of the human eye, and its correct development depends on the activation of transcription factors including epigenetic factors in a timely manner, as demonstrated by Chen et al. [81], which in turn controls the expression of retinal cell structure proteins. Genetic control of retinal development has been well recognised [82]. However, basic knowledge regarding the control of retinal development (cell fate, proliferation and neurogenesis) via epigenetic factors, especially m6A methylation, remains limited. Accumulating evidence suggests that m6A modification is correlated with embryonic development in mammals [83]. Therefore, retinal development may also be subjected to the regulation of m6A methylation [53, 84]. A study showed that METTL3 and METTL14 are expressed in amacrine, bipolar and ganglion cells but not in the rod and cone photoreceptors and horizontal glial cells of the mouse retina [53]. The expression of these two methylases peaked on post-natal day 6 and the proliferation and differentiation of neuronal precursor cells and the maturation of neuronal synapses of the retina peaked on post-natal day 8, implying that m6A methylation may actively regulate retinal development [85]. In addition, the global expression of m6A RNA was significantly lower in adult mouse retina than in newborn mouse retina, which may be associated with a reduction in the expression of m6A writers such as METTL3, METTL14, and WTAP [84]. Similarly, Huang et al. [86] found that the expression of m6A modification factors such as METTL3, METTL14 and WTAP was high in the developing eye of zebrafish, indicating that these factors were involved in retinal development in zebrafish. Functional analysis verified the critical role of m6A methylases in the regulation of eye development in zebrafish. Moreover, deletion of the m6A methylation complex resulted in impairment of eye formation and retinal progenitor cell differentiation, suggesting that the opportune retinal development is m6A-dependent. These studies suggest that m6A modification occurs earlier than cell proliferation and differentiation. The eye, especially the retina, is considered a part of the neural system. Although the abovementioned studies suggest that m6A modification regulates retinal development, further investigation is warranted to examine the mechanism of action of m6A methylation factors, such as writers, erasers and readers, in regulating retinal development during the embryonic and post-natal stages.
Whether the balance between extraocular muscles, intraocular pressure maintenance, lens transparency, vision acuity and the metabolism of vision cycle proteins is controlled by m6A modulation remains unclear. However, the expression of m6A modification factors is observed in the cornea, sclera, iris, lens, retina and optic nerve, and protein expression indicates its function in the tissue. Therefore, we speculate that m6A participates in the maintenance of normal vision function. A recent study showed that a majority of genes expressed in the mouse retina exhibit circadian rhythmic fluctuations [87]. Retinal cell response and hypoxia-related pathways are more active at night, whereas the expression of genes involved in photoreceptor function is higher during the day [87]. These studies suggest that m6A modification is important for maintaining normal vision, rhodopsin metabolism, vision adaptation and memory effects on colour vision especially perception and highlight the possible influence of circadian rhythm disorders on vision function.
m6A modification is controlled by methyltransferases and demethylases, which should be balanced under normal conditions because the loss of balance may result in disease occurrence. Several ocular haemostasis mechanisms have been reported, including maintenance of the delicate balance among tear production, evaporation and drainage; corneal endothelial cell-dependent thickness maintenance; accommodation of the eye; aqueous humor production and drainage; blood–retinal barrier (BRB) intake maintenance and molecular trafficking between the retina and retinal microvessels; innate immune response and subretinal immune privilege; retinol metabolism; photoreceptor outer segment shedding and RPE phagocytosis; and nutrient and waste material transportation between the retina and choroid. Ocular haemostasis may be regulated by m6A modification factors. Although the following data were obtained from systemic studies, the principle explained by them may also apply to ocular cells. m6A methylases/demethylases are essential for producing a normal hypertrophic response in cardiomyocytes [88]. However, reduced levels of m6A methylation/increased levels of demethylases are related to cardiomyocyte dysfunction, implying that m6A modification regulates cardiac homeostasis. In addition, a study showed that METTL3 played a critical role in maintaining mouse T-cell homeostasis, and T cells failed to undergo homeostatic expansion under METTL3-deficient conditions [89]. Therefore, extremely high or low expression of either methyltransferases or demethylases may break the balance between them. As discussed in previous sections, m6A modification factors, especially the major enzymes of m6A methylation/demethylation, namely, METTL3/FTO, can be found in most ocular tissues. The balance between m6A methylation and demethylation may be critical for maintaining normal cell function, and its loss may contribute to the development of numerous diseases including eye diseases. These studies suggest a novel mechanism of m6A methylation/demethylation underlying the maintenance of cell homeostasis [89, 90], therefore, whether m6A modification participates in maintaining and regulating the homeostasis of vision function should be investigated further.
The eye has both innate and adaptive immune responses, which are critical for
maintaining normal vision function [91, 92]. The ocular innate immune system
comprises tears (containing lysozymes, lactoferrins, lipocalin, beta-lysine and
immunoglobulins), conjunctival epithelium, corneal epithelium, the complement
system, RPE [93], interferons (IFNs), toll-like receptors (TLRs) [94] and
transforming growth factor-beta (TGF-
Ocular infectious diseases may be caused by viruses, bacteria, fungi or parasites. Such eye diseases may become difficult to control if the infectious agent becomes resistant to commonly used drugs or if there is no cure for the infection, such as infection with deadly bacteria or severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Therefore, it is essential to recognise the complexity and diversity of ocular microorganisms. To facilitate diagnosis and understand the pathogenesis of ocular diseases related to microorganism infections, genomic approaches are highly recommended for precise detection of the diversity of ocular surface microorganisms. However, the interruption of homeostatic microbiota may contribute to some ocular infections, and the gut and oral microbiota may be closely associated with the pathogenesis of glaucoma, uveitis and AMD [107].
Research into the role of m6A in infectious diseases may help to improve our understanding of the pathogenesis of systemic and ocular infectious diseases and their treatment. Among ocular microorganism infections, the alteration of m6A modification has been reported in experimental fungal keratitis [43], which was the first evidence of the relevance of m6A modification in ocular microorganism infections. Increased m6A methylation, especially METTL3 expression, contributes to the pathogenesis of fungal keratitis [43], whereas aberrant m6A methylation is associated with the production of inflammatory cytokines. In terms of the interaction between m6A and bacterial infections, a recent study revealed that the production of inflammatory cytokines is increased in bacterial infections through demethylation of histone H3 lysine 27 trimethylation (H3K27me3) owing to the loss of YTHDF2. Specifically, YTHDF2 silencing promoted bacterial infection-induced inflammatory responses [108]. Therefore, the study revealed a new mechanism of cross-talk between m6A and H3K27me3 in bacterial infections [108]. In a study on the effect of gut microbiota on the expression of m6A modification factors, the expression of the methyltransferase METTL16 was found to be reduced in microbiota infections, and the methylation of METTL16-targeted mRNA encoding S-adenosylmethionine synthase MAT2a was also inhibited [109]. These studies reveal that m6A modification is a novel mechanism underlying the interaction between bacterial infections and the host.
The relevance of viral infection to m6A modification has attracted significant attention from biomedical researchers, and viral mRNA modified by m6A regulatory factors has been found in prototypic polyomavirus simian virus 40 (SV40) [110], influenza virus, Rous sarcoma virus, human immunodeficiency virus type 1 (HIV-1), Flaviviridae [111], dengue virus, Zika virus, West Nile virus and hepatitis C virus [112]. Herpes simplex virus 1 (HSV1) infection is the most common viral infection in the cornea, and persistent HSV1 infection in the cornea often results in serious vision loss or blindness. The recurrence of herpetic keratitis is owing to the activation of latent HSV1 in trigeminal ganglia cells [113]. The lifelong establishment of the latent infection of HSV1 in the host cell nucleus may occur after the first HSV infection, and the transcription of activation stimulated by environmental factors (such as ultraviolet [UV] light and stress) is limited by the latency-associated transcript; therefore, lytic-related genes are silent [113]. In addition, activation of the latent viral genome may be regulated by post-translational modifications besides DNA methylation [113]. Therefore, active and inactive transcription of HSV1 may be regulated by m6A modification. More recently, a study showed that SARS-CoV-2 infection triggers a global response in m6A methylome of the host, and m6A negatively regulates SARS-CoV-2 infection [114]. SARS-CoV-2 enters the human body mainly through respiratory cells; however, it may enter through the eye as well. Troisi et al. [115] showed that the positive rate for conjunctival swabs was as high as 72% in patients with viral infection; in addition, corneal epithelial defects were reported in some patients. The study indicated that keratoconjunctivitis caused by viral infection occurs if the eye is unprotected, and the infected eye may transmit the virus. Therefore, eye protection may be one of the measures for preventing the spread of a virus. These studies suggest that viral infection may alter the m6A modification of antiviral mRNAs in the host to regulate infection [112]. In addition, m6A modification of viral RNA may represent a new mechanism of host–pathogen interactions during viral infection [111]. Therefore, in addition to anti-microbial agents, m6A regulatory factors related to microorganism infections may also lead to infection control. In addition, the mechanism of action of m6A in regulating the establishment and activation of HSV1 latent infection and its recurrence in the cornea warrants further investigation.
The purpose of understanding the pathogenesis of eye diseases is to transfer the bench-top knowledge to clinical application. With the advancement of technology, many new therapies have emerged for tumour treatment; among which, oncolytic viruses (OVs) have received increasing scientific and industrial attention because of their ability to specifically replicate in tumour cells and cause tumour cell lysis without affecting normal cells. OVs are a class of natural or genetically engineered viruses that can selectively kill or lyse tumour cells but have no killing effect on normal tissues [116]. In addition, OVs can be combined with inhibitors of DNA methyltransferases and histone deacetylases as well as microRNAs [117], which may be a promising treatment strategy for some complex eye diseases including ocular microorganism infections.
Ocular inflammatory responses caused by pathogens, injuries and toxins are
characterised by the release of pro-inflammatory factors from activated
inflammatory cells such as macrophages, monocytes, neutrophils and lymphocytes.
The primary inflammatory factors are cytokines such as interleukin-1
Wei et al. [124] reported that ocular inflammatory diseases are
regulated by genetic and epigenetic factors. Recent studies have demonstrated
that m6A plays a crucial role in controlling inflammatory responses, and several
studies have shown that TNF-
The findings of the above mentioned studies, which state that m6A is involved in
the regulation of inflammatory responses in the eye, have been supported by a
systemic study. An interesting finding was that METTL3 knockdown downregulated
the expression of LPS-induced inflammatory cytokines, including IL-6, IL-8 and
growth-regulated oncogene-alpha (GRO-
In addition to METTL3, the role of the m6A eraser FTO and reader YTHDF2 in
inflammation has been demonstrated in a mouse model of alcohol-induced kidney
injury. Aberrant modification of peroxisome proliferator-activated receptor-alpha
(PPAR-
Altogether, these studies reveal an important role of METTL3 in regulating inflammatory responses and provide a novel therapeutic target for inflammation-related eye and systemic diseases. However, the role of m6A modification factors in specific inflammatory ocular diseases should be investigated further.
Many degenerative diseases, including ocular diseases (keratoconus, cataract,
retinitis pigmentosa and AMD) and systemic diseases (Alzheimer’s disease,
multiple sclerosis, arthritis, Parkinson’s disease, muscular dystrophy,
Huntington’s disease and cystic fibrosis), share some common characteristics of
sustained degenerative response and cell death (apoptosis, necroptosis,
autophagy, ferroptosis and pyroptosis) [140]. m6A modification is closely related
to the pathogenesis of ocular cell degeneration. Age-related cataract is a
typical ocular disease associated with lens epithelial cell degeneration. A study
reported that the expression of the key element of m6A demethylases was
significantly increased in lens epithelial cells in patients with age-related
cataracts [51]. Furthermore, the formation of diabetic cataracts induced by high
glucose levels is also associated with aberrant m6A modification, and METTL3
mediates lens epithelial cell apoptosis in patients with diabetes [141]. RPE cell
dysfunction is closely related to the pathogenesis of AMD, and RPE degeneration
may lead to photoreceptor death. Amyloid-
The role of NLRP-3 and IL-1
 Fig. 4.
Fig. 4.The proposed role of
abnormal m6A modification contributes to the pathological process of cellular
degeneration. m6A modification may either upregulate IL-1B or downregulate FTO to
increase the expression of METTL3, which may inhibit PI3K/Akt/GSK-3
The development of ocular diseases (corneal dysfunction, cataract, glaucoma,
uveitis, AMD and DR) [147, 148, 149, 150, 151, 152] and systemic diseases, including obesity,
diabetes, autoimmune diseases, neurodegeneration, ischaemia/reperfusion injury
and cancer, is closely associated with oxidative stress-induced damage caused by
pollutants, smoking, UV rays, radiation and toxic chemicals [153, 154, 155, 156]. One of the
most common features of oxidative stress is the accumulation of reactive oxygen
species (ROS). Excess ROS production induces cell damage and leads to cell death,
including but not limited to corneal endothelial cells [156, 157], lens
epithelial cells [158], RPE cells [159] and retinal ganglion cells [149, 160],
and photoreceptor dysfunction [161, 162], which may be regulated by m6A
modification [163, 164]. Li et al. [51] reported that m6A is involved in
the pathogenesis of age-related cataracts through its target gene of circRNA in
lens epithelial cells, and the m6A methyltransferase ALKBH5 may majorly
contribute to aberrant m6A methylation in lens epithelial cells. The pathogenesis
of AMD involves oxidative stress-induced damage in RPE [165, 166].
Amyloid-
Although the role of m6A modification in inducing oxidative stress in ocular
diseases remains unclear, several studies have shown that oxidative
stress-induced damage is closely related to mRNA modification, especially m6A
[137, 163, 169]. The involvement of m6A modification in redox homeostasis is
evidenced by the following findings. The level of m6A methylation is increased
globally in response to oxidative stress [170, 171]. Hypoxic preconditioning
treatment-induced protection of myoblast heart cells (H9c2) from H
In addition to m6A writers (METTL3), m6A demethylation factors may also regulate
oxidative stress [137, 173, 174]. Recent studies have reported that increased
PGC-1
m6A modification can regulate the oxidative stress response through lncRNAs [176]. Oxygen–glucose deprivation/re-oxygenation enhances free radical production, induces METTL3-dependent lnc-D63785 m6A methylation, resulting in miR-422a accumulation, and reduces the expression of the downstream gene MEF2D-MAPKK6, leading to neuronal cell apoptosis [177]. In addition, the increased expression of m6A induced by oxidative stress is associated with an enhanced expression of p21, which may regulate apoptosis [164]. These studies show that oxidative stress may be regulated by m6A methylation, and m6A expression may also be affected by oxidative stress, indicating that m6A and oxidative stress may regulate each other.
The purpose of understanding the pathogenesis of ocular diseases is to interfere with or cure a disease. Resveratrol, which stimulates the expression of SIRT1, is a well-recognised agent with broad-spectrum anti-oxidative and apoptosis-inhibiting properties [178]. Intravitreal injection of resveratrol can prevent retinal ganglion cell death caused by high intraocular pressure [179]. In a study, dietary resveratrol inhibited ROS production, reduced the expression of m6A and downregulated the expression of cleaved caspase-3 in a mouse model of AFB1-induced liver injury [180], suggesting that resveratrol is useful for treating systemic and ocular diseases related to oxidative stress through the regulation of m6A methylation.
Although ROS has been traditionally considered harmful to the human body, this
concept is increasingly challenged because the dynamic modification of redox
proteins by ROS is similar to the phosphorylation and acetylation modification of
other proteins in various physiological processes [181]. ROS is required for
cytokine, insulin growth factor (IGF), AP-1 and NF-
Angiogenesis is a complex process mediated by the overproduction of growth factors, cytokines and ECM proteins and the loss of balance between angiogenic inducers such as vascular endothelial growth factor (VEGF) and anti-angiogenic factors such as pigment epithelium-derived factor (PEDF). In addition to PEDF, the second factor of VEGF negative regulator, vascular endothelial growth inhibitor (VEGI; known as TNFSF15), plays an important role in vascular homeostasis, which specifically targets VEGF production, VEGF receptor (including R3) activation, endothelial cell proliferation and apoptosis [187, 188, 189]. The significance of VEGFI in ocular angiogenesis has been demonstrated in our previous study and other studies as well [190, 191]. In a study, VEGFI treatment prevented the leakage of retinal blood vessels in a diabetic rat model [191]. Many ocular diseases such as corneal neovascularisation, neovascular glaucoma, neovascular AMD, DR, central retinal vein occlusion and other systemic diseases (tumours, heart diseases, skin diseases, cardiovascular diseases and stroke) are associated with angiogenesis. VEGF is known to be a critical mediator of angiogenesis. However, maximal angiogenesis cannot be induced by VEGF alone, and resistance to VEGF treatment often presents in clinical practice, implying that more currently unknown factors may participate in the underlying pathogenesis of angiogenesis. m6A modification may play a critical role in regulating angiogenesis, including ocular angiogenesis [52, 75, 192, 193, 194]. To date, m6A methyltransferases (METTL3, METTL14 and WTAP), the demethyltransferase FTO and the m6A-binding protein IGF2BP3 have attracted greater attention in different cell types and animal models of angiogenesis [52, 192, 193, 194]. In DR, a break in the outer BRB is caused by RPE cell dysfunction or apoptosis induced by high glucose levels [195, 196, 197]. A recent study demonstrated that reduced expression of METTL3 may respond to the RPE barrier breakdown, whereas overexpression of METTL3 can prevent RPE cell death [42]. Furthermore, VEGF contributes to inner BRB breaks and the progression of DR [198]. The most impressive finding in this field was the relevance of m6A modification to VEGF during the formation of new vessels [194]. VEGF mRNA expression and VEGF secretion are regulated by the m6A-binding protein IGF2BP3 and YTHDF1 and 2 [194]. IGF2BP3 silencing suppresses VEGF mRNA stability and expression [194]. Furthermore, an IGF2BP3 binding site is present in the VEGF gene, and mutations in this abrogate the binding of IGF2BP3. Moreover, VEGF expression is enhanced by overexpression of IGF2BP3 [194]. These studies suggest that angiogenesis inducers may participate in VEGF-induced angiogenesis through m6A methylases, IGF2BP3 and YTHDF1 and 2, indicating that interfering m6A modification factors may be a potential therapeutic target for inhibiting ocular and systemic angiogenesis (Fig. 5).
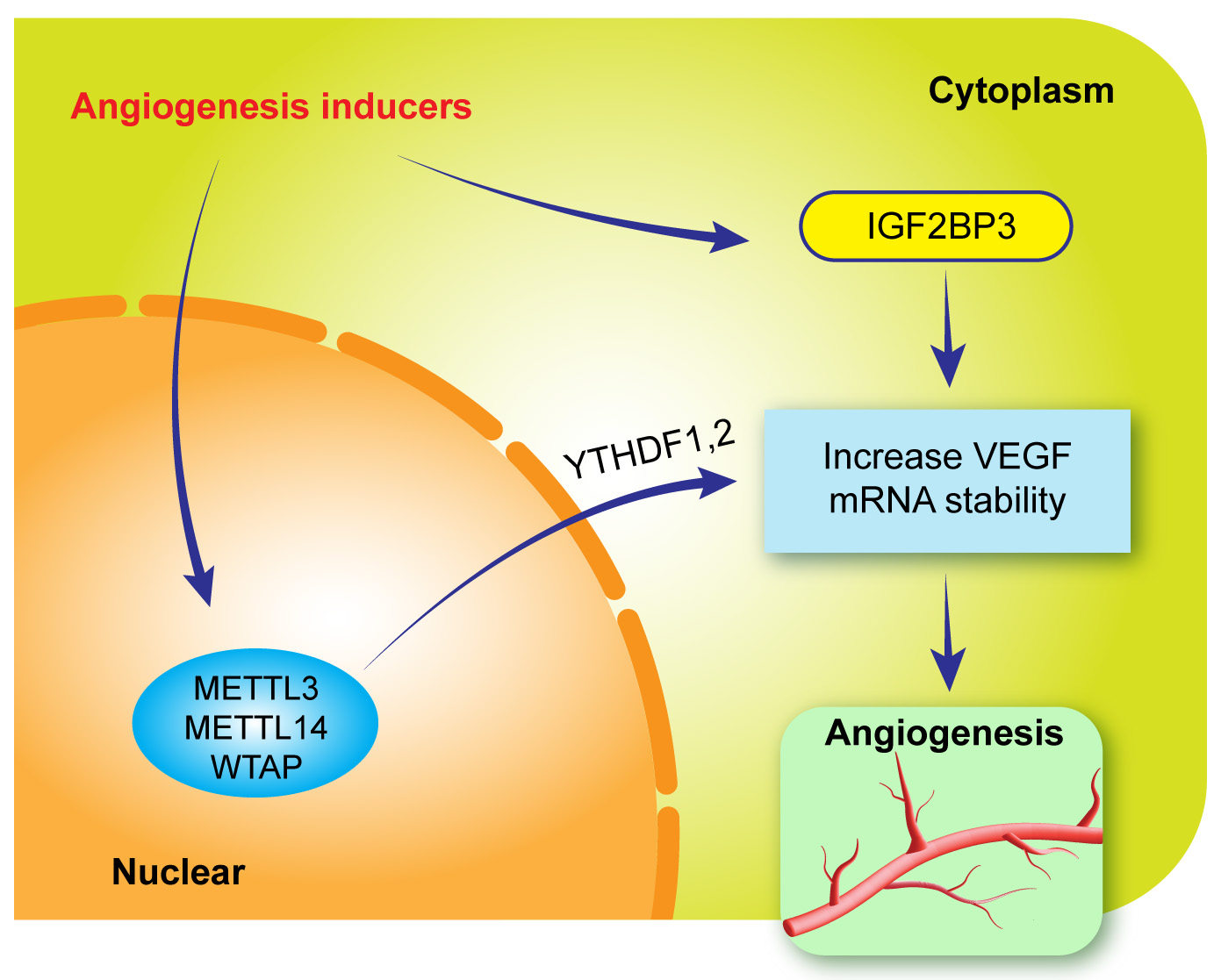 Fig. 5.
Fig. 5.Proposed role of m6A modification in the pathogenesis of angiogenesis. Angiogenesis inducers upregulate the expression of m6A methylases and IGF2BP3, through which YTHDF1 and 2 increase VEGF mRNA stability and promote angiogenesis.
The relationship between methyltransferases and VEGF during angiogenesis has
also been revealed. In an oxygen-induced retinopathy model (retinal
neovascularisation) and alkali burn-induced corneal neovascularisation model, in
which VEGF serves as a major player, neovascularisation was significantly
inhibited by METTL3 knockout [52]. The importance of METTL3 in angiogenesis has
been emphasised in several studies on different systems [199, 200]. METTL3
knockdown reduces the expression of pro-angiogenic factors such as hepatocyte
growth factor (HGF), TGF-
One of the important factors for m6A demethylation is FTO. FTO expression was found to be increased in a model of corneal neovascularisation [74]. A study reported that FTO knockdown inhibited the growth and survival of von Hippel–Lindau (tumour suppressor)-deficient cells [208]. In another study, inhibition of FTO reduced the survival of both HIF-wildtype and HIF-deficient mice with tumours, indicating that FTO is an important mediator of angiogenesis [209].
Although the role of m6A in regulating angiogenesis has been extensively investigated, no studies have reported the role of m6A in maintaining the balance between VEGF and PEDF and VEGF/VEGI under normal and pathological conditions of the eye. In addition, neither dynamic changes in m6A methylation during overall angiogenesis nor specific changes in m6A methylation during angiogenesis of specific ocular tissues have been reported. Therefore, this field should be investigated further. Therefore, understanding the role of m6A modification in the regulation of ocular and systemic angiogenesis will provide insights into the pathogenesis of ocular angiogenesis.
During epithelial–mesenchymal transition (EMT), epithelial cells transform into
mesenchymal cells in response to physiological and pathological changes in the
microenvironment of cells. EMT is involved in the initial steps of pathogenesis
of many ocular fibrotic diseases, such as advanced dry eye, corneal fibrosis,
glaucoma, filtration surgery, posterior capsule opacification after cataract
surgery, PVR, proliferative DR (PDR), macular neovascularisation, ocular wound
healing and many systemic fibrotic diseases (pulmonary fibrosis, cystic fibrosis,
cirrhosis, retroperitoneal fibrosis, myocardial fibrosis, kidney fibrosis and
idiopathic pulmonary fibrosis). However, the aetiology of EMT and fibrosis is not
well understood. There are three types of EMT. Type 1 EMT refers to embryonic
development. Type 2 EMT is often associated with wound healing response and
fibrosis after trauma, inflammation and angiogenesis. Type 3 EMT occurs with
tumour invasion and metastasis [210]. Most ocular EMTs belong to type 2. In
addition to EMT, another pathological process called endothelial–mesenchymal
transition (EndMT) also contributes to subretinal fibrosis, including PDR [211, 212], macular neovascularisation [213] and retinopathy of prematurity [214].
EndMT is characterised by the loss of endothelial markers, including vascular
endothelial (VE)-cadherin, CD31, platelet–endothelial cell adhesion molecule
(PECAM) 1, TIE-1, TIE-2 and von Willebrand Factor (vWF) [215], and the
upregulation of N-cadherin, vimentin and
Altogether, these studies suggest that TGF-
Senescence is a complex process that involves various cellular and tissue
changes throughout the body [226, 227]. All cells and tissues undergo senescence,
which is characterized by reduced response to stress, declined homeostasis, and
age-related diseases. Senescence is an important predisposing factor in several
ocular diseases. The effect of senescence in ocular disease is often seen in
corneal endothelial cell dysfunction [228], glaucoma [229], age-related cataracts
[230, 231], and age-related macular degeneration (AMD) [232, 233, 234]. Recent
discoveries have shown that the m6A RNA methylation modification plays an
important role in age-related disorders [235, 236]. A study of the effects of m6A
modification on the pathogenesis of age-related cataracts by m6A-IP-seq and
RNA-seq analysis showed that major enriched and meaningful host circRNA genes are
involved in the processes of autophagy, DNA repair, and aging in age-related
cataracts [51, 237, 238, 239]. In addition, A
There is no tissue-specific biomarker of senescence. Therefore, the following parameters may be used as indicators of senescence in ocular cells to study its relevance to m6A: loss of genomic stability, telomere shortening, aberrant epigenetic factors [241], mitochondrial dysfunction [242], increased senescence-associated beta-galactosidase [243] and senescence-associated secretory phenotype (SASP) [244], accumulation of ROS in cells, and activation of P53 [245] and p16 [246].
It would be interesting to determine whether m6A modification factors could be
epigenetic biomarkers of senescence. Importantly, the global reduction in m6A
methylation and METTL3 levels have been shown to be associated with the process
of senescence [236, 247, 248]. The level of m6A methylation in human peripheral
blood mononuclear cell (PBMCs) and human diploid fibroblasts was significantly
reduced in older individuals compared with that in younger individuals [247].
Importantly, mRNA expression of the m6A methylated gene argonaute 2 (AGO2) in the
older PBMC subjects was decreased, and senescence-associated
RNA modifications associated with cell senescence are not limited to m6A. The synergistic effect of RNA 5-methylcytosine (m5C) and m6A on senescence regulation was recently reported. Moreover, NSUN2 (an RNA methyltransferase) can accelerate oxidative stress-induced cellular senescence by upregulation of P21 through m5C and METTL3/METTL14-mediated m6A methylation [163].
Furthermore, the possible signalling mechanisms through which m6A mediates cell
senescence induction have not been characterized. Therefore, precise
understanding of the mechanism underpinning m6A and senescence warrants further
research. The most recent hypothesis states that Ras activation induces ROS
overproduction which inhibits the expression of m6A reading protein YTHDF2 and
subsequently increases the activation of MAP2K4 and MAP4K4, leading to SASP
expression. Importantly, it was found that the senescence process mediated by the
ROS-YTHDF2-MAPK-NF-
The key elements of m6A modification in regulating the senescence process in ocular tissues need to be determined. Senescence is not a static endpoint but rather reflects diverse cellular abnormalities with a series of progressive cellular and behavioral changes [249]. Importantly, there is no m6A dynamic analysis in vivo from human tissues during ocular tissue senescence. Furthermore, no large-scale longitudinal cohort studies have been conducted to determine the specific transcripts that are modified by m6A in young and old people. Therefore, in this review, we discuss the key themes of m6A modification, with a focus on their role in vision function and eye diseases.
There are two types of immune responses, innate and adaptive immune responses, which play an important role in maintaining ocular cell homeostasis by eliminating damaged cells and protecting the ocular tissue from pathogen infection. The innate immune response acts rapidly and can identify molecules from numerous pathogens. In contrast, adaptive immune response is slower but more specific. Aberrant immune responses are linked to many ocular diseases, including immunodeficiency due to diabetes [250, 251], HIV infection/ocular involvement, and overreaction of the immune system to the native existing cell, such as in autoimmune diseases like Sjögren’s syndrome [252], keratoconus [253], multiple sclerosis [254], rheumatoid arthritis, Graves’ disease [255], uveitis [256], and autoimmune retinopathy [257]. Of the ocular autoimmune diseases, Graves’ ophthalmopathy (GO) is a typical autoimmune disorder that causes ophthalmopathy due to hyperthyroidism and excess autoantibodies production [255, 258]. In a study of the role of m6A RNA modification in the pathogenesis of GO, we demonstrated that the expression of genes related to immune response and inflammatory processes such as lymphocyte activation and inflammatory pathways, which are associated with aberrant m6A modification factors, including m6A methylase WTAP, m6A demethylase ALKBH5, and m6A readers YTHDF2, YTHDF3, and YTHDC2, are significantly high in surgically excised extraocular muscles from patients with GO compared with that of normal subjects [45]. The results imply that m6A modification participates in the regulation of the pathogenesis of the autoimmune response in GO.
Many ocular immune-related diseases occur secondary to systemic disease. Therefore, the ocular immune diseases caused by the aberrant immune response systemically, cannot be ignored. Both systemic and ocular innate immune responses are characterized by the expression of interferon during viral infection. In addition to METTL3, the m6A reader YTHDF3 regulates innate immunity by binding to the transcription corepressor forkhead box protein O3 (FOXO3) mRNA [259]. Importantly, the innate immune response during hematopoietic development is closely related to m6A methylation, and deletion of METTL3 results in an abnormal innate immune response, which manifests as endogenous double-stranded RNA production, and activation of pattern recognition receptor pathways.
The role of m6A modification in the regulation of adaptive immunity has recently been recognized. The most impressive discovery is a new function of the m6A modification in tumor immune response [260], which shows that antigen presentation from dendritic cells to CD8+ T-cells is regulated by m6A, specifically by the m6A reader protein YTHDF1, which enhances the translation of lysosomal cathepsins, leading to reduced tumor antigen presentation. Tumor growth was significantly inhibited in mice with silenced YTHDF1 compared with wild-type mice because more CD8+ T-cells and natural killer cells were recruited in YTHDF1-deficient mice [260], suggesting that control of YTHDF1 expression may be a new approach for the treatment of tumors.
The regulation of the immune response mediated by m6A occurs through multiple mechanisms. One of the critical functions of m6A modification is the control of T-cell homeostasis. In the last few years, several studies have validated that the suppressor of cytokine signaling (SOCS) family proteins SOCS1 and SOCS3, and cytokine-inducible SH2 containing protein (CISH) SOCS genes control IL-7 signaling and play critical roles in adaptive immunity [89]. SOCS gene activation induces the proliferation and differentiation of naïve T-cells, which are tightly regulated by m6A [89]. Modification of m6A in IL-7-induced naïve T-cells boosts the inactivation of SOCS (JAK-STAT) [261, 262]. Deletion of METTL14 induces colitis in mice, suggesting that m6A methylation is critical for maintaining immune homeostasis stability of T regulatory cells [139] and alleviating various autoimmune diseases [262, 263, 264]. Nevertheless, whether these principles can be applied to ocular immune homeostasis is unclear.
Finally, the clinical value of m6A in the regulation of immune response in the prognosis and treatment of tumors has also been recognized [265, 266]. It was found that m6A regulators are related to malignancy and antitumor immune response in cancer cells [266] and that the effectiveness of anti-PD-1 therapy could be highly enhanced by knocking down METTL3 or METTL14 due to increased infiltration of CD8+ T-cells, production of IFN, monokine, chemokine (C-X-C motif) (CXCL9), and interferon-gamma induced protein CXCL10 in tumors. METTL3 and METTL14 may therefore be used as potential therapeutic targets in anti-cancer immunotherapy, including ocular melanoma [42, 73, 265].
Collectively, m6A modification is involved in numerous ocular pathologic processes (Fig. 6), and its understanding may be valuable in improving our understanding of the pathogenesis of ocular diseases.
 Fig. 6.
Fig. 6.Roles of m6A modification in the pathogenesis of eye diseases. m6A may play a significant role in various pathological processes of eye disease such as microorganism infection, inflammation, autoimmune response, senescence, angiogenesis, degeneration, EMT, fibrosis, and ocular tumorigenesis.
Through reprogramming, we can obtain pluripotent stem cells with re-established epigenetic marks [267]. The discovery of cell reprogramming has changed the traditional view of how cells function and opens up exciting possibilities for the treatment of eye diseases [268]. Different types of cells can be genetically and epigenetically reprogramed. For example, Nrl knockdown prevents retinal degeneration [269], and reprogramming of photoreceptors can be used for vision rescue in mice [268]. Therefore, cell reprogramming has emerged as a promising strategy for the treatment of ocular diseases. However, several problems concerning cell reprogramming persist, including low efficiency, tumorigenic potential, and instability of the reprogrammed cell, Therefore, the molecular mechanisms underlying cellular reprogramming are still needed to be further investigated. The study of m6A methylation may improve our understanding of the control of cell reprogramming.
Recent studies suggest that reprogramming from one cell type to another may occur through the modulation of m6A modification. Chen et al. [270] mapped m6A modification in four cell types with different degrees of pluripotency and found that reprogramming of mouse embryonic fibroblasts to pluripotent stem cells was boosted under increased m6A levels, whereas it was hindered under reduced m6A levels. Metabolic reprogramming in hepatocellular carcinoma cells was also mediated by m6A modification [171], suggesting that m6A modification regulation may restore normal cell functions under various pathological conditions.
Stem cells, including ocular stem cells, are pluripotent cells that can differentiate into different types of cells. Normal cell differentiation is an important process in biological development. Ocular stem cells are one of the earliest stem cells used in regenerative medicine [271]. Ocular stem cells comprising the iris, sclera, photoreceptors, corneal limbus, cornea and conjunctiva, iris pigment epithelium, ciliary body epithelium, trabecular meshwork, retina, RPE, and orbital stem cells can be potentially used to treat eye diseases [272], including corneal disease, cataract, glaucoma, retinitis pigmentosa, and age-related macular degeneration AMD. However, understanding the precise mechanisms controlling single stem cell differentiation warrants further research. For example, the transcription factors that control ocular stem cell differentiation still need to be ascertained. m6A methylation plays a fundamental role in stem cell self-renewal and differentiation [68, 270]. It has been shown that the expression of pluripotency genes SOX2, NANOG, and DPPA3, was compromised when METTL3 and METTL14 were silenced. This phenomenon was associated with an increased expression of FGF5, CDX2, and SOX17, genes related to development in mouse embryonic stem cells [273]. Furthermore, METTL3 depletion prevented the self-renewal of porcine-induced pluripotent stem cells [83, 274]. The stability and expression levels of transcripts encoding developmental regulatory factors are negatively correlated with m6A because of the inhibition of the methyltransferases METTL3 and METTL14 compromise the expression of pluripotency genes such as SOX2 and NANOG [273]. The importance of methyltransferases and the m6A modification is supported by a recent report that showed that the self-renewal of porcine iPSC was inhibited by METTL3 knockdown [275].
Stem cells have been used for cell therapies, the development of new drugs, and for understanding the regulation of stem cell differentiation, including that of ocular stem cells is critic important [276, 277, 278, 279]. However, directional control of the differentiation of stem cells into specific organs or tissues, including ocular tissues, is still under investigation [280]. In addition to physical, chemical, mechanical, and biological (DNA methylation, histone modification, and non-coding RNA) approaches, regulation of m6A modification may be promising for the control of stem cell directional differentiation. Mapping of m6A modifications during embryo development in zebrafish revealed that the key gene notch1a controls the direction of endothelial–hematopoietic transformation through the cooperation of YTHDF2 with the activation of the Notch signaling pathway [281]. These results indicate the role of mRNA m6A modification in regulating the fate determination of stem cells and explain the key role of m6A in directional stem cell differentiation [281]. The expression of m6A modification factors has been illustrated in ocular cells, as mentioned previously. It has also been demonstrated that transcriptional factors, such as NANOG, PAX6, and SOX2, can control stem cell differentiation in numerous ocular stem cells [271, 282].
In conclusion, m6A RNA methylation provides an additional regulatory mechanism in regulating stem cell differentiation and pluripotency.
Biomarkers are crucial in the study of the pathogenesis and the early detection, diagnosis, and prognosis of diseases. Moreover, biomarkers are closely observed for monitoring treatments [283, 284]. However, few biomarkers are translated into clinical practice. Biologic stability and specificity are the major issues in using some biomarkers in clinical practice. The use of m6A as a biomarker could be challenging as m6A modification factors are likely to vary in different cells, and therefore, may be tissue-specific [11, 39, 57, 285]. As RNA and m6A modification factors are universally expressed in all eukaryotic cells, including ocular cells, plasma, serum, aqueous humor, vitreous, and any solid ocular tissue can be used to identify m6A biomarkers for the screening, diagnosis, and therapy of ocular diseases.
In the last few years, the value of RNA modification, especially m6A, for its potential roles in tumors [286], neovascular disease [74], degenerative disease [287], diabetes [288], and fibrotic diseases [220, 289] has been recognized. Potential biomarkers for ocular disease, particularly for uveal melanoma, have also been suggested [42, 73, 290]. In particular, the aberrant expression of METTL3 in melanoma cell lines and tissues may be considered a new biomarker for tumor diagnosis and treatment. Notably, reduced m6A is suggested as a possible biomarker of type 2 diabetes [291]. ZFAS1 expression regulated by METTl14 has been shown to be significantly increased in patients with atherosclerosis compared with those without atherosclerosis, suggesting that m6A modification factors may be a novel biomarker for monitoring atherosclerosis susceptibility [292, 293, 294]. Moreover, the prognostic value of m6A methylation has also been reported in patients with uveal melanoma [290], bladder cancer [295], and colon cancer [296]. In addition, METTL3 is considered a promising biomarker for determining the progression of gastrointestinal cancer and therapeutic response prediction [297]. Thus, collectively, the alteration of m6A modification may serve as a potential diagnostic biomarker for human diseases, including ocular diseases. However, the m6A signature in normal ocular tissues needs to be determined before m6A modification factors could be used as a biomarker for the screening of ocular disease.
Autophagy clears degenerated cells and reuses cellular components to maintain cell hemostasis. Without exception, autophagy participates in maintaining homeostasis of the cornea, aqueous humor, RPE, neural retina, and lens. Abnormalities in autophagy function contribute to retinal degeneration [298, 299], retinal injury [300], light-induced stress [301, 302], hypoglycemia [303, 304], lipofuscin accumulation in the RPE, and the pathogenesis of AMD [305]. Moreover, dysfunction of autophagy may be linked to photoreceptor degeneration [183, 220]. Furthermore, inhibition of autophagy induces retinal ganglion cell death [271, 306]. The implication of m6A modification and autophagy in ocular disease also was reported recently, a study investigating the relationship between m6A and the pathogenesis of age-related cataracts using gene ontology analysis revealed that autophagy-related genes are associated with altered m6A-circRNAs and methyltransferases in lens epithelium cells [51]. Systemic studies have shown that m6A plays a critical role in regulating macro autophagy/autophagy by targeting ATG5 and ATG7 [307], which are key proteins in autophagic vesicles. Interestingly, knockdown of FTO inhibited autophagosome formation and autophagy, suggesting that the expression of autophagy-related genes and autophagy is regulated by m6A modification, and m6A modification is required for autophagy [308]. Data from ocular and systemic studies on the relevance of m6A to autophagy imply the importance of autophagy in ocular homeostasis and diseases. Nevertheless, several fundament questions need to be answered before linking m6A and autophagy to vision function and eye diseases. For example, whether autophagy in ocular tissues is consistent with the expression of m6A modification factors and what is the role of the m6A and autophagy in the maintenance of the structure and/or normal physiological function of the eye (from the cornea to the vision pathway), need to be determined.
Exosomes provide intercellular communication and transmission of macromolecules between cells and they are enriched in proteins, lipids, mRNA, miRNA, and DNA. Therefore, they may contribute to the maintenance of normal cell functions, including cellular proliferation and differentiation, extracellular matrix (ECM) production, wound healing, immune response, and metabolic waste removal [309, 310]. In addition, there is a growing interest in using exosomes as biomarkers and therapy carriers [309]. Exosomes have been found in most ocular tissues, including corneal epithelial cells, RPE, trabecular meshwork, iris, and ciliary body epithelial cells, tear fluid, vitreous humor, human iPSC-RPE, retinal astrocyte, and uvea [311]. RNA is shuttled from one cell to another via exosomes, which is known as “exosomal shuttle RNA”. Interestingly, depletion of METTL3 reduced cellular and extracellular levels of miRNAs containing m6A consensus sequences [312]. To date, no study has demonstrated the direct regulation of the mRNA in exosomes by m6A. Nevertheless, it is known that exosomes are enriched in mRNA and miRNA, and it is likely that RNA is subject to m6A regulation and could potentially affect gene expression and protein translation. The roles of m6A modification factors derived from exosomes of ocular tissue in the maintenance of corneal and lens transparency, normal intraocular pressure, retinal blood barrier integrity, and ocular immune tolerance, the prevention of ocular autoimmune diseases and the retinal protective effects are largely unknown. Therefore, the precise role of exosomes mediated by m6A in maintaining normal cellular function and its effects on the pathogenesis of diseases, including ocular disease, should be determined.
Cell signalling is complex and essential for ocular cell differentiation,
proliferation, migration, survival, cell polarity, stem cell renewal, apoptosis,
and the development of many pathological conditions. Common cell signalling
pathways include TGF-
Besides the PI3K/Akt signalling pathway involved in m6A modification in ocular cells, MAPK signalling is also an important signalling pathway in the lens epithelium of age-related cataracts modulated by m6A methylation [51]. Importantly, PI3K and FAK have a cross-talk with MAPK [318], suggesting that activation of PI3K and FAK signalling may also activate MAPK [319].
Protein kinase A (PKA), commonly known as cAMP-dependent A kinase, is responsible for cAMP-mediated stimulation. It was found that FTO can downregulate PKA expression by demethylating PKA mRNA in RPE cells. Importantly, RPE cell death is increased by activation of the PKA/cyclin AMP-responsive element-binding (CREB) signalling pathway on FTO inhibition [72].
The NF-
It is currently challenging to treat ocular tumors, such as RB and melanoma, using a non-surgical approach. The genetic background may be quite different in people with the same tumor, therefore, different people may respond differently to the same treatment. After decades of research, the application of precision medicine wherein treatment is tailored based on the understanding of the genetic variability of patients’ tumors, has gained impetus. Precision medicine aims to target disease with patient-specific treatment. Interestingly, m6A modification may facilitate precision medicine. For example, the status of m6A methylation in uveal melanoma differed between patients with uveal melanoma in two separate reports [42, 73], suggesting that m6A modification is individual-specific. Thus, m6A RNA methylation may be dynamic and relevant to precision medicine. m6A methylation signatures may thus improve our understanding of the mechanism of many complex eye diseases. According to the Precision Medicine Initiative (https://obamawhitehouse.archives.gov/precision-medicine), precision medicine emphasizes the variability of the genetic background, environment, and lifestyle. However, precision medicine has not been widely applied in clinical practice, especially in relation to ocular tumor therapy.
Many studies have shown that m6A modifications are characteristic of tumor
development. Remarkably, the m6A modification patterns in the pathogenesis of
tumors, such as uveal melanoma [42, 73, 321], retinoblastoma [317], and
glioblastoma [322, 323], female reproductive system cancers [324], acute myeloid
leukemia [325], breast cancer [326], prostate cancer [327], liver cancer [57, 328], lung cancer [329], pancreatic cancer [330], colorectal cancer [331], kidney
cancer [332], and gastric cancer [333] are highly variable. The role of m6A
modifications in cancer genesis is controversial. The effects of m6A
modifications on cellular function vary frequently. Some methylated genes promote
tumor development, while the demethylation of other genes enhances tumor growth.
For example, m6A methylation can promote or inhibit uveal melanoma [42, 73].
Elevated levels of METTL3 or METTL14 (suggesting increases in RNA methylation)
facilitate the translation of the oncogene c-MYC and BCL2 in AML. In contrast,
demethylation of tumor suppression genes, ankyrin repeats, and SOCS box protein 2
and retinoic acid receptor alpha (RAR
The role of m6A methylation in tumorigenesis is variable in different tumors, and the interplay between methylation and demethylation in specific tumors and eye diseases and specific target genes seems to be critical for tumorigenesis and tumor treatment [42, 58]. However, studies that have used the parameters of m6A modification factors as a specific target (implying precision medicine) for cancer, including ocular tumor and complex eye disease treatments, are limited. A biopsy procedure might be required to determine which m6A modification factor is unique in a specific cancers and individuals, followed by a test to obtain the profiling of m6A regulatory factors in the ocular tumor. Through this, ophthalmologists may be able to develop different treatments tailored to different individuals.
Owing to the extensive involvement of m6A modification in the pathogenesis of diseases, the targeting of m6A regulatory factors may be a treatment strategy for human diseases by promoting or inhibiting m6A methylation.
Small molecular compounds such as m6A activators may be useful in the treatment of certain diseases associated with the decreased expression of m6A methylase [335]. A small molecular compound was found to bind the active site of the METTL3–METTL14–WTAP complex and promote the activation of m6A RNA methylation [335], suggesting a new treatment for the regulation of m6A modification externally and promoting the development of more such small molecules for possible applications in ocular disease. In conclusion, m6A activators may be considered potential therapies for the treatment of some ocular diseases, such as melanoma, in which m6A methylation is reduced [42].
The aberrant expression of m6A modification factors has been shown to be related to many ocular diseases [42, 45, 51, 72, 73, 74, 75]. The altered expression of m6A modification factors in ocular diseases has also been demonstrated. Therefore, inhibition of m6A modification factors may be promising for the treatment of ocular diseases, especially complex diseases (macular neovascularization, uveitis, DR, glaucoma, and myopia) and diseases resistant to regular therapy. For example, the success of anti-VEGF treatment for macular neovascularization is variable; 15–45% of patients do not respond well to anti-VEGF treatment [336, 337]. In a review paper, Garbo et al. [338] summarized the most recent updates on m6A related modulators and their potential treatment in human diseases, including 15 m6A modulators targeting METTL3, FTO, ALKH5 [338]. The review discussed the potential of FTO inhibitors, including dihydroxyfuran sulfonamides, inhibitors of hypoxia-inducible factor prolyl-hydroxylases (PHDs), chlororesorcinol analogs, meclofenamic acid (MA) and entacapone, in inhibiting FTO. Furthermore, imidazobenzoxazin-5-thione MV1035 as a specific ALKBH5 inhibitor and its ability to modulate the immune response in tumors were discussed [338]. There are several approaches to inhibit or regulate m6A modification factors. These include the new use of old drugs. We now realize that drugs, such as meclofenamic acid, can regulate m6A modification factors. Meclofenamic acid is a non-steroidal anti-inflammatory drug that can inhibit FTO function. The use of meclofenamic acid in ocular diseases has not yet been reported. Thus, research is being conducted to identify specific inhibitors of FTO for application in clinical settings [339]. Furthermore, the use of general methyltransferase inhibitors is also being explored. Currently, no m6A writer, eraser, and reader-specific inhibitors are available commercially, except for the general nucleoside analog Sinefungin-SAH [340, 341], which is a nucleoside S-adenosyl-1-methionine analogous to methyltransferase inhibitors for protein, DNA, and RNA methyltransferases [342] and lacks specificity. Next, the application of small molecular compounds with a relatively specific function for easy penetration into tissue has drawn great attention from researchers in the field. Two studies have been conducted using small molecular compounds to treat acute myeloid leukemia in vitro and in experimental animals. One study used the m6A writer METTL3 [343], while the other study used an FTO inhibitor [344, 345]. Both studies initially aimed at assessing the effects of METTL3 and FTO inhibition on the treatment of acute myeloid leukemia. The FTO inhibitor FB23-2 inhibited cell proliferation and induced cell apoptosis in myeloid leukemia cell lines and primary cells in xeno-transplanted mice by specifically binding to FTO molecules [344]. Later, the other two small molecular compounds were named CS1 and CS2 (or Brequinar), both of which could selectively inhibit FTO. Importantly, they have much higher efficiency and are stronger than the previously reported FTO inhibitors [309]. The compounds not only significantly inhibited tumor growth but also improved the survival of experimental animals. Mechanically, these compounds can enhance the sensitivity of cancer cells to T-cell cytotoxicity and immune evasion. Other FTO and ALKBH5 inhibitors, 2-oxoglutarate (2OG), N-oxalylglycine (NOG), pyridine-2, 4-dicarboxylate, Rhein, MA, IOX3, and R-2-hydroxyglutarate (R-2HG), have been shown to inhibit tumor cell growth and induce apoptosis [346, 347]. Recently, the METTL3 inhibitor STM2457 was reported to suppress myeloid leukemia cell growth in vitro and in animals through the selective inhibition of METTL3 and induction of tumor cell apoptosis [348]. Bedi et al. [349] also identified two derivatives of S-adenosyl-L-methionine (SAM) that show a high biding efficiency to METTL3 and may be considered as a candidate for a small molecular compound for the inhibition of METTL3. Moroz-Omori et al. [350] showed that an inhibitor of METTL3, UZH1a, which is selective and cell permeable, can function for up to six days and shows no negative effects on other RNA modifications (m1A, m6Am, m7G). Other consideration of the control of the expression of m6A modification factors is the application of CRISPR/Cas9, the CRISPR/Cas9 techniques can not only enable precise and target-specific modifications of DNA but also modify RNA to edit m6A at a specific site on mRNA [209, 351, 352, 353]. A study by Barbieri showed that the growth of acute myeloid leukemia cells was significantly inhibited by the deletion of METTL3 using the CRISPR technique [209]. Furthermore, specific deletion of ALKBH5 using the CRISPR technique led to the demethylation of oncoproteins of epidermal growth factor receptor and MYC, and inhibited proliferation of tumor cells [353]. Therefore, targeting m6A modification factors such as writers, erasers, and readers as a potential treatment for tumors is gaining attention in m6A research [346, 354].
Besides the progress in the study of the inhibitors of m6A modification factors, a number of manufacturers are also developing drugs targeting m6A modification factors, such as METTL3 inhibitors, which are also inching closer toward clinical trials [355]. The majority of these compounds, for the manipulation of m6A modification, focus on tumor treatment, which may be beneficial for patients who do not respond well to traditional cancer therapy or develop drug resistance [354, 355]. These developments in tumor treatment will help drive the development of potential therapies for ocular and systemic diseases, Notably, we are still at an early stage of the use of m6A-related drug in the treatment of human diseases.
In addition to m6A modification, the importance of other RNA methylations such
as N1-methyladenosine (m1A), 5-methylcytosine (m5C), pseudouridine (
The regulation of RNA methylation, specifically m6A methylation, may provide novel therapeutic strategies for the treatment of eye diseases, particularly for diseases that produce resistance to traditional therapies, as well as metabolic, degenerative, inflammatory, and vascular eye diseases. Determination of RNA methylation patterns may be facilitated by the application of single-cell sequencing and third-generation sequencing and will enable and improve prognostic analysis, early screening, diagnosis, and targeted therapy for eye diseases.
Nevertheless, the importance of m6A modification in biomedical research has been recognized relatively recently and its potential clinical applications in the treatment of eye diseases is a relatively new concept. Some of the challenges and questions we face going forward are as follows:
(1) Do m6A writers, erasers, and readers participate in the regulation of other cellular functions besides RNA transcription, processing, translation, and metabolism in ocular cells? Do different m6A modification factors play the same function in different ocular cells? Are they tissue- or cell-specific?
(2) The role of RNA methylation in vision function and the pathogenesis of eye diseases is complex and variable. The most challenging question in the study of m6A modification in vision function and eye diseases is to monitor dynamic (real-time) m6A changes under physiological and pathological conditions. For example, what is the role of m6A methylation in vision and eye embryonic development, maintenance of corneal transparency, maintenance of aqueous humor homeostasis, lens epithelial cell metabolism, blood–retinal barrier intake, subretinal vascularization, immune privilege, RPE, photoreceptor cell, glial cell function, extraocular muscles, maintenance of intraocular pressure, lens transparency, vision acuity, and the metabolism of vision cycle proteins? What is the pattern of m6A methylation after birth and what changes occur during a person’s life in specific ocular tissues and cells, and what is the relevance to vision function? Importantly, what are the dynamic changes in m6A in the pathogenesis of all eye diseases?
(3) The discovery of glycoRNAs on the surface of multiple cell types in different organisms, such as humans, mice, hamsters, and zebrafish, was a revolutionary finding in biological research [356]. The RNA molecules modified by glycosylation may exert some important cellular functions and insight into the mechanism of human diseases. If RNA methylation occurs in glycoRNAs, what is its significance in the regulation of cellular function and its relationship to ocular disease?
(4) What is the relationship between RNA methylation and extrachromosomal circular DNA (eccDNA)? EccDNA is widely present in various eukaryotes, including humans, carries complete genes and special regulatory elements and can replicate independently [357, 358, 359, 360]. The genes carried by eccDNA can actively transcribe into mRNA and play an important role in regulating cell function and in numerous human diseases as independent DNA molecules. Therefore, it would be interesting to determine if RNA from eccDNA is modified by m6A modification factors. The most recent study led by Zhang showed that mRNA m6A modification can also contribute to human disease heritability [361]. This concept in vision function and eye research remains to be elucidated.
m6A, N6-methyladenine; RPE, retinal pigment epithelium; AMD, age-related macular
degeneration; TGF-
XL, SH and HY carried out the conception of this review; XL, SH and HY designed this review; XL, BM and SH drafted the manuscript and prepared the Figs; ML, LL, XZ, MD and JY edited the manuscript and collected the related references; XL, SH, HY and BM revised the manuscript and improved the language.
Not applicable.
Not applicable.
This work was supported by National Natural Science Foundation of China (81873681, 81770952), the Basic Research Program of Henan Eye Hospital (20JCQN005) and the “23456” project of Henan Provincial People’s Hospital (ZC20200296).
The authors declare no conflict of interest. SH is serving as one of the Editorial Board members of this journal. We declare that SH had no involvement in the peer review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to GP.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.






