1 INEB-Instituto Nacional de Engenharia Biomédica, 4200-135 Porto, Portugal
2 i3S, Instituto de Inovação e Investigação da Universidade do Porto, 4200-135 Porto, Portugal
Academic Editors: Anna Gałązka and Graham Pawelec
Abstract
The preservation of natural ecosystems, as well as the correct management of human societies, largely depends on the maintenance of critical microbial functions associated with soils. Soils are biodiversity rich pools, and rhizosphere soils can be associated with increased plant functions in addition to the regulation of nutrient cycling, litter decomposition, soil fertility and food production by agriculture systems. The application of biocontrol agents or plant growth-promoting bacteria has been tested in order to colonize roots at initial stages and offer advantages by promoting healthier and higher-yielding crops. In this review we describe the efforts to develop more sustainable systems that seek to minimize environmental disruption while maintaining plant health. Particular emphasis is given in this review to soil improvement strategies and the taxonomic groups involved in plant growth and protection against biotic stresses. It is important to define the impacts of land management and crop production practices on the structure and composition of soil bacterial communities. By promoting, monitoring and controlling the plant microbiome, and understanding the role of certain biocontrol agents within the plant throughout the lifecycle of the plant, we may substantially improve nutritional and environmental standards and reduce the negative impact of some agrochemicals. The integration of biological alternatives with traditional strategies may be critical to improve the sustainability of agriculture systems.
Keywords
- biocontrol
- biofertilizer
- composting
- plant growth promoting bacteria
- review
- rhizosphere
- seed coat
- soil engineering
- soil microbiome
- sustainable agriculture
It is well known that bacteria can easily live and flourish in the soils, making soils extremely diverse in microbial organisms. One of the first studies evaluating the microbiology of 1g of boreal forest soil revealed up to 10,000 different bacterial species [1]. And further studies demonstrated that the amount of microorganisms per gram of soil can be even higher, reaching the surprising value of 1 million genomes in such a small sample [2]. These studies represent the first steps of an amazing and fruitful journey to understand the soils and how its diversity emerges and is largely affected by physical, chemical and biological variables in the environment.
The preservation of natural ecosystems, as well as the correct management of human societies, largely depends on the maintenance of critical microbial functions associated with the soils and plant productivity. The taxonomical and functional distribution of bacteria in soils are influenced by multiple factors, including soil features (e.g., texture and pH), soil types, water availability, climatic conditions, geogenic factors, competition and other inter-relationships among living creatures, and even anthropogenic activities (soils used for disposal/dumping sites, other land uses, building and constructions, agriculture) [3, 4, 5, 6, 7, 8, 9, 10]. Nevertheless, the distribution of soil microbial communities across the landscape is still far from being completely understood.
The presence of plants is known to affect the structure of soil microbial communities and generate uneven distribution of microbes in soil that are affected by the surrounding environment, conditions and nutrient availability [3, 11]. Plant roots secrete large amounts of photosynthetically fixed carbon, in a wide range of molecules such as carbohydrates, amino acids, organic acid ions, and vitamins, providing a unique niche for microbes. Subsequently, soil in close proximity to plant roots harbours an increased number of microorganisms [12]. Soils are biodiversity-rich pools, and areas under the direct influence of the plant roots, commonly denominated as rhizosphere soils, showed even higher values of biodiversity due to the amount of nutrients and other chemical complexes released by plant cells [13, 14, 15, 16, 17]. Interestingly, the first studies on bacterial communities near plant roots showed a significant increase in microbial diversity, paralleled by an increase in soil functions such as regulation of nutrient cycling, litter decomposition, soil fertility and food production by terrestrial ecosystems. Due to the enormous genetic pool, microbes dominate matter and energy transference between above- and below-ground communities, as well as the availability of essential nutrients for plant growth, as some of these communities are capable of participating in processes such as nutrient mineralization [18, 19, 20]. These critical soil processes then modulate the stability of the plant microbiome [21, 22, 23, 24] and support plant life as we know it, and, consequently, its productivity [25, 26, 27, 28].
It is well known that long-term monoculture results in a constant decrease of plant performance due to reduction of bacterial and fungal richness and diversity over time and increase of soilborne diseases [29, 30]. Such results were recently observed in soils used for coffee and tobacco plantations, specifically, a reduction in diversity of Proteobacteria, Bacteroidetes and Nitrospira, and fungal Ascomycota phyla over time, most likely associated with nutrient deficiencies and soil degradation [27, 31]. Actinobacteria, Bacteroidetes and Proteobacteria dominate the bacterial communities in the rhizosphere soils surrounding the plant roots and are critical for plant growth and productivity [32, 33, 34, 35, 36, 37, 38]. Soil manipulation can be used to improve degraded, exhausted or diseased soils and therefore positively impact the plant growth. This strategy can play a significant role by adding or improving the way plants sustain and get nutrients and water from the soil, as it was the case demonstrated by Sellitto and colleagues [39] in tomato plants.
Traditional strategies, such as tillage, can affect Oomycete and fungal agents causing disease by interfering with their distribution in the soils, disrupting hyphae and soil clumps, increasing air flow and lowering the potential for disease in the soils [13, 27, 29, 30, 31, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62] (Table 1). This is well known by farmers and used prior to planting for enhanced protection of seedlings. Other strategies include the removal of the green bridge with herbicide timing [40, 63], crop rotation [64], in-furrow chemical treatments [40], stubble burning and integrated control; however, some strategies result in soil erosion or imply soil constraints, making them unproductive. Composting has been suggested as an alternative to correct or complement soils, in addition to reducing the environmental cost of organic waste disposal [65]. But compost effects can sometimes be unpredictable, depending on the feedstock and composting process that is employed. Both can be very different in nature and complexity, and subsequently associated with very different microbial communities [66, 67].
| Alpha and beta diversities | Relationship and effects on taxonomic groups | Cost | Other information | References | |
| Monoculture | reduction of alpha diversity | reduction of diversity within Proteobacteria, Bacteroidetes and Nitrospira, and Ascomycota fungi; it affects soil Acidobacteria, Verrucomicrobia and candidates phyla Rokubacteria, formerly known as SPAM (Spring Alpine Meadow), and WS3 (Wurtsmith aquifer Sequences-3) | low | reduction of bacterial and fungal richness | [27, 29, 31] |
| Rotation of crops | increase of bacterial alpha diversity (at a slow pace), low effects on fungal population | increase of Bradyrhizobium, Rhizobium, Pseudomonas, Flavobacterium, Sphingomonas, Rhizobacter, Arthrobacter, Streptomyces and Allostreptomyces in soils | low | reduction of pesticides usage, gradual replacement of taxa in single cropping season, correction of nutrient deficiencies and soil erosion, improvements in soil carbon sequestration, structural stability and organic matter | [30, 44, 45, 47, 48] |
| Tillage | reduction of alpha and beta diversities | low | reduce Oomycete and fungal agents causing disease, it increases air flow in the soils | [62] | |
| Chemical treatments | reduction of alpha and beta diversities | medium | soil erosion, high plant productivity | [40] | |
| Addition of wheat straw | increase of alpha diversity | increase of Proteobacteria (namely Bradyrhizobium and Rhizobium) and reduction of Acidobacteria | low | [46] | |
| Manure and sewage sludge | increase of alpha diversity | increase of Actinobacteria; Rhodanobacter, Terrimonas and Chitinophaga populations | low | increase of microbial biomass | [49, 50, 51] |
| Organic farming | reduction of alpha and beta diversities | Actinobacteria, Firmicutes, candidate phyla radiation Saccharibacteria (TM7), Planctomycetes and Verrucomicrobia are higher in conventional farming | low | [52] | |
| Biochar | increase soil bacterial and fungal diversities and activities | increase of Bradyrhizobiaceae and Hyphomicrobiaceae | medium | improve soil quality, plant performance and nutrition, nutrient cycling and tackle the progression of soilborne diseases | [56, 57, 58, 59, 60] |
| Compost | increase of bacterial and fungal alpha diversity | increase of bacteria such as Pseudomonas, Streptomyces, Bacillus, Burkholderia, Enterobacter, Lysobacter and Pantoea, as well as the fungi Gliocladium and Trichoderma | low to medium | [53, 54, 55] | |
| Soil transplantation | increase of bacterial and fungal alpha diversity | introduction of new microbiota into soil specially rare taxa, such as Massilia, Dyadobacter, Terrabacter, Arachidicoccus and Dyella, as well as pathogen-suppresors such as Pseudomonas, Bacillus, Gp16 (Acidobacteria), Spartobacteria genera incertae sedis, Hyphomicrobium and Sphingomonas; regarding fungal communities, Podospora, Chaetomium, Mortierella and Phialemonium are the most affected taxa | high | addition of nutrients and water availability to degraded soils, improvement of plant productivity | [39, 61] |
| Plant growth promoting bacteria (PGPB) | slight increase of alpha diversity | improving the interaction of rhizosphere-root microbiomes | medium | promote health, nutrition, resistance to biotic and abiotic stresses and higher yields | [13, 41, 42, 43] |
More recently, the application of biocontrol agents or plant growth-promoting bacteria (PGPB), added for example as coated seeds or by using sprays, has been tested in order to colonize roots at initial stages and offer an advantage in terms of promoting healthier and higher-yielding crops [41, 42, 43]. PGPB act inside the plant by promoting growth and facilitating nutrient acquisition, phytohormone production, induction of defence responses, removal of contaminants and competition with plant pathogens. Efforts to develop more sustainable systems that seek to minimize environmental disruption while maintaining plant health are contingent upon defining the impacts of land management and crop production practices on the structure and composition of soil bacterial communities. PGPB may play an additional role by improving the interaction of rhizosphere-root microbiomes, therefore providing extra benefits to the plant [17, 24, 68, 69, 70]. These microbes, once adapted to the plant and environmental conditions, support the plant development, growth, health and nutrition and enhance its ability to resist biotic and abiotic stresses, such as soils with water or salinity deficiencies [71, 72]. Recently, Bano and colleagues [73] suggested the concept of a microbiome-mediated smart agriculture system (MiMSAS) that integrates technology and information regarding practices, the application of synthetic microbiome and plant genome editing/engineering for agricultural benefit.
But can these strategies change the soil microbiome in a sustainable and economically feasible manner? Can the soil microbiome be changed dramatically by implementing strong and integrated systems? Or are the soil microbiome dynamics only affected punctually and in a very limited manner that needs constant investment and new strategies to address the problems? Which are the taxonomic groups more helpful for plants and how do they change in multiple systems? In this review, the most recent studies on soil microbiome, namely the rhizosphere, and the impacts of using organic amendments and other strategies to change the microbial populations present in the soils will be addressed. In addition, the role of particular taxa and bacterial strains in protecting plants and limiting the dissemination of pathogens in productive soils will be discussed.
Rotation of crops is used worldwide as a simple strategy to reduce, and sometimes even correct, nutrient deficiencies and erosion in soils (Table 1). By changing the crops in a particular terrain over time, it is possible to prevent soil exhaustion and manage the microbial communities present in the soil in a more sustainable manner than by using monoculture systems. Multiple crop associations have been tested in different world regions and some plants described as more productive under such conditions. The microbial community composition of both bulk soil and rhizosphere can be affected by distinct agricultural management practices, and crop rotation is among the most common of the traditionally employed practices, due to its productivity and reduced costs [30]. It also has the added benefit of reducing soil diseases that are dependent on a particular crop and lack the ability to proliferate when other crops are promoted. By shifting and modifying the composition, and subsequently the suppressiveness of natural soil microbiomes, crop rotation can reduce the burden of diseases and avoid the use of pesticides [44]. Specific genera and species can be gradually replaced by the indigenous soil taxa, and this change can happen in a single cropping season [45]. Some taxa can be more relevant in maintaining rhizosphere microbial networks. The application of crop rotation to eroded cropland planted with perennial forage showed an increase of soil organic matter, nutrient cycling, and soil conditions [74, 75].
The increase of plant productivity following crop rotation strategies is
frequently reported. The wheat-oilseed rape rotation is commonly used in Europe
[45] and, by returning straw to soils, crop yields can also be increased [46].
Crop-pasture rotation is frequently used in temperate regions and it has been
associated with improvements in soil carbon sequestration, structural stability,
organic matter and biological diversity, supporting a more sustainable
agriculture in these areas [74, 76, 77, 78]. Maize–rice crop rotation has also shown
soil improvements regarding microbial communities compared to rice-rice systems
[30]. The effect of crop rotation appears to be mainly limited to bacterial
populations, as only small differences in community composition of fungi have
been observed in both the rhizoshpere and bulk soil [32, 46, 79]. The addition of
wheat straw to soils can significantly improve the
Crop rotation may also bring advantages to infected soils. In addition to displaying improved growth in soils preceded by sunflower or pea, maize seedlings showed significantly less root damage following the exposure to Fusarium graminearum and western corn rootworm [82]. The relative abundance of Glomeromycota was also higher in soils preceded by other cultures. Infestation with western corn rootworm affected mostly Acinetobacter, Smaragdicoccus, Aeromicrobium and Actinomucor, while F. graminearum affected fungal endophytes, including Trichoderma and Endogone. Gong and colleagues [83] described paddy-upland rotation as an efficient strategy to promote cucumber growth. The potential of Fusarium and Monographella spp. pathogens was reduced by regulating the soil water content under 100% soil water content in a single cress. Simultaneously, under 80% of cress cultivation could promote the colonization of beneficial microbes such as Roseiflexus and Pseudallescheria spp. [83].
An effective approach for soil fertility recovery has long been the use of biofertilizers or organic amendments such as animal and green manure, organic wastes, composts, or, more recently, biochar [84] (Table 1). These are nutrient-rich composites that can be used to correct and manage soils by adding nutrients, especially nitrogen, phosphorus and carbon, in a much slower, targeted and sustainable manner compared to chemical fertilizers. Different biofertilizers or amendments promote and potentiate distinct microbial communities, resulting in distinct consequences on soil fertility and health [85, 86, 87]. The positive effects of the application of biofertilizers or organic amendments on crop yields is related to the microbial structure, composition and richness of the soil, as well as the variety of functions made available by the microbial communities [86, 88, 89]. These communities also play a role in nutrient cycling, conversion of soil organic matter and improvement in soil physical conditions [84, 87]. The repeated use of organic amendments over several years, such as manure and sewage sludge, increased the microbial biomass and changed its microbial community structure [49, 50]. But the quality of the organics can produce different results and the response to soilborne pathogens can also differ according to the type of biofertilizer that is employed and the crop being tested [66]. Soil microorganisms are involved in multiple processes fundamental to soil fertility and health, plant nutrition and the production of natural products that can add protection against pathogens and pests. Generally, the addition of biofertlizers has been associated with an increase of Actinobacteria in soils [51], similarly to what was described for conventional soils versus soils with organic farming [52]. Actinobacteria are well known producers of secondary metabolitesthat enable them to control some diseases more efficiently [13, 85]. In addition to Actinobacteria, Firmicutes and candidate phyla radiation Saccharibacteria (TM7) were more abundant in the conventional farm, while Planctomycetes and Verrucomicrobia were less abundant in organic-farmed soils [52]. Organic amendments could change rhizosphere microbiome, mainly Rhodanobacter, Terrimonas and Chitinophaga populations, also improving disease suppression [28]. Actinobacteria and Proteobacteria were also more abundant in soils treated with synthetic fertilizers [84].
Composting is a controlled biological decomposition process resulting in the degradation of organic materials into humic substances by successive groups of microorganisms [90]. Composting has been used in farming to improve soil fertility, nutrient levels and crop health for centuries, and it helps reduce soil diseases and pathogen attacks by promoting microbial population growth and activity [53, 67, 91, 92]. The role of Pseudomonas, Streptomyces and Trichoderma strains against soil diseases is well known, as multiple strains are frequently described with plant growth promoting activity [43, 53, 54, 67, 93]. These bacteria are frequently found in high abundance in compost, in addition to other bacteria such as Bacillus, Burkholderia, Enterobacter, Lysobacter and Pantoea, as well as the fungus Gliocladium [55]. Other examples of composting utility can be found in studies describing the employment of vineyard pruning waste that showed higher relative abundance of Ascomycota, mainly of the orders Sordariales and Hypocreales, in soils affected by Phytophthora nicotianae [94], or the use of composted agro-waste for controlling diseases on a broad range of horticultural crops [95]. In addition, there are studies describing the employment of compost to control diseases caused by Pythium spp. [96], Rhizoctonia solani [97], Fusarium oxysporum [53], Verticillium [98], Sclerotinia sclerotiorum [99] and Sclerotinia minor [100] by testing multiple plants. The multi-suppressive properties of composts from agricultural residues, agro-industrial co/by-products, and plant green waste shows a very complex microbiome structure [98]. The profits associated with the circular economy is expected in the coming years to promote the employment of composts coming from agro-wastes, agro-industrial residues or bio-energy co/by-products in agriculture systems [101].
Biochar, a carbon-based product, can also improve soil quality, plant performance and nutrition and nutrient cycling, and tackle the progression of soilborne diseases [56, 57, 58]. Review of the literature showed that direct application of biochar can be advantageous in tomatoes and asparagus against Fusarium [102, 103] and foliar diseases caused by Botrytis or Phytophthora in tomatoes and strawberries [104, 105, 106], but no effects were observed against Rhizoctonia or Pythium [59]. The addition of extra beneficial soil microorganisms to biochar may increase soil bacterial and fungal diversity and activity [59]. Biochar with wider surface area and pores may offer potential sites for soil microbiota, such as Pseudomonas, Azotobacter, Rhizobium and Azospirilium, as it offers carbon, nutrient and water for microbial proliferation, also benefiting the plants [107, 108, 109]. In addition, biochar application may promote growth of denitrifying rhizobials, such as Bradyrhizobiaceae and Hyphomicrobiaceae [60].
Intensive agriculture can result in the degradation and erosion of soils that may take a long time to be corrected . As the purchase of new and vast terrains for crops can be very expensive, and sometimes such options are not even available, transplantation of new soils not used for intensive agriculture to cover or replace the topsoil in unproductive or diseased fields was considered. The replacement of topsoil in soils needing restoration has been tested in Europe [110, 111] and some of these soils did return to being productive. In 2016, Wubs and colleagues [112] tested this strategy with the objective of introducing new microbiota into soil and proved its efficiency for ecosystem restoration. The results were positive when the soil inoculant was introduced into intact topsoil, but the largest impact was observed when the topsoil layer was exchanged with the soil inoculant. Such strategy could be used to improve plant productivity and offer new agriculture usefulness to degraded soils. Nevertheless, such works can be technically expensive and sometimes carry high environmental costs [112]. It may be very important to monitor the transplanted soils as changes in the microbial communities may happen during the process. Zhao and colleagues [61] reported considerable changes in soils from donor sites to destination, namely in fungal communities, being Podospora, Chaetomium, Mortierella and Phialemonium the most affected taxa. Fungal communities could easily adapt to the destination environment, while bacterial populations remained stable 6 years after soil transplantation.
It has been suggested that controlling plant disease outbreaks can be achieved by increasing the relative abundance of naturally protective bacteria using enriched-soil transplantation, thereby managing the composition of soil microbiome as a whole [44]. The transplantation of rhizosphere microbiota from resistant tomatoes helped to suppress disease symptoms in susceptible plants, supporting the idea that in some cases the transplantation of rhizosphere microbiota may work as a probiotic for promotion of plant functions [25]. It has been suggested that the composition of the initial soil microbiome may predetermine a plants resistance to certain diseases. The identification of flavobacteria, such as TRM1, were more abundant in the rhizosphere microbiome of Ralstonia-resistant tomatoes than in the rhizosphere collected around the susceptible plant. Rare taxa, such as Massilia, Dyadobacter, Terrabacter, Arachidicoccus and Dyella, highly pathogen-suppressing Pseudomonas, Bacillus, Gp16 (Acidobacteria), Spartobacteria genera incertae sedis, Hyphomicrobium and Sphingomonas may play a relevant role on supporting plants against certain diseases [24, 44, 93, 113, 114]. In addition, the high abundance of genes encoding antimicrobial compounds can also be associated with plant protection [19, 20, 89, 115, 116].
Microbiome composition is certainly a critical factor for plant protection and the availability of a large pool of rare taxa in the soil may help to correct microbial dysbiosis and tackle disease. Beneficial microorganisms could be added at seed or seedling stages in order to enrich plant growth and better conduct soil management [11, 13, 117]. The relevance of specific soil taxa was demonstrated when contaminated soils treated with gamma-irradiation resulted in modified growth of the willow plant, although microbial communities tend to converge after 100 days [118].
A current challenge in agriculture is to improve yield per hectare, considering arable soil, and the nutrients available within, are finite resources [119]. In light of this, as the human population grows, it is important to be able to increase plant productivity and yields to meet demand. One of the most sustainable ways is to improve natural abilities of plants, especially in soils rich in microbes that can already be associated with increased plant health [44]. Agrochemicals have been overused worldwide to improve yield per hectare substantially at the cost of negative effects to human health and the environment [120, 121]. The addition of nitrogen agrochemicals to soils had always been the immediate solution to improve crop yields. Its use is estimated at ~1011 kg/year, but ~60% of this synthesized nitrogen cannot be absorbed by plants and reaches the groundwater [119, 122]. Nitrogen and phosphorus are normally the limiting nutrients for cyanobacterial and algal blooms, and blooms in rivers, lagoons and the sea are known to be harmful to aquatic life [121, 123].
Biopromoters based on specific strains of PGPB or fertilizers enriched with such strains have been introduced as an alternative that reduces the use of chemicals and improves nutrient utilization by plants [43, 124, 125]. Nevertheless, evidence of the phytoprotective effect of microbes needs to be evaluated under agricultural conditions, because certain microbes may be detrimental in such conditions [126]. Some soil bacteria have the ability to use nutrients that are in excess in the soil, potentially making these available to plants. In addition to nitrogen and phosphorus, iron is another element which plants can acquire via soil microorganisms. Some PGPB are capable of sequestering the insoluble ionic compounds from the rhizosphere environment, reducing iron availability in the soil and slowing the growth of microorganisms opportunistic toward the plants [119, 127, 128]. An example of this can be seen in iron-poor soil where plants grow better in the presence of some microorganisms, supporting the idea that these microbes may increase the bioavailability of scarce nutrients for use by plants [129].
Based on the limitations described above, it is currently not feasible to expect large improvements in plant yields in the coming years. The main challenge will be replacing the presently damaging agriculture systems with new and more sustainable alternatives while maintaining current levels of productivity in the most productive soils. In fact, the most likely solution for increasing yields worldwide in a short timeframe may come from controlling and tackling plant diseases—these are still responsible for substantial yearly losses, and direct research and investment is needed for the recovery of these less productive soils.
Microbiome management is particularly relevant nowadays to predict correct soil degradation, improve plant growth and productivity and understand the network of microbes working on the production systems [11, 130, 131, 132, 133]. Multiple studies have shown predictive taxa with activity against certain diseases, as is the case for Streptomyces and Trichoderma for R. solani, Pseudomonas and Streptomyces for Pythium sp., and Aspergillus, Pseudomonas and Streptomyces for F. oxysporum [13, 24, 53, 93, 95, 134, 135]. Specific strains have more impact than others, so it is important to select for strains with an ability to interfere and add protection to the plants. Multiple strains belonging to diverse bacterial phyla, and even some fungi have been described (see Table 2, Ref. [24, 78, 86, 136, 137]).
| Actinobacteria | Bacteroidetes | Firmicutes | Proteobacteria | Fungi | |||||
| Brevibacterium | Chryseobacterium | C. balustinum | Bacillus | B. amyloliquefaciens | Achromobacter | Aspergillus | |||
| Corynebacterium | C. agropyri | Sphingobacterium | B. cereus | Acinetobacter | Claroideoglomus | C. claroideum | |||
| Curtobacterium | B. circulans | Agrobacterium | Trichoderma | T. harzianum | |||||
| Microbacterium | B. licheniformis | Azospirillum | T. koningiopsis | ||||||
| Streptomyces | S. fulvissimus | B. pumilus | Bradyrhizobium | B. japonicum | |||||
| S. thermocarboxydus | B. subtilis | Burkholderia | B. cepacia | ||||||
| B. velezensis | Chromobacterium | ||||||||
| Paenibacillus | P. lentimorbus | Comamonas | |||||||
| P. peoriae | Janthinobacterium | ||||||||
| Kosakonia | |||||||||
| Leclercia | |||||||||
| Lysobacter | L. enzymogenes | ||||||||
| Pantoea | |||||||||
| Pseudomonas | P. aeruginosa | ||||||||
| P. chlororaphis | |||||||||
| P. fluorescens | |||||||||
| P. libanensis | |||||||||
| P. putida | |||||||||
| P. stutzeri | |||||||||
| Rhizobium | R. leguminosarum | ||||||||
| R. etli | |||||||||
| Serratia | S. plymuthica | ||||||||
| Stenotrophomonas | S. maltophilia | ||||||||
| S. rhizophila | |||||||||
| Xanthomonas | |||||||||
Microorganisms artificially inoculated in seeds, plants or soils have to compete with a highly diverse microflora [43, 138, 139]. Introduced strains may establish well in the new community, or very poorly, with rapid decline in numbers to complete disappearance in just a few days. Therefore, it is critical to know the microbial communities and how the introduced strains fit into the established population. Beneficial strains can be added to plant seeds as seed coats or by using microbial formulations sprayed on flowers of crop plants, where the bacteria become incorporated into the progeny seed upon colonising the flower [43, 138, 140]. The first option has been tested more often as the new microbial strains help the plant in early stages of development [13, 24]. As seeds develop and plants grow in the soil, bacterial populations tend to increase in the early stages, while fungi dominate at mid and later stages [13, 141]. One major challenge is the selection of strains that do not interfere with indigenous endophytic populations, as these may be valuable in later stages of plant growth. After a few weeks, the inoculant can disappear or establish as part of the plant microbiome where it may even colonize the next generation of plants if incorporated into the seeds. Inoculated strains should be capable of competing and interacting with rhizosphere microbes. A correct management of plant cropping should take into account the full microbial consortia in soil, namely in the rhizosphere, and the capability of beneficial strains to survive under such conditions.
Among the microbial consortia available in the soil, the group of nitrogen fixers is usually mentioned as one of the most relevant as it may enrich soils with nitrogen, a critical nutrient for increasing plant biomass and productivity. Nitrogen fixers can be found in symbiotic relationships with the plants (e.g., legumes), as is the case for the well described genus Rhizobium, but also Allorhizobium, Azoarcus, Azorhizobium, Bradyrhizobium, Burkholderia, Frankia, Mesorhizobium and Sinorhizobium [72, 142, 143]. Additional groups of free-living nitrogen fixers include Azoarcus, Azotobacter, Azospirillum, Gluconacetobacter and Herbaspirillum [144]. These organisms use organic molecules to produce nitrogen at an average cost of 16 moles of ATP per mole of nitrogen fixed [72]. And if some productive plants, such as legumes, do not require additional nitrogen for their development due to interaction with nitrogen-fixer microbes, the same cannot be observed for other plants. Tomato inoculants can benefit from the addition of PGPB, reducing the plants dependence on chemical fertilizers by up to 25% [145]. Maize is another plant that may benefit from PGPB addition as it may increase yield and biomass production by 12 and 18%, respectively, in addition to reducing chemical dependency by up to 20% [146].
These strategies can be seen as innovative for implementation of new approaches for sustainable agriculture. For example, Bacillus circulans strain GN03 was described as effective for promotion of growth-related hormones (indole acetic acid, gibberellic acid, and brassinosteroid) and disease resistance-related hormones (salicylic acid and jasmonic acid) in cotton seedlings through the upregulation of genes related to synthesis (EDS1, AOC1, BES1, and GA20ox), auxin transporter (Aux1) and disease-resistance (NPR1 and PR1) [147]. Stenotrophomonas rhizophila strain SR80 enhanced wheat growth, both below and above ground, and induced strong disease resistance by boosting plant defence in the above-ground plant parts when the pathogen Fusarium pseudograminearum was present [148]. But there are several strains described with beneficial effects proved in cereals, such as wheat, rye or rice, as well as in other cultures such as barley, tomato, cucumber, pepper or Chinese cabbage (see reviews [43, 72, 149, 150]). Organic amendments can also be supplemented with PGPB, and their effects on plants extended even further when compared to controls. Bibi and colleagues described four agro-industrial wastes, black gram husks (15–22% crude protein), rice bran (10–15% crude protein), peanut shell (6–7% crude protein), and dry leaves, supplemented with a specific strain of Burkholderia cenocepacia to carry solid-state fermentation of organic wastes [151]. The results showed increased maize germination, promptness, and seedling vigour, as well as chlorophyll, water, protein and amino acid contents, compared to controls.
Soilborne plant diseases represent a serious threat to global food security as they are associated with a large reduction in plant productivity and decline of yields worldwide. There are strategies to effectively control such threats as described above, but they are not continually consistent or predictable, as it depends on the soil, culture and other conditions [43, 66, 84, 131, 152]. The mechanisms underlying the suppressive effect on diseases by particular soils or, for example, in soils treated with organic amendments, are not fully understood, but the biological activity of the microbiomes is critical, as well as the interactions established between the soil and the plants [67, 98]. Bacillus, Pseudomonas and Streptomyces strains are well known producers of secondary metabolites that may act as potential biocontrol agents and PGPB [43, 93, 153]. The relative abundances of certain bacteria, such as Acidobacteria (Acidobacterium), Actinobacteria (Streptomyces), Firmicutes (Bacillus) and Gamma-Proteobacteria (Pseudomonas), and fungal groups, namely Eurotiales (Aspergillus and Penicillium), Hypocreales (Trichoderma and Fusarium) and Mortierellales (Mortierella), should be monitored based on what is known of their activities and functions for plant protection [98].
Dignam and colleagues [54] identified Pseudomonas species diversity and
richness (Margalef’s) as the primary parameters explaining the greatest
proportion (
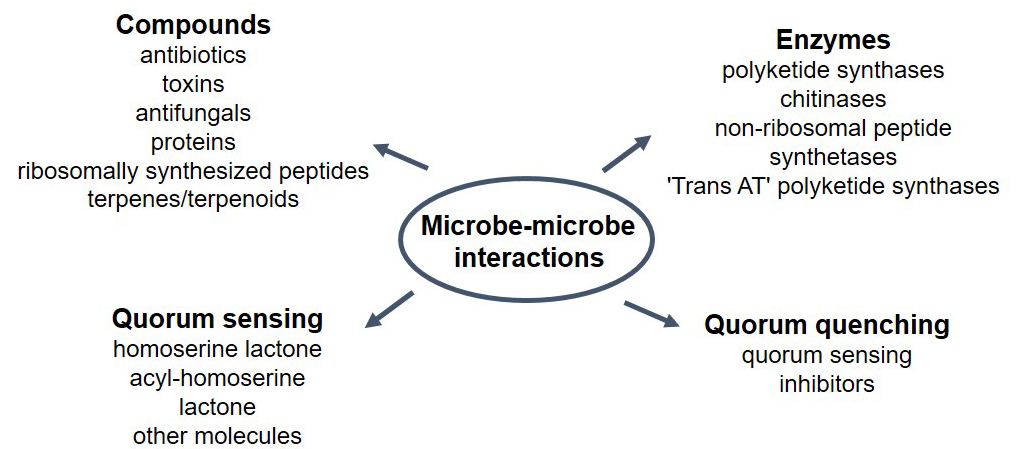 Fig. 1.
Fig. 1.Multiple fuctions have been described in soils related to microbe-microbe interactions. These are mostly associated with the production and different compounds and enzymes released to the environment, as well as molecules participating in quorum sensing and quorum quenching.
The fungal genus Mortierella was suggested as an indicator and enhancer of Fusarium wilt disease suppression in vanilla [135]. The suppressive soil was enriched by fungi belonging to Zygomycota and Basidiomycota, and bacteria belonging to the phyla Acidobacteria, Verrucomicrobia, Actinobacteria and Firmicutes, and the interactions between these groups were enhanced in suppressive soils. Similar effects were observed when watermelons were exposed to F. oxysporum [85]. The rhizosphere soils showed the highest relative abundance of potential antagonists against F. oxysporum, and some chemical properties were associated with the inhibition of Fusarium wilt disease. Actinomycota were increased in rhizosphere soils exposed to Fusarium wilt and enhanced activities of defence enzymes were observed in the leaves of melons [85].
The relationship between the endogenous rhizosphere microbiota, endophytes with potential biocontrol agents (internal or externally added) and the plant host, is essential to improve strategies for more productive and less degraded soils. Some bacterial endophytes have shown to impact plant growth and provide protection to the plant. Although the effects initially tend to be small for multiple plants [43, 156, 157, 158], these valuable organisms can be used as a basis for developing safer ecological approaches to disease management. Nevertheless, such tests are difficult to conduct, as the results of field and greenhouse trials often disagree, which can only be partly explained by climatic conditions [159, 160, 161]. Different cultivation systems affect the soil biodiversity and promote changes in the microbial profile, even when the same starting soil is used [24]. These effects tend to be more visible among the less common taxonomic groups found in the soils, designated as rare taxa. These rare taxa tend to change drastically among systems (field versus greenhouse), soil types and even among replicates. It is possible that environmental conditions and the variability found among rare operational taxonomical units explain the differences found in field versus greenhouse trials.
The productivity and yield of cereal cropping increased exponentially after the nineteenth century with the introduction of machinery and technology, but this increase was interrupted in recent decades, and instability prevailed due to soil degradation, soil infestations, other plant diseases and abnormal climatic events [162, 163]. Cereals are critical for feeding world populations, contributing decisively to the global food security in the last half century. Understanding cereal crop system dynamics is critical, and the rhizosphere soil and its microbial communities may be the easiest way to tackle such challenges by monitoring, preserving and adding beneficial organisms and enhancing plant protective interactions.
Wheat and other cereal seeds are rich in microorganisms that may work as a protective layer during the early stages of plant development, but soil bacteria have also been reported to act as adjuvants in these stages, namely in infested soils. Burkholderia and Caulobacter can be acquired from the rhizosphere soil early on and help on plant protection, as these bacteria are rarely found in the seed microbiome but commonly found in the root microbiome during the first weeks of plant growth [11, 13, 133, 164]. Some of these helpful bacteria are also capable of promoting plant health in soils with early disease pressure by priming the plant defence response [165]. The proliferation of beneficial microbes at the right stage of the any cereal crop, both in the plant and rhizosphere soil, may represent a highly efficient barrier against the spread of fungal infestations, keeping the plant and the biodiversity of the soils stable [11, 43, 166]. Such organisms can also be used to punctually correct degraded soils. The addition of selective treatments to seeds, either as inoculants or components that promote the growth of specific microbes, can also improve the productivity of crops in several consecutive years [167]. The integration of microbiome function using machine learning algorithms into precision farming strategies has the potential to revolutionise cereal cropping practices [168]. The identification of taxa within communities that impact the phenotype of the host is very important and may enable the prediction of plant traits from microbiome data [169, 170]. Conversely, selecting the required microbial inoculants to correctly manage soil infestations and soil degradation, and quickly respond to plant needs, will become routine. Defined predictive models have the potential to improve cereal cropping and productivity based on data from specific soils and farming practices. Such information can be valuable to attenuate harmful effects on crops by naturally increasing microbial diversity and functions that add sustainability, predictability and productivity to the cereal crop system.
Microbial populations present in the soil are relevant to completely understand the interaction between plant and rhizosphere. Revealing the ability of microbes to provide essential nutrients to plants is an important goal of rhizosphere plant–microbe studies. Nevertheless, it is also relevant to monitor all the interactions between soil and plants across space and time. Some microbes “communicate” more efficiently with certain plants than others and are better integrated within the plant or the rhizosphere environments. It is well known that some strains are more advantageous to plants than others, even comparing closely related strains. The description of PGPB strains that add value to relevant crops is fundamental to the development of sustainable alternatives of microbial communities in the soils, especially in degraded soils or soils affected by devastating diseases. Better understanding of soil-related microbial communities and processes can help to better manage our ecosystem and subsequently improve plant productivity, environmental sustainability, human health, food security and mineral wealth.
We are entering the era of biological sustainability strategies, and it is possible to act at multiple levels to reduce and limit the consequences of productivity loss and yield reduction. By promoting, monitoring and controlling the plant microbiome and understanding the role of certain biocontrol agents within the plant at each period of development, we may substantially improve nutritional and environmental standards. Then it can be attention can be given to developing breeding programmes to select genotypes that favour more beneficial interactions and higher productivity levels. Climate change may also play a decisive role in the near future, as certain soil populations may drive considerable consequences for ecosystem-scale carbon cycling [61]. It is increasingly clearer that any loss or gain in microbial biodiversity as a consequence of environmental changes on climate, land use or nutrient enrichment alter the capacity of microbes to sustain ecosystem functions. Therefore, correct management of microbial populations may be the only solution for appreciating soils exposed to extreme conditions and global changes during the next century.
PGPB, plant growth promoting bacteria.
RA wrote, read and approved the final manuscript.
Not applicable.
The author thanks to Steven Myers for the revision of this manuscript.
RA was supported by Individual Call to Scientific Employment Stimulus—Second Edition (grant number CEECIND/01070/2018).
The author declares no conflict of interest.

