1 Department of Infectious Diseases, The First Affiliated Hospital of Anhui Medical University, 230022 Hefei, Anhui, China
2 Department of Gynecologic Oncology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, 100021 Beijing, China
†These authors contributed equally.
Academic Editor: Cheng-Maw Ho
Abstract
Background: Ubiquitination is one of the most common post-translational modifications in cells and dysregulation is closely associated with the development of cancer. However, a comprehensive analysis of the role of ubiquitination in hepatocellular carcinoma (HCC) is still lacking. In this study we analyzed expression and prognostic value of Ubiquitin-Specific Proteases (USPs) in HCC, and the immunological role of USP36 in HCC. Methods: Expression data, prognostic data, and DNA methylation data in cases of HCC were obtained from the cancer genome atlas (TCGA). Overexpression of USP36 in HCC was confirmed in the gene expression omnibus (GEO) database and verified by quantitative PCR in 10 pairs of HCC samples. ULCAN was used to analyze the correlation between USP36 and clinicopathological features. TIMER2.0 and DriverDBv3 were used to analyze the USP36 mutational profile. GSEA analysis explored the potential signaling pathways of USP36 affecting HCC. The immune and stromal scores of HCC samples were calculated using the ESTIMATE algorithm. TIMER1.0 was used to explore the correlation between USP36 and immune cell infiltration. Finally, we analyzed the correlation of USP36 expression with immune checkpoint molecules and determined the IC50 values of 6 chemotherapeutic drugs using the pRRophetic software package. Results: Most USPs are abnormally expressed in HCC, among which USP36 and USP39 are most closely associated with HCC prognosis. We also found that USP36 is associated with TP53 mutational status. GSEA analysis indicated that USP36 may affect HCC progression through the dysregulation of various pathways such as ubiquitin-mediated proteolysis. USP36 expression positively correlated with both macrophage infiltration levels and multiple immune checkpoint molecules. Finally, chemosensitivity analysis indicated that chemosensitivity was lower in cells within the USP36 high expression group. Conclusions: Most USPs are abnormally expressed in HCC. Overexpression of USP36 in HCC is closely related to poor prognosis. In particular, the unique immunological role of USP36 may have potential clinical application value.
Keywords
- Ubiquitin-Specific Proteases (USPs)
- USP36
- HCC
- immune
- bioinformatics analysis
Primary liver cancer (PLC) displays high morbidity and mortality in patients with this disease. The latest cancer statistics report shows that 905,677 new cases are diagnosed, and 830,180 patients die from this disease, every year. These figures place PLC as the sixth highest incidence and third highest mortality rate among all cancers [1]. The 3-year survival rate of patients diagnosed with primary liver cancer is approximately 50% [2]. Hepatocellular carcinoma (HCC) is the major form of PLC observed clinically. In early stages, curative treatments for HCC, such as tumor resection, ablation, and liver transplantation can be used for treatment of this disease [3]. However, a large proportion of HCC patients initially present with advanced disease and thus have limited therapeutic options. Systemic drug therapy is one of the approaches to treat advanced HCC, but first-line treatments for advanced HCC, such as sorafenib or lenvatinib, only extend survival by approximately 3 months [4]. Despite tremendous advances in treatment in recent years, the overall prognosis of liver cancer remains poor. Therefore, it is crucial to explore novel biomarkers with prognostic value for prognosis HCC patients.
Ubiquitination is an important post-translational modification that involves the coordinated activity of ubiquitin activating enzymes (termed E1), ubiquitin conjugating enzymes (E2), and ubiquitin ligases (E3). These molecules are responsible for covalently conjugating single or multiple ubiquitin molecules to target proteins [5]. Ubiquitination alters important properties of target proteins, including target protein activity, cellular localization, half-life, receptor endocytosis, and protein-protein interactions [5, 6, 7]. Similar to other post-translational modifications, ubiquitination is a reversible process that is dynamically regulated by ubiquitinases and deubiquitinases (DUBs). Thus, dysregulation of the ubiquitination process can lead to impairment of many important cellular processes and contribute to serious diseases, including cancer [8].
DUBs cleave ubiquitin chains from protein substrates, and owing to structural
differences within their catalytic domains, DUBs can be divided into 5 families:
Ubiquitin C-Terminal Hydrolases (UCHs), Ubiquitin-Specific Proteases (USPs),
Machado-Joseph Disease Protein Domain Proteases (MJDs), Ovarian Tumor Proteases
(OTUs), and JAMM Motif Proteases [9]. Among these, USPs is the largest family and
has the most diverse structures. Moreover, USPs also belong to the cysteine
protease family [9]. Growing evidence suggests that DUBs act as important
regulators of immune response, making the development of new drugs targeting USPs
a promising therapeutic avenue for the treatment of cancer [10]. Loosdregt
et al. [11] found that USP7 can control Treg cell number and
function by regulating Foxp3 expression in this type of T lymphocyte. Liu
et al. [12] found that USP18 plays a key role in T cell
activation and Th17 generation, and further affects NF-
RNA-seq data, clinical information, and DNA 450k methylation data obtained from HCC tumors were downloaded from The Cancer Genome Atlas (TCGA, https://tcga-data.nci.nih.gov/tcga/), and missing data were removed from the data set. In addition, RNA-seq data of HCC patients were downloaded from three data sets (GSE14520, GSE36376 and GSE64041) in the public database Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/).
Tumor and adjacent normal tissues from 10 primary HCC patients the First Affiliated Hospital of Anhui Medical University. Ethical approval was obtained from the Ethics Committee of the First Affiliated Hospital of Anhui Medical University (No. Quick-PJ2020-16-23). All patients provided written informed consent and complied with the Declaration of Helsinki.
Total RNA was extracted from frozen tissue using an RNA extraction kit (ShangHai
YiShan Biotechnology), and cDNA was synthesized and amplified using
PrimeScript™ RT Master Mix (Takara Bio). USP36 mRNA was quantified
by TB-Green qPCR (Takara Bio) and normalized to GAPDH. The USP36 primer
sequence used in this study is: Forward: 5
Using the Limma package, differential gene expression analysis was performed to determine if USPs expression differed between HCC and normal tissue groups. In this study, we initially focused on 48 USPs (USP1-USP48). In this group, we found that 4 USPs members (USP9, USP17, USP23 and USP27) were missing in the database, and the average expression of 5 USPs members (USP6, USP26, USP29, USP41 and USP44) was lower than 0.2. As a result of these initial efforts, we analyzed the differential expression of 39 USPs. Survival analysis was conducted to determine their relative prognostic value using Kaplan-Meier analysis. Prognostic endpoints included overall survival (OS), disease-free survival (DFS), disease-specific survival (DSS), and progression free survival (PFS).
UALCAN (http://ualcan.path.uab.edu/analysis.html) is a portal for
straightforward exploration, analysis, and visualization of data derived from
TCGA [14]. We used UALCAN to explore the expression of USP36 in
different genders, ages, lymph node metastasis, tumor grade and stage, and
TP53 mutation status. Significant differences in gene expression levels
between groups were calculated by t-test. p
We explored USP36 gene mutation spectrum using TIMER2.0
(http://timer.comp-genomics.org/) and DriverDBv3
(http://driverdb.tms.cmu.edu.tw/). These analysis packages can be obtained at no
charge, and provide the mutation modules of genes [15, 16]. Using the DNA 450k
methylation data of liver cancer samples downloaded from the TCGA database, the
degree of methylation was determined by calculating the
The potential USP36 signal transduction pathways that affect HCC progression were analyzed using gene set enrichment analysis GSEA (version 4.2.3). The 5 pathways most significantly enriched in the high-expression group and the low-expression group are shown.
We used the HCC gene expression signature to first infer the proportion of stromal and immune cells in HCC tumor samples using the ESTIMATE algorithm [17], and predicted the Immunescore and Stromalscore in the tumor tissue. In addition, we also used TIMER 1.0 (https://cistrome.shinyapps.io/timer/) to explore a potential correlation between USP36 expression and immune cell infiltration. TIMER 1.0 is an open platform that can be used to explore tumor immune interactions [18]. Due to a unique correlation between TP53 and USP36, we also investigated immune cell infiltration in HCC tumors with TP53 mutation.
We calculated the 50% inhibitory concentration (IC50) of 6 commonly used chemotherapeutic drugs using the pRRophetic package to evaluate the sensitivity of HCC samples to chemotherapeutic drugs. Wilcoxon signed-rank test was used to compare the difference in IC50 between high and low expression groups.
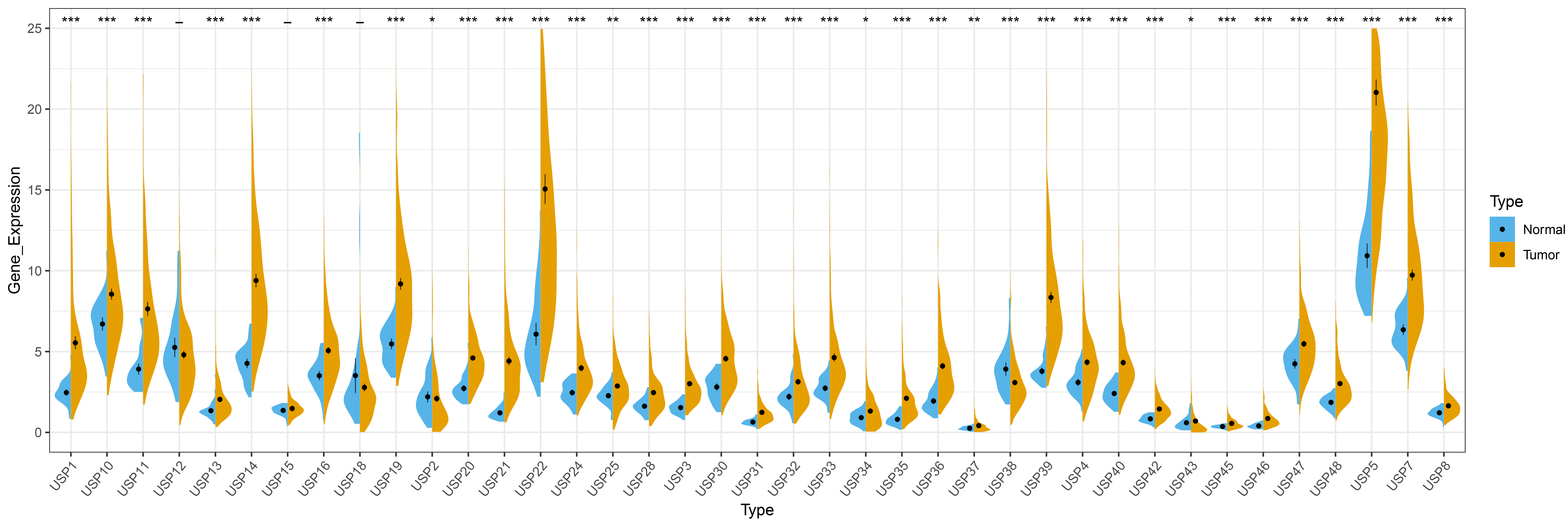
Among the 39 USPs family members, except for USP12, USP15 and USP18, the remaining 36 USPs are abnormally expressed in HCC. Specifically, 34 USPs (USP1, USP3, USP4, USP5, USP7, USP8, USP10, USP11, USP13, USP14, USP16, USP19, USP20, USP21, USP22, USP24, USP25, USP28, USP30, USP31, USP32, USP33, USP34, USP35, USP36, USP37, USP39, USP40, USP42, USP43, USP45, USP46, USP47 and USP48) are highly expressed in HCC, and 2 USPs (USP2 and USP38) display reduced expression in HCC, results of this analysis are shown in Fig. 1.
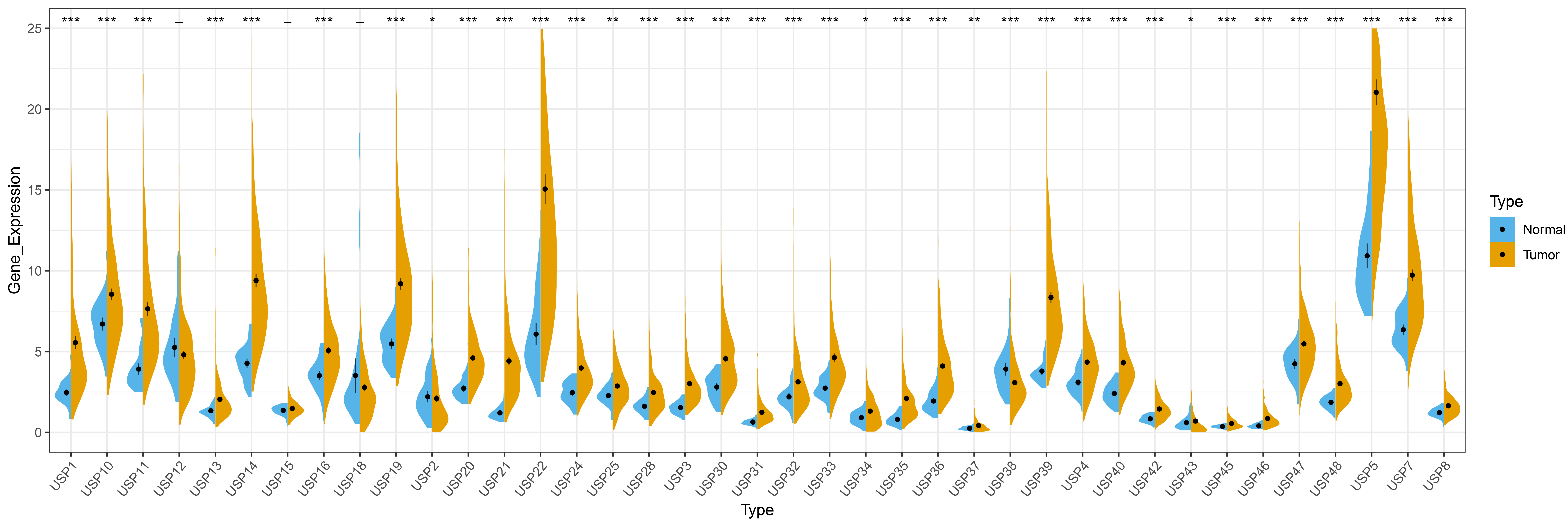 Fig. 1.
Fig. 1.Differential expression of USPs members in HCC. “*” represents
p
We also analyzed the relationship between abnormally expressed USPs and HCC patient prognosis. According to median expression of these USPs, HCC patients were divided into a high expression group and a low expression group, and Kaplan-Meier survival analysis was used to analyze survival among HCC patients in these two groups. The results showed that only 10 of 35 abnormally expressed USPs, specifically USP1, USP11, USP13, USP14, USP21, USP24, USP36, USP39, USP46 and USP48, affected the overall survival (OS) of HCC patients with high expression of these USPs indicating poorer OS (Fig. 2A).
 Fig. 2.
Fig. 2.Kaplan-Meier plot for prognostic analysis of USP36 and USP39 in HCC. OS (A), DFS (B), DSS (C) and PFS (D) of HCC patients grouped by median expression of USPs members. OS, overall survival; DFS, disease-free survival; DSS, disease-specific survival; PFS, progression free survival.
In addition to OS, we also explored other important prognostic indicators such as disease-free survival (DFS), disease-specific survival (DSS), and progression-free survival (PFS). The results showed that expression of four USPs (USP19, USP36, USP39 and USP44) affect HCC patient DFS. Except for high expression of USP44 which correlated with favorable DFS, high expression of the remaining three USPs was associated with poorer DFS (Fig. 2B). Expression of seven USPs, specifically USP1, USP13, USP21, USP36, USP39, USP42 and USP48, is correlated with altered HCC patient DSS. This analysis indicated that high expression of all of these USPs indicate poorer HCC patient DSS (Fig. 2C). Similarly, seven USPs, USP1, USP3, USP13, USP14, USP21, USP24, USP32, USP36, USP39, USP42 and USP48, affected HCC patient PFS. Specifically, high expression of all of these USPs correlated with poorer PFS (Fig. 2D). Further Kaplan-Meier analysis of expression of other USPs in HCC patients was conducted (Supplementary Fig. 1).
These analyses lead us to conclude that USPs expression is commonly closely related to HCC patient prognosis, and USP36 and USP39 can simultaneously affect OS, DFS, DSS and PFS in these patients.
Several studies have suggested an important role for USP39 in HCC [19, 20, 21], but USP36 remains largely unexplored in this tumor type. It has been reported that USP36 participates in the progression of non-small cell lung cancer and ovarian cancer [22, 23]. Importantly, USP36 can increase c-myc stability by altering ubiquitination of this important oncogenic transcription factor [24]. A prominent pro-tumorigenic effect of c-myc in HCC is supported by observations that overexpression of c-myc in mouse liver drives induction of HCC [25]. Therefore, we speculated that USP36 has strong hepatocarcinogenic potential and, given the unique expression pattern of USP36 within HCC tumors as well as its biological effects on c-myc levels, we selected USP36 for further investigation. We verified the expression of USP36 in 3 GEO datasets (GSE14520, GSE36376 and GSE64041) and by qRT-PCR in 10 matched pairs of HCC tumors and adjacent pathologically normal tissues. The results indicated that expression of USP36 was consistent with the results obtained from our analysis of the TCGA database (Supplementary Fig. 2).
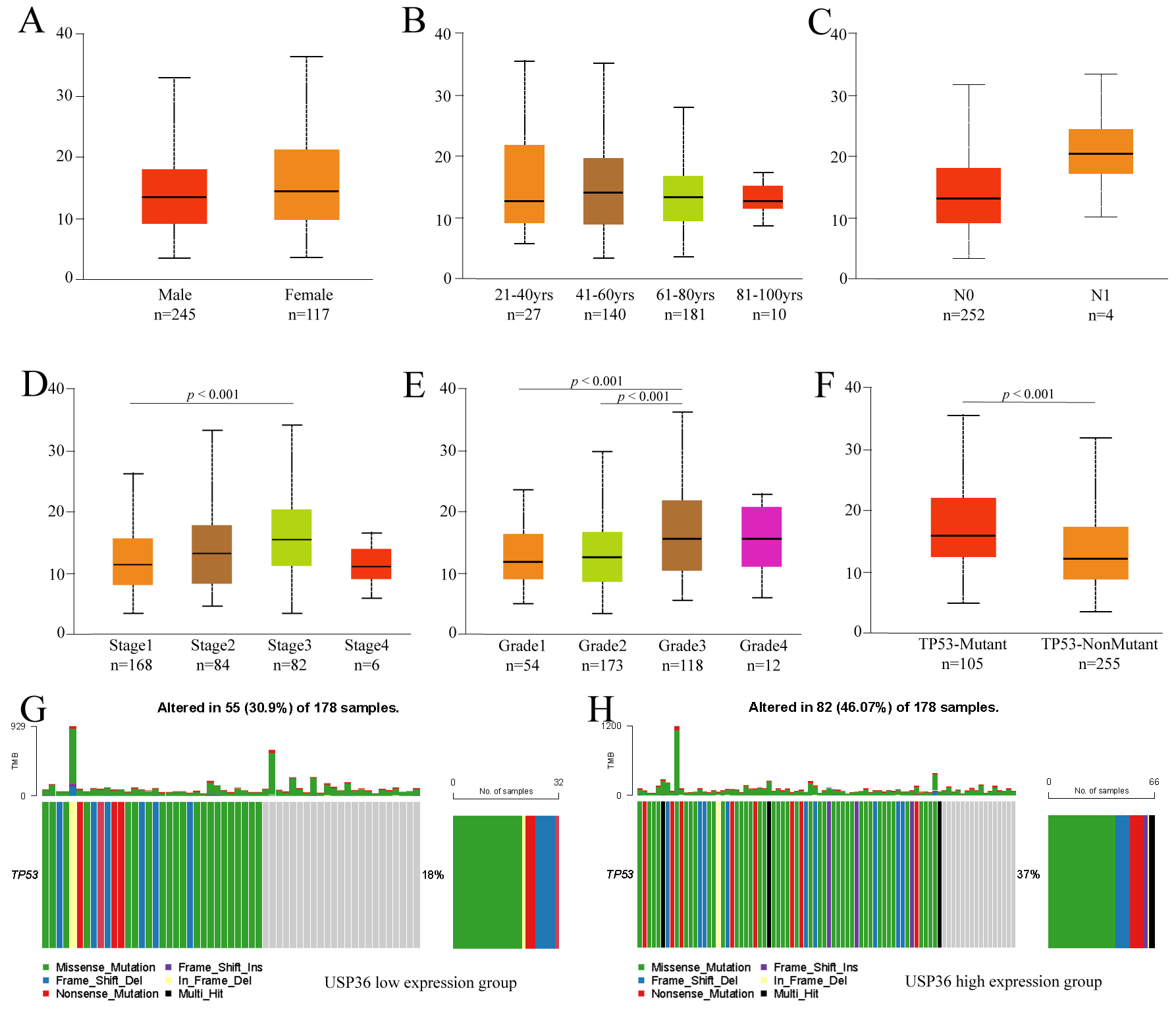
To further explore the clinicopathological parameters of USP36 in HCC,
we performed subgroup analysis using UALCAN. This analysis uncovered no
differences in USP36 expression when patient data was grouped by gender,
age, or lymph node metastasis (Fig. 3A–C). The results did show, however, that
USP36 was highly expressed in Stage3 when compared to Stage1 HCC tumors
(p
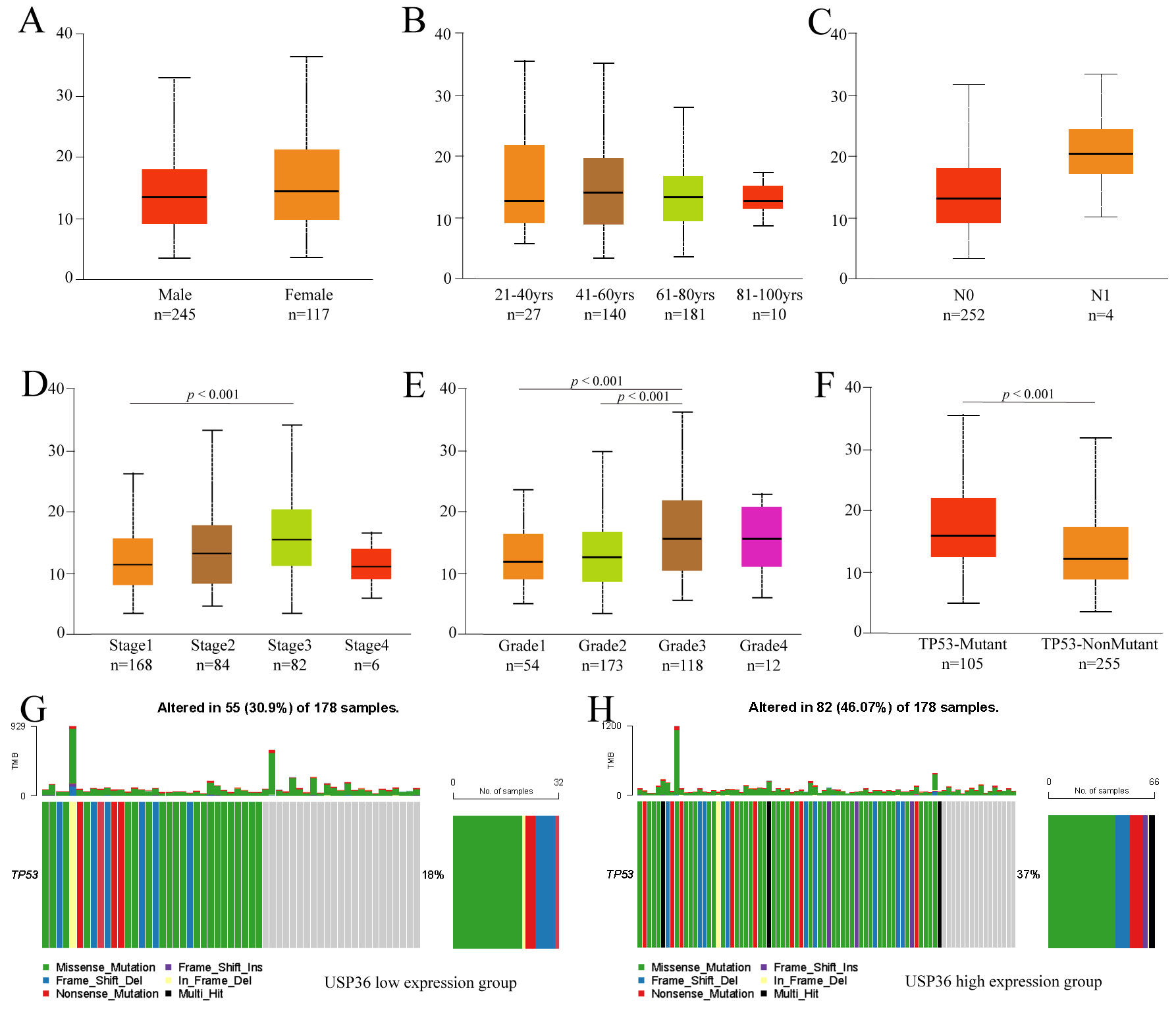 Fig. 3.
Fig. 3.Subgroup analysis of clinicopathological features of USP36 in HCC. Expression of USP36 in different gender (A), age(B), lymph node metastasis status (C), tumor stage (D), tumor grade (E) and TP53 mutation status (F) grouping. TP53 mutation in USP36 high expression group (G). TP53 mutation in USP36 low expression group (H).
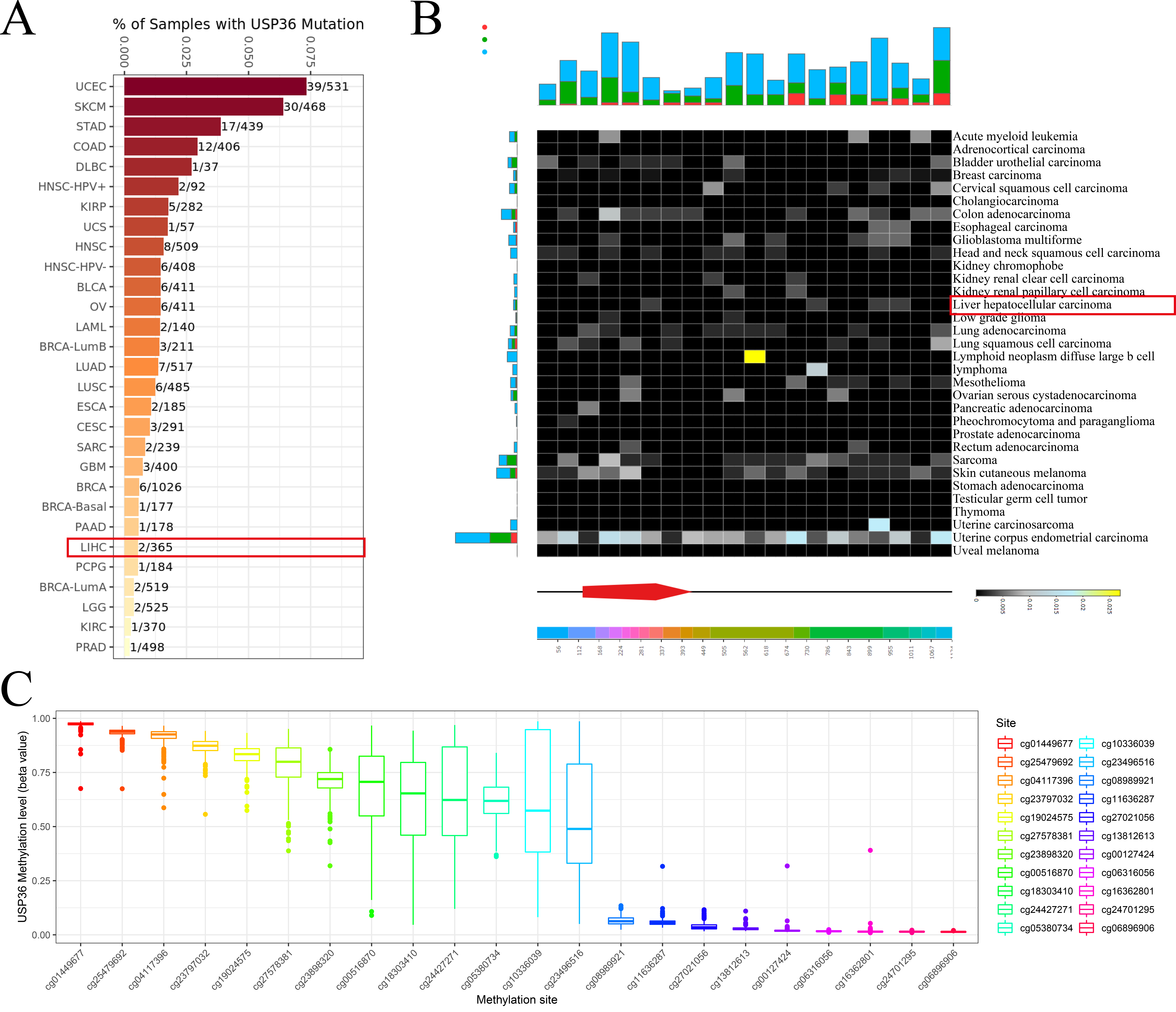
To comprehensively analyze the mutational spectrum of USP36, we explored the mutation status of USP36 in all cancer types in the TCGA database using TIMER2.0 and DriverDBv3. Results obtained using TIMER2.0 showed that USP36 had the highest mutational frequency in uterine corpus endometrial carcinoma (UCEC) (39/531), and the least USP36 mutations were noted in prostate adenocarcinoma (PRAD) (1/498), whereas the mutation frequency of USP36 in HCC was 2/365 (Fig. 4A). The DriverDBv3 database indicated that the mutational rate of USP36 in HCC was 0.003 (Fig. 4B). We next analyzed the DNA methylation level within the USP36 gene in HCC tumors. The results of this analysis showed that the USP36 gene was methylated at multiple CpG dinucleotides (11/22) (Fig. 4C). Detailed methylation beta values for each sample provided in Supplementary Table 1.
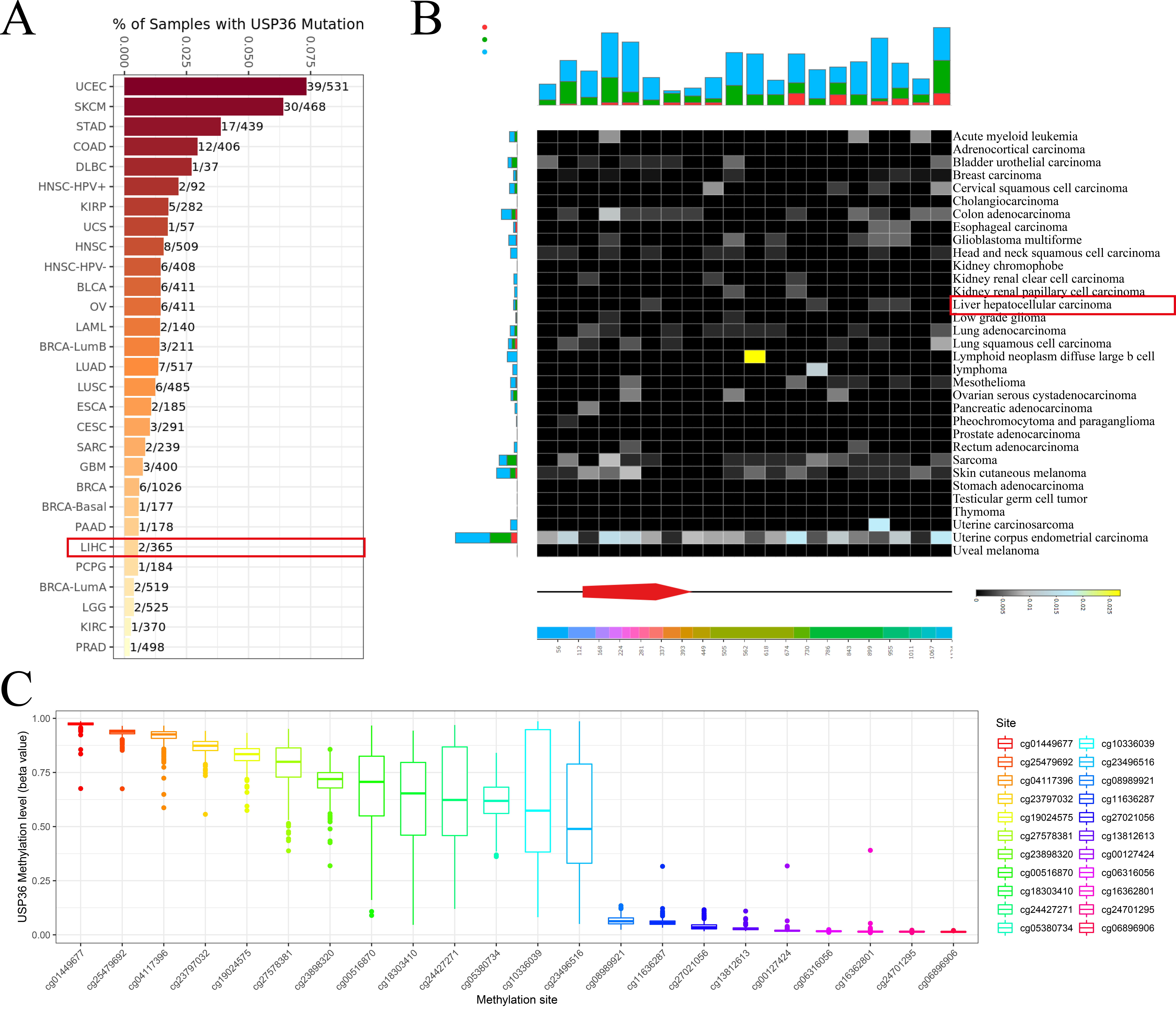 Fig. 4.
Fig. 4.Mutation and methylation analysis of USP36 in HCC. (A) The TIMER 2.0 database shows the USP36 mutation in various tumors. (B) The mutation status of USP36 in various tumors according to the DriverDBv3 database. (C) Methylation analysis of each methylation site of USP36 in HCC.
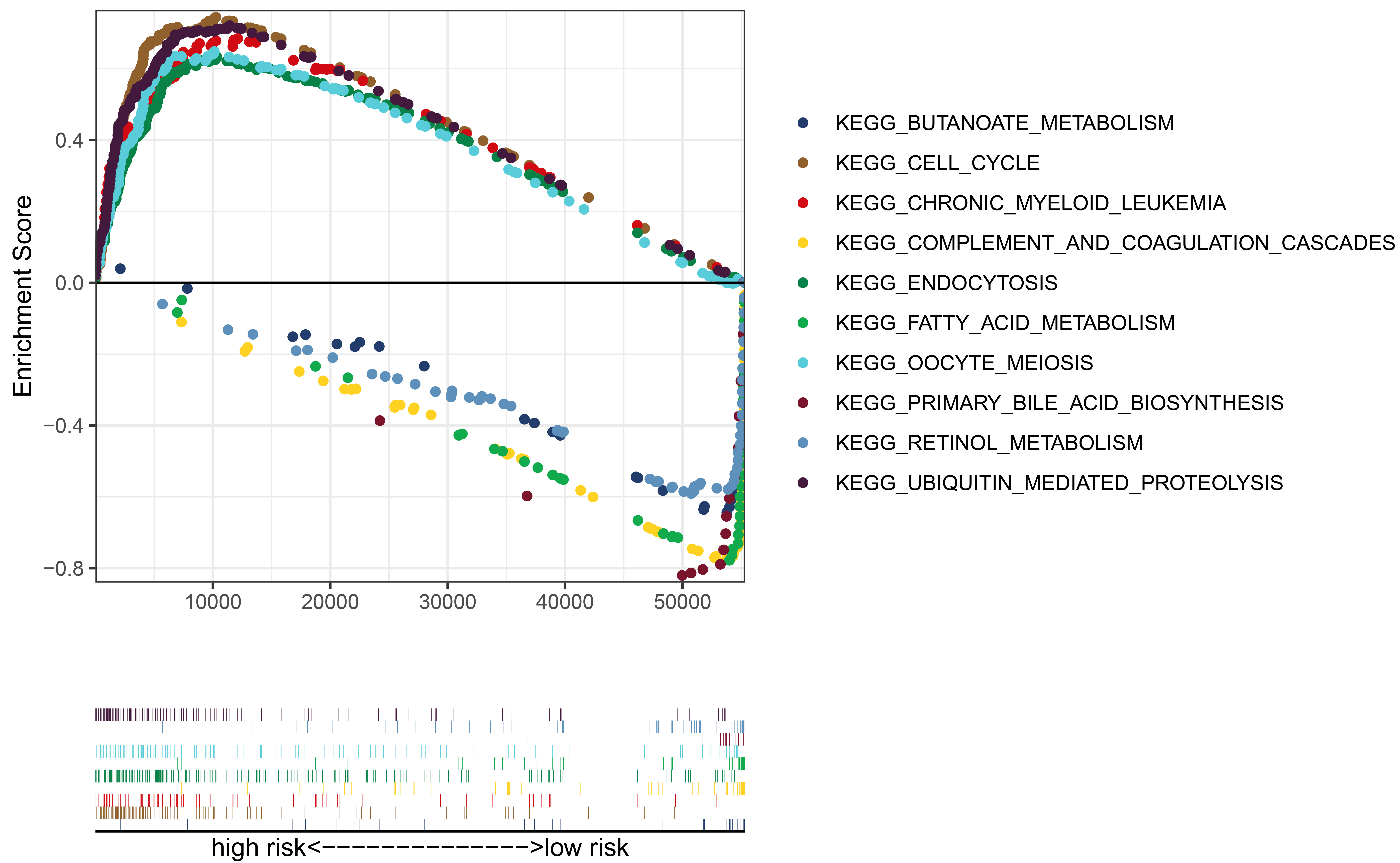
Given the importance of USP36 within the USP family, we next decided to explore potential mechanisms of USP36 dysregulation in HCC. We divided HCC patients into high and low-expression groups according to median USP36 mRNA expression (median: 3.75302) in the HCC cohort in TCGA and subsequently conducted GSEA. The five most significantly affected pathways in tumors with high USP36 expression included KEGG CELL CYCLE, KEGG CHRONIC MYELOID LEUKEMIA, KEGG ENDOCYTOSIS, KEGG OOCYTE MEIOSIS and KEGG UBIQUITIN MEDIATED PROTEOLYSIS. In the USP36 low-expression group, the five most significantly altered pathways included KEGG BUTANOATE METABOLISM, KEGG COMPLEMENT AND COAGULATION CASCADES, KEGG FATTY ACID METABOLISM, KEGG PRIMARY BILE ACID BIOSYNTHESIS and KEGG RETINOL METABOLISM. Results of GSEA are shown in Fig. 5.
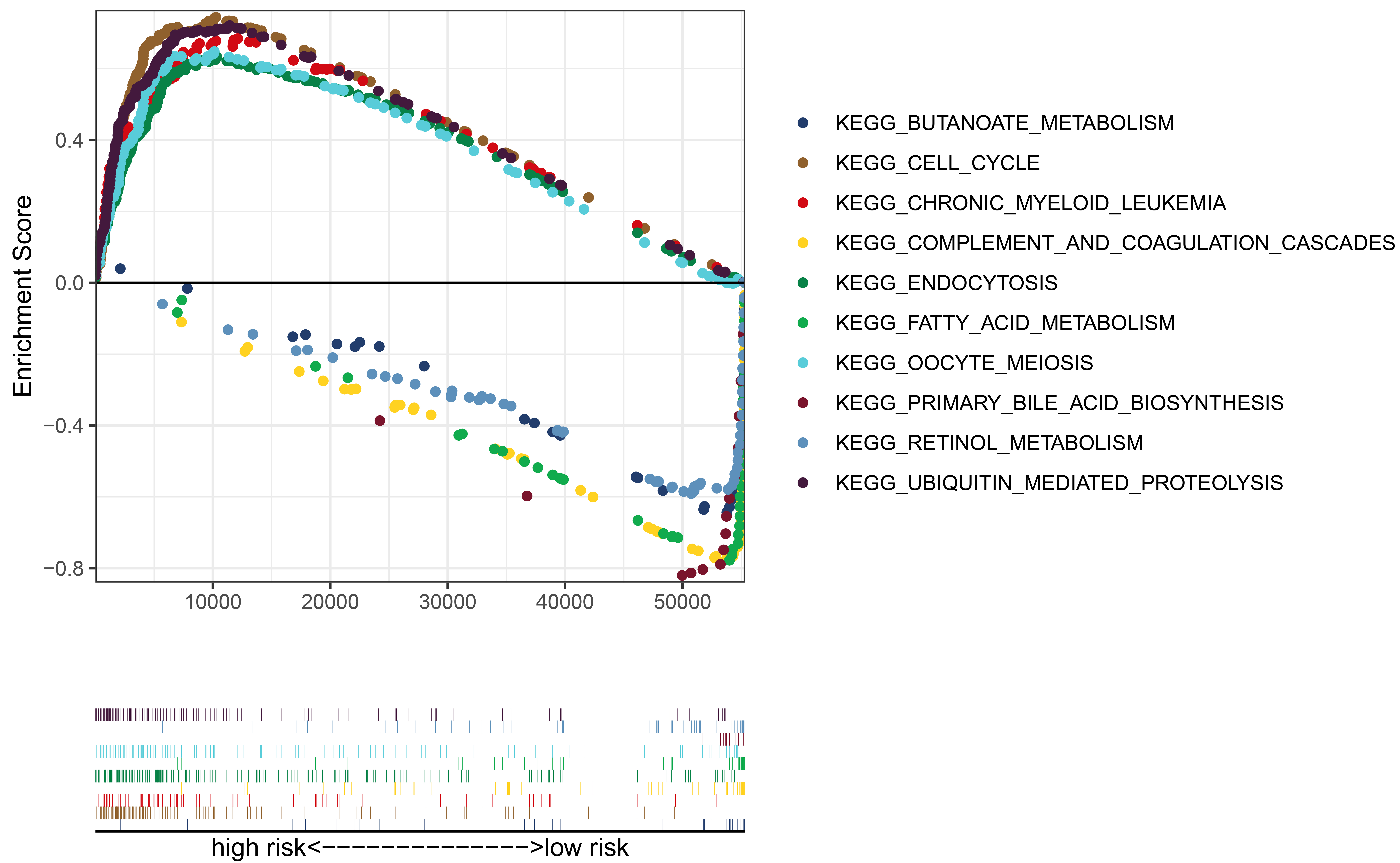 Fig. 5.
Fig. 5.Gene Set Enrichment Analysis of USP36 in HCC. The five pathways most significantly enriched in the USP36 high-expression group and low-expression group.
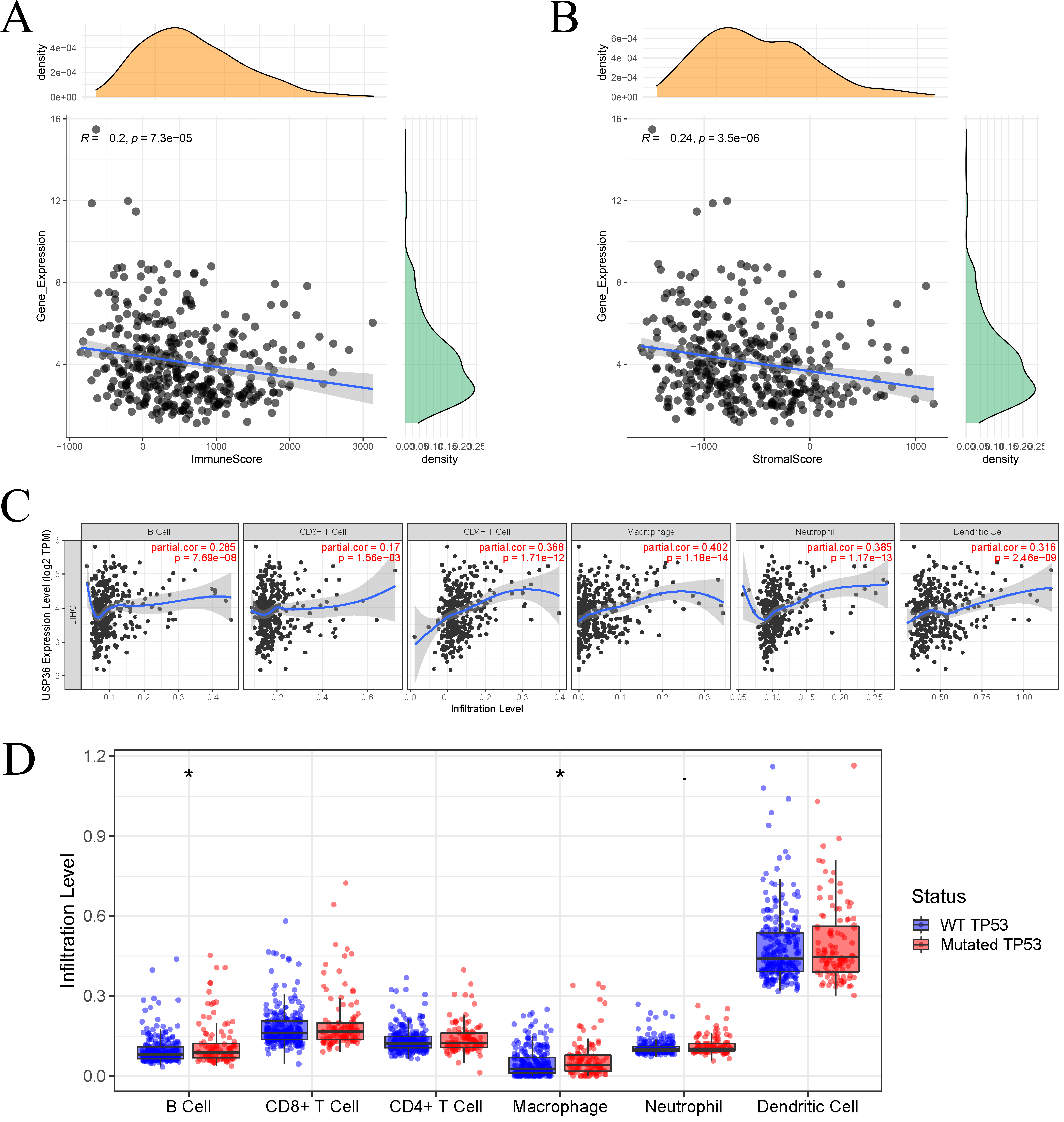
Since ubiquitination and the immune microenvironment are closely related [8, 26], we next explored the relationship between USP36 and the tumor immune microenvironment. As a first step, we analyzed the correlation between USP36 expression and the tumor ESTIMATE score. Results showed a weak negative but statistically significant correlation between USP36expression and Immunescore (Relevance = –0.20, p = 7.3e-05) (Fig. 6A) as well as Stromalscore (Relevance = –0.24, p = 3.5e-06) (Fig. 6B). In addition, we further explored the correlation of USP36 expression with the tumor infiltration of six types of immune cells. The results showed that the expression of USP36 was positively correlated with infiltration of B cells (Cor = 0.285, p = 7.69e-08), CD8 + T cells (Cor = 0.170, p = 1.56e-03), CD4 + T cells (Cor = 0.368, p = 1.71e-12), macrophages (Cor = 0.402, p = 1.18e-14), neutrophils (Cor = 0.385, p = 1.17e-13), and dendritic cells (Cor = 0.316, p = 2.46e-09) (Fig. 6C).
We subsequently examined the relationship between TP53 mutation and
immune cell infiltration in HCC. The results showed that B cells and macrophages
were significantly increased in HCC tumors that contain TP53 mutations
(p
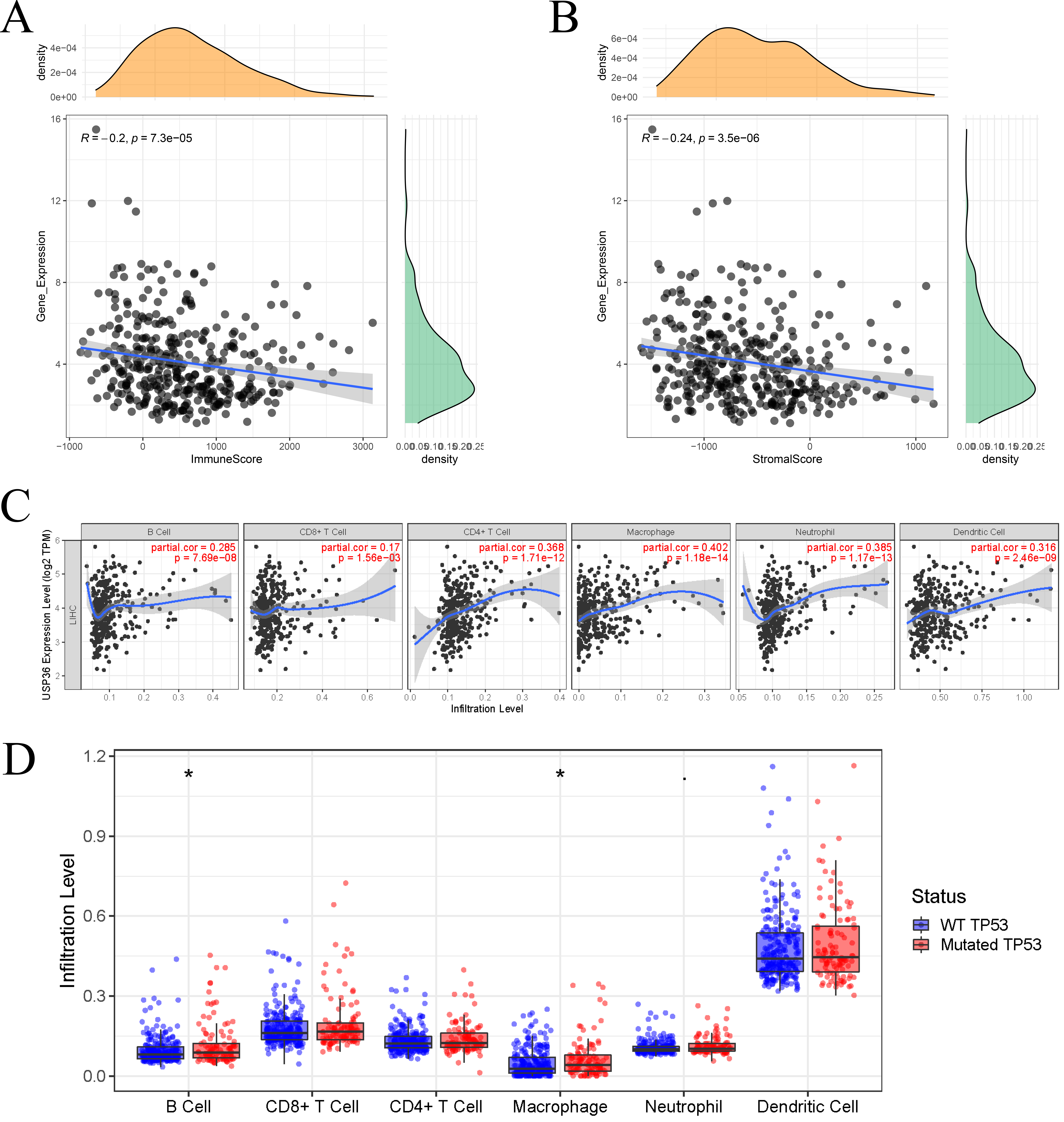 Fig. 6.
Fig. 6.Correlation between USP36 expression and immune microenvironment. The Spearson test analyzed the correlation between USP36 expression and Immunescore (A), Stromalscore (B). Correlation between USP36 expression and infiltration levels of six types of immune cells (C). Levels of immune cell infiltration in different TP53 mutation states (D).
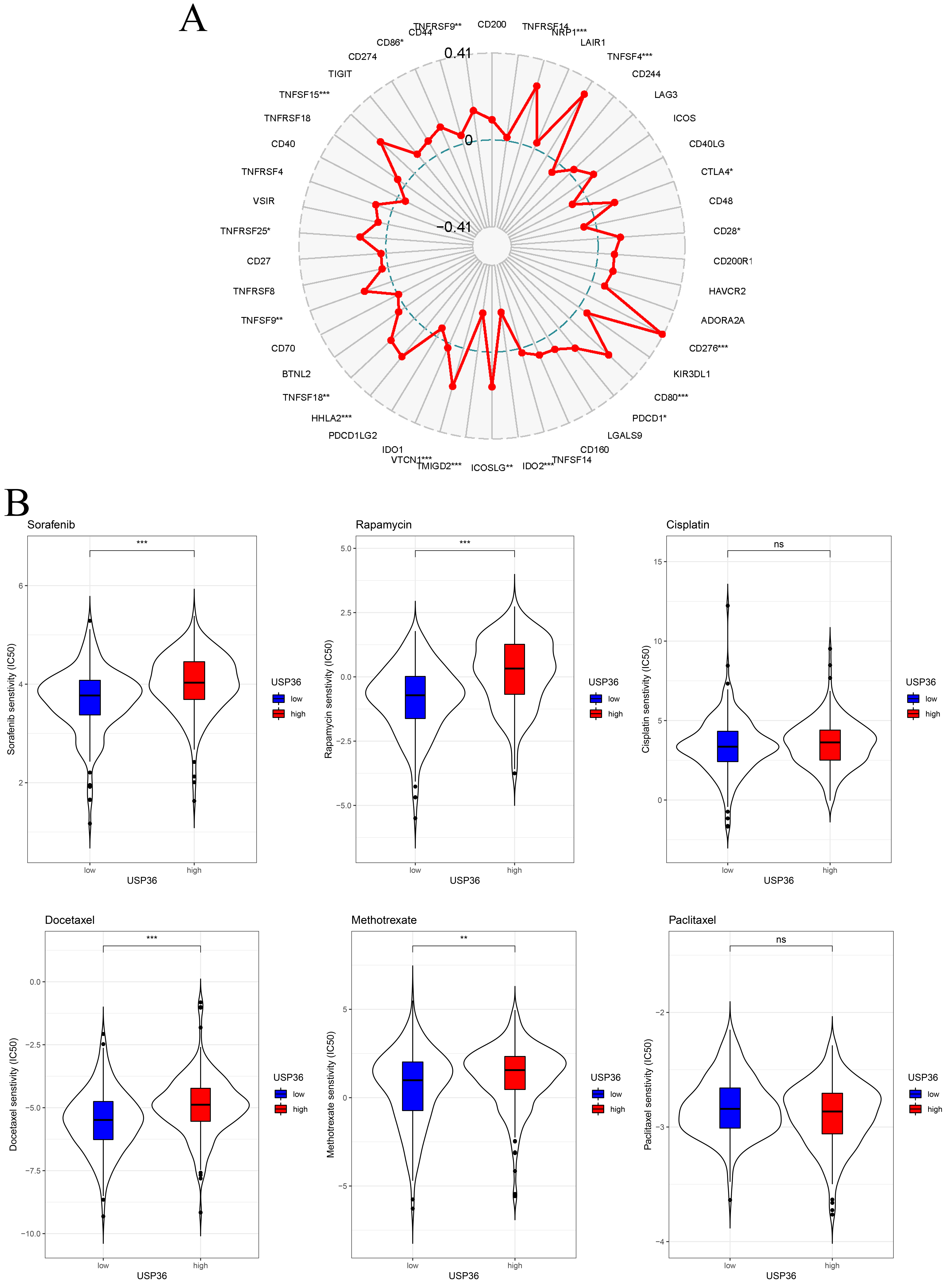
In addition, given the substantial progress in cancer treatment in the resent past, immunotherapy targeting immune checkpoint molecules is a promising therapeutic approach for liver cancer patients. We analyzed the correlation between USP36 expression and 48 immune checkpoint molecules using Spearman correlation analysis. The results showed a positive correlation between USP36 and 16 immune checkpoint molecules, specifically, NRP1, TNFSF4, CTLA4, CD28, CD276, CD80, PDCD1, ICOSLG, TMIGD2, VTCN1, HHLA2, TNFSF18, TNFSF9, TNFSF25, TNFSF15, CD86 and TNFSF9. Of note, 2 immune checkpoint molecules, IDO2 and TMIGD2, were negatively correlated with USP36 expression (Fig. 7A).
Finally, we explored the relationship between USP36 expression and sensitivity to commonly used HCC chemotherapeutics. The results of this analysis showed that high expression of USP36 was associated with higher IC50 concentrations for sorafenib, rapamycin, docetaxel, and methotrexate (Fig. 7B).
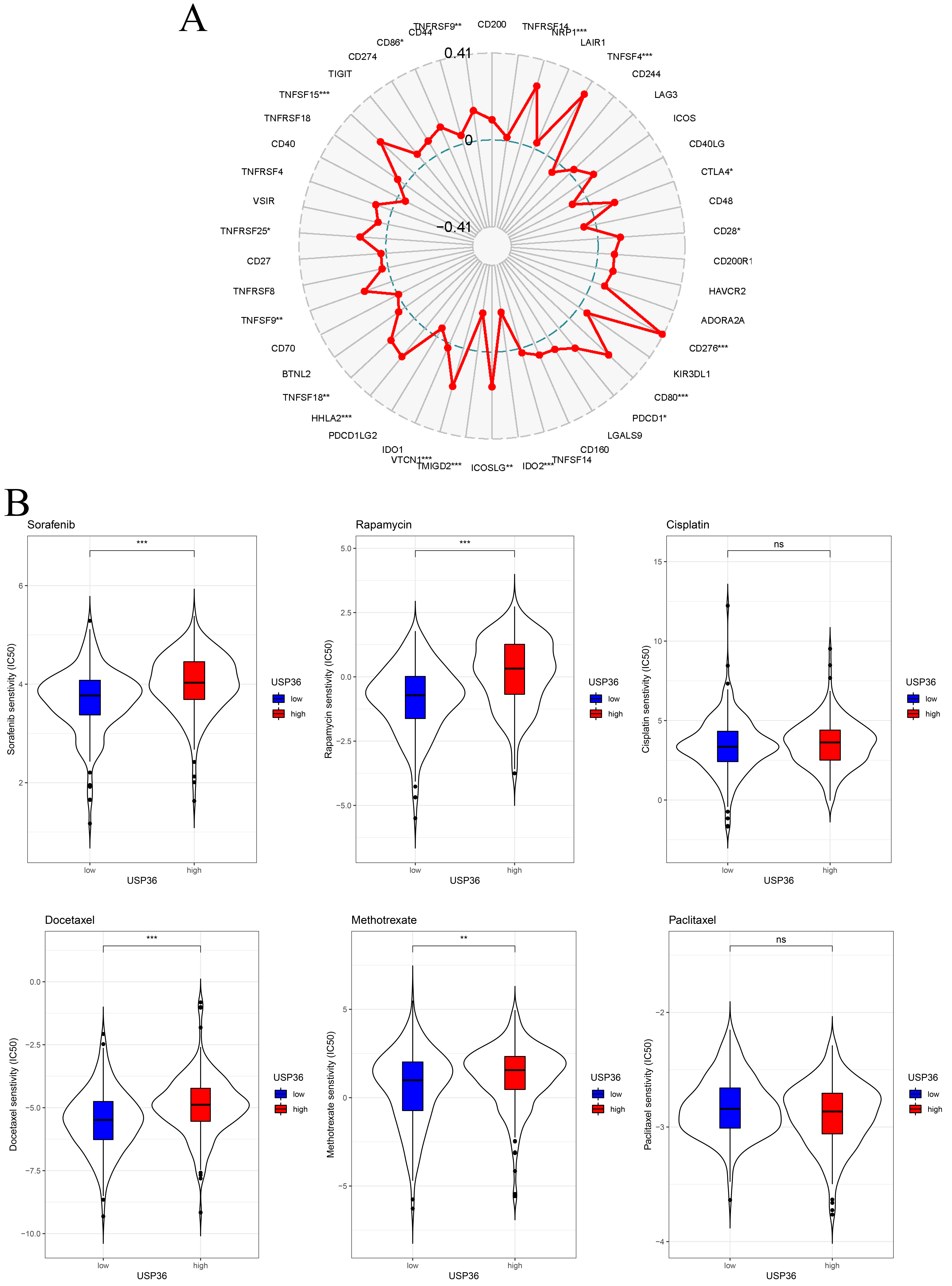 Fig. 7.
Fig. 7.Correlation analysis of USP36 expression with immune
checkpoint molecules and chemotherapeutic drug sensitivity. (A) Spearson test to
analyze the correlation between USP36 expression and 48 immune
checkpoint molecules (B) Difference analysis of the sensitivity of 6
chemotherapeutic drugs in USP36 high expression group and low expression
group. “*” represents p
Ubiquitination is one of the most common and important post-translational protein modifications [27, 28], and the ubiquitin-proteasome system is involved in the degradation of more than 80% of proteins in cells [29]. In addition, ubiquitination is also involved in diverse processes such as regulation of apoptosis, cell cycle progression, endocytosis, and transcription [5, 27]. Ubiquitination and deubiquitination have been observed to be dysregulated in cancer and these processes play multiple roles in cancer-related pathways [30]. Interestingly, ubiquitination is a dynamically reversible process. DUB can effectively downregulate the ubiquitination of key target proteins, and thus further serve to control cell signaling events and cell fate.
At present, a large number of studies have shown that dysregulation of USPs are
common molecular events in a variety of cancer types [31] and several studies
have sought to clarify the role of USPs in liver cancer. For example,
USP7 can stabilize the E3 enzyme TRIP12 and form a complex that
promotes HCC cell growth and is significantly associated with HCC malignant
phenotype [32]. USP22 promotes hypoxia-induced HCC stemness by
regulating HIF-1
Our analysis showed that the vast majority (36/39) of USPs were differentially expressed in HCC, suggesting that aberrant protein ubiquitination is prevalent in HCC. We further analyzed the prognostic value of these abnormally expressed USPs in HCC and found that more than ten USPs were linked to HCC prognosis. Importantly, this analysis indicated that USP36 and USP39 can simultaneously, and independently, affect OS, DFS, DSS and PFS in HCC patients, and support our conclusion that increased expression of these two genes are poor prognostic factors in HCC.
Previous studies explored a potential USP39 mechanism of action in HCC. For example, USP39 and TRIM26 co-regulate ZEB1 ubiquitination which controls HCC progression [19]. USP39 promotes HCC progression by deubiquitinating SP1 protein and stabilizing SP1 [21]. In addition, high expression of USP39 was significantly associated with age, tumor status, advanced pathological stage, T stage and higher histological grade in HCC [34].
Previous studies have shown that USP36 can participate in the progression of a variety of cancer types [22, 23], and USP36 can stabilize the protein stability of the hepatoma promoting protooncogene c-myc through dysregulation of its ubiquitination [24, 25]. However, USP36 has not been studied in HCC. This lead us to explore the clinical significance and potential mechanism of USP36 in HCC. We found that in HCC, USP36 was highly expressed in three GEO databases (GSE14520, GSE36376, GSE64041) and validated this finding in 10 matched HCC tumor/normal tissue pairs. These studies revealed that USP36 mutations are uncommon in HCC samples, but that abnormal gene methylation may be one of the reasons for altered USP36 expression in this tumor type.
We also explored the correlation between USP36 and 6
clinicopathological features. Results obtained showed no significant difference
in USP36 expression linked to different genders, ages, or lymph node
metastasis. However, a significant correlation was noted with tumor stage and
tumor grade as increased USP36 expression is more common in higher tumor
stage and grade in HCC. This finding prompts us to speculate that USP36
may be involved in HCC progression and, more importantly, expression of
USP36 was significantly increased in HCC tumors with TP53
mutation. Previous studies have shown that TP53 is the most frequently
mutated gene in HCC and is associated with poor prognosis [35, 36, 37]. Moreover,
previous studies have explored a potential interaction between USPs and
TP53 gene mutations. For example, the
USP22/HIF1
We also analyzed the potential signaling pathways that are affected by USP36 in HCC by using GSEA. Results indicated that the USP36 high-expression group there was significantl enrichment in KEGG UBIQUITIN MEDIATED PROTEOLYSIS, KEGG ENDOCYTOSIS, and KEGG CELL CYCLE and other ubiquitination-related and cancer-related pathways [39, 40]. Studies have shown that ubiquitination is closely related to the cell cycle and endocytosis and that this dysregulation can contribute to cancer progression [41, 42, 43]. Therefore, we speculate that USP36 may affect HCC progression by affecting certain key cyclin expression or receptor endocytosis by dysregulating deubiquitination.
Tumor cells and immune cells, as well as many other cellular and acellular components, make up the complex tumor microenvironment (TME). The TME is highly heterogeneous and dynamic, and can ultimately impact the course of tumor progression [44]. Many studies have shown that TME plays a significant role in HCC and is a very promising potential therapeutic target [45, 46, 47, 48, 49]. Previous studies have shown that ubiquitination plays a crucial role in various physiological processes, including innate and adaptive immunity [8]; thus, the crosstalk between ubiquitination and TME deserves further exploration. It has been reported that USP36 plays an important role in microbial immunity [50] leading us to explore a potential correlation between USP36 and the TME. We used TIMER1.0 to explore the correlation between USP36 expression and six immune cells found in the HCC TME. Notably, USP36 expression showed the strongest correlation with macrophages in the TME. Previous studies have shown that hepatic macrophages contribute to the formation of HCC by suppressing antitumor immunity [51], and that high macrophage infiltration is generally linked to poor HCC prognosis [52]. The potential role of macrophages in HCC is consistent with our analysis of the possible tumor promoting effect of USP36 in this tumor type, suggesting that USP36 may promote HCC by affecting TME components. In addition, our results suggest that USP36 is positively correlated with 16 immune checkpoint molecules, including PDCD1 and CTLA4. Moreover, the IC50 of chemotherapy drugs such as sorafenib, rapamycin, docetaxel and methotrexate were significantly increased in the USP36 high expression group. In conclusion, these results provide some theoretical basis for the use of USP36 expression to help guide therapeutic regimen choices in HCC patients.
However, we must acknowledge some limitations in the current study. First, although we used the authoritative TCGA database, some members of the USPs were not included in the discussion of this study. Second, most of the results of this study were obtained by bioinformatics methods, and there were no in vitro and in vivo experiments and clinical data to verify. In the future, we will focus on the role of USPs members not included in the discussion in HCC. Third, our analyses are all based on the TCGA database, which may reduce the reliability of our findings.
In conclusion, we comprehensively analyzed the expression and prognostic value of USPs in HCC, and analyzed the clinicopathological features of USP36 as well as the potential mechanism of action in detail. We found that USP36 expression is closely related to the tumor microenvironment, which has potential value in clinical application. Future studies should focus on the oncogenic mechanism of USP36 in HCC as a means to independently validate our findings regarding USP36 in this cancer type.
This study is the first to comprehensively analyze the gene characteristics and prognostic value of USPs members in HCC, and focus on the potential mechanism and immunological role of USP36 in HCC. These findings provide new horizons for future research.
ESTIMATE, Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data; qRT-PCR, Real-Time Quantitative Reverse Transcription PCR.
YG and LL contributed to the study design. YG and WS were responsible for the writing and proofreading of the article. JS is responsible for data analysis and download. JL and KH took care of the experimental part and graphic editing. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Ethical approval was obtained from the Ethics Committee of the First Affiliated Hospital of Anhui Medical University (No.Quick-PJ2020-16-23).
Not applicable.
This research received no external funding.
The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/j.fbl2706190.