1 Institute of Chemical and Biological Technology António Xavier (ITQB NOVA), NOVA University Lisbon, 2780-157 Oeiras, Portugal
2 Associate Laboratory i4HB – Institute for Health and Bioeconomy, NOVA School of Science and Technology, NOVA University Lisbon, 2829-516 Caparica, Portugal
3 UCIBIO – Applied Molecular Biosciences Unit, Chemistry Department, NOVA School of Science and Technology, NOVA University Lisbon, 2829-516 Caparica, Portugal
Academic Editor: Rosa Alduina
Abstract
Extracellular electron transfer is a key metabolic process of many organisms that enables them to exchange electrons with extracellular electron donors/acceptors. The discovery of organisms with these abilities and the understanding of their electron transfer processes has become a priority for the scientific and industrial community, given the growing interest on the use of these organisms in sustainable biotechnological processes. For example, in bioelectrochemical systems electrochemical active organisms can exchange electrons with an electrode, allowing the production of energy and added-value compounds, among other processes. In these systems, electrochemical active organisms exchange electrons with an electrode through direct or indirect mechanisms, using, in most cases, multiheme cytochromes. In numerous electroactive organisms, these proteins form a conductive pathway that allows electrons produced from cellular metabolism to be transferred across the cell surface for the reduction of an electrode, or vice-versa. Here, the mechanisms by which the most promising electroactive bacteria perform extracellular electron transfer will be reviewed, emphasizing the proteins involved in these pathways. The ability of some of the organisms to perform bidirectional electron transfer and the pathways used will also be highlighted.
Keywords
- extracellular electron transfer
- bioelectrochemical systems
- biogeochemical cycling of elements
- electroactive organisms
- reduction potential
- iron
- multiheme cytochromes
Extracellular electron transfer (EET) is the process by which microorganisms sustain their metabolism by capturing or delivering electrons to donors or acceptors, respectively, which do not permeate the cell boundaries, such as metal compounds in their natural environment [1] or electrodes in the case of bioelectrochemical systems (BES) [2]. The thermodynamic driving force for this process, typically defined by the nature, abundance, and bioavailability of the species, determines if the electrons are delivered or collected from the microbial metabolism.
EET is essential for the biogeochemical cycling of elements on earth, given the ability of microbes in changing the redox state of metals, and therefore their bioavailability [3, 4]. For example, microorganisms are able to oxidize and reduce iron. Ferric iron (Fe(III)) is poorly soluble at circumneutral pH, and is mostly found in nature as iron (hydro)oxides, such as hematite and goethite, while ferrous iron (Fe(II)) is more soluble and therefore more available [5]. Fe(II) oxidizing microorganisms can couple the oxidation of iron to intracellular reduction of oxygen, carbon dioxide and nitrate [5], a process that occurs extracellularly to avoid cell encrustation. Such event can be seen in banded iron formations, where photoferrotrophic organisms are thought to be responsible for the massive depositions of iron that predate by billions of years the emergence of oxygenic photosynthesis [6] (Fig. 1a). In the case of iron reduction, Fe(III) reducing organisms are able to couple the reduction of iron with the oxidation of organic or inorganic matter by performing EET [1] (Fig. 1b). This process occurs in almost every anoxic environment on Earth by various microorganisms, including archaea and bacteria [5]. EET is also responsible for the mobility of metals, called leaching, which has been used at industrial scale in biomining operations for the recovery of copper and gold from low grade ores [7]. The opposite effect of biological precipitation and immobilization of metals upon biological EET activity has also been used in bioremediation operations of sites and aquifers contaminated with radioactive metals, including uranium [8] (Fig. 1c).
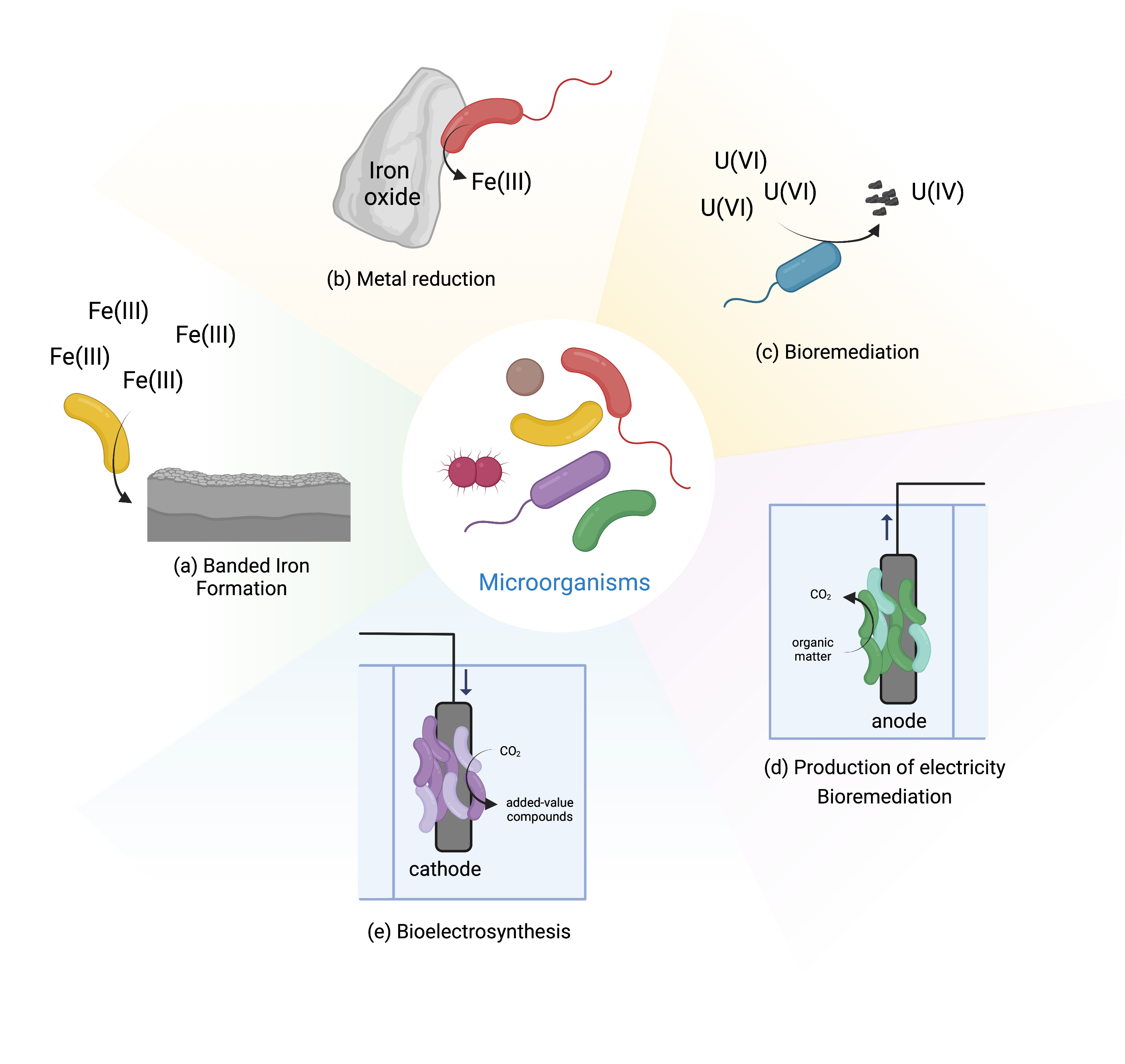 Fig. 1.
Fig. 1.Microbial activities mediated by EET.
In addition to redox active minerals, some microorganisms can also exchange electrons extracellularly with electrodes. These electrochemically active organisms are called electroactive [2]. This has led to the development of BES, that collect the electrons generated in the catabolic metabolism of microorganisms (named electrogens), or that provide electrons for the anabolic metabolism of the microorganisms (named electrotrophs). In BES the microbial metabolism is linked to electrical circuits and can be operated in diverse modes (Fig. 1d and e). For example, in microbial fuel cells, one of the most studied BES, electricity can be generated by the microbial degradation of organic matter of low added value, including wastewater. This system offers a method for wastewater treatment with lower energy demand than traditional methods [9]. Another example is bioelectrosynthesis that has received tremendous attention in recent years [10]. In this case, electricity from the grid is used with cheap substrates to drive the production of added value compounds by the bacteria [11]. Within the last two decades, BES have seen tremendous improvement and developments, and today are being explored for a multitude of other applications, including biosensing, microbial desalination and decontamination (for a review see [10] and [9]).
Many microorganisms are able to transfer electrons extracellularly and electroactive microorganism from different classes (i.e., bacteria, algae and fungi) have been identified to work on BES (for a review see [12]). From these, Gram-negative bacteria constitute the major group of electroactive bacteria and those producing the highest power density in BES described so far [13]. However, recently, there has been great interest in Gram-positive bacteria, given their ability to work better in some extreme conditions (e.g., high temperatures) and by being found in the gut of many animals [14, 15, 16]. The genomes of novel organisms are frequently sequenced [17, 18].
Over the past decade, substantial progress has been made in the understanding of microbial EET mechanisms [12]. However, elucidation of EET processes is not a straightforward process, since it requires a multidisciplinary approach that makes use of different fields of expertise, including genomics, molecular biology, biochemistry, biophysics, electrochemistry and structural biology [19]. It is now well recognized that in some electroactive organisms, multiheme c-type cytochromes (MC) form conductive electron transfer pathways responsible for the exchange of electrons with extracellular electron donors and acceptors, and that several of them play a key role in both direct and indirect electron transfer processes [20, 21]. MC contain several heme cofactors that are covalently bound to the polypeptide chain. This enables a close proximity of the hemes, necessary to modulate the thermodynamic properties of the redox protein and define its functional role [21, 22]. This review will focus on the MC that participate in the EET pathway of electroactive organisms, in particular on those for which a detailed thermodynamic characterization exists. These include MC that participate in the EET pathways of Shewanella oneidensis MR-1, Geobacter sulfurreducens, Rhodopseudomonas palustris TIE-1, Sideroxydans lithotrophicus ES-1, and Gram-positive bacteria that belong to the Thermincola sp. While S. oneidensis MR-1, G. sulfurreducens and Thermincola spp. perform EET to reduce extracellular electron acceptors, R. palustris TIE-1 and S. lithotrophicus ES-1 are recognized to oxidize extracellular electron donors. A critical overview on the properties of these proteins will be given, to start unravelling molecular factors that may be limiting electron transfer rates to and from electrodes.
S. oneidensis MR-1 is a facultative anaerobe that proliferates mainly in aquatic environments [23]. Under anoxic conditions this bacterium can utilize a variety of terminal electron acceptors, including metals, either as soluble complexes or solid mineral (hydr)oxides [24]. The anaerobic bioenergetic metabolism of this Gram-negative bacterium uses organic compounds more oxidized than acetate to reduce the quinone pool in the inner-membrane. This is composed by one or more menaquinones, and their derivatives, which participate in anaerobic metabolism, or ubiquinones that participate in aerobic metabolism. Disruption of the menaquinone biosynthesis was shown to prevent EET [25]. Several MC were shown to contribute to EET (Fig. 2).
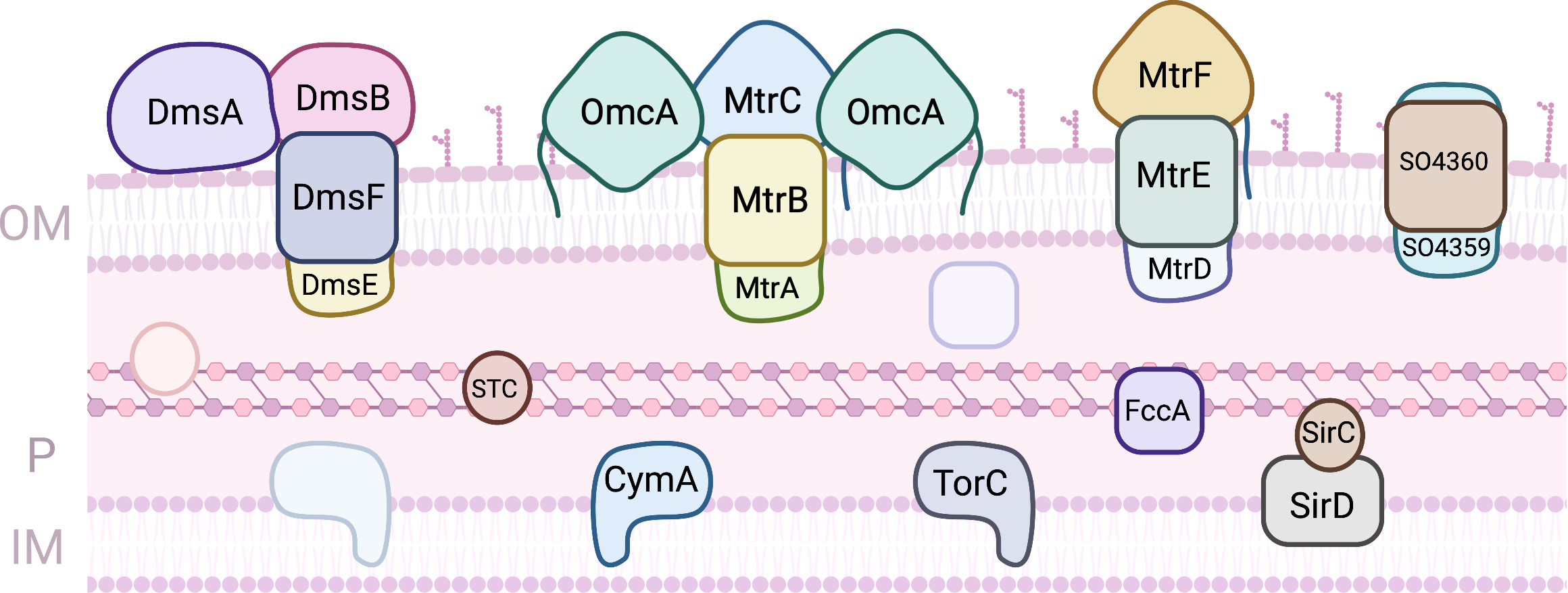 Fig. 2.
Fig. 2.Proteins involved in EET processes of S. oneidensis MR-1. (OM, outer-membrane; IM, Inner-membrane; P, periplasm).
On the periplasmic face of this membrane, three quinol oxidoreductases that collect the electrons and distribute them onwards have been identified: TorC, SirD and CymA. TorC is a pentaheme cytochrome that is encoded by the torECAD operon. It contains one transmembrane alfa helix that serves as anchor to the protein on the periplasmic side. The proteins encoded by the tor operon perform the dissimilatory reduction of trimethylamine N-oxide (TMAO), contributing to the respiratory versatility of Shewanella [26]. SirD is encoded within a 10 gene operon that contains an octaheme cytochrome c sulfite reductase. Together with the periplasmic iron sulfur protein SirC these two proteins can functionally replace cymA deletion mutants in the reduction of several anaerobic terminal electron acceptors, including extracellular electron acceptors [27]. CymA is by far the most studied of the three quinol oxidoreductases, but despite numerous efforts no molecular structure has been obtained thus far. Like TorC it has one transmembrane helix that serves to anchor the protein to the inner-membrane and a heme domain exposed to the periplasmic space. CymA is a tetraheme cytochrome c where three of the hemes have bis-histidine axial coordination of the iron and one of the hemes has an open coordination position, being proposed to be the site of quinol oxidation. The hemes have all negative potentials and CymA is redox active in a broad window of potentials that spans 250 mV centered at –200 mV (vs Standard Hydrogen Electrode - SHE) at pH 7, and that shows substantial pH dependence [28]. CymA shows little selectivity with respect to downstream redox partners, and contributes to several respiratory pathways, leading to the reduction of metallic minerals, nitrate, fumarate, dimethylsulfoxide (DMSO), and the delivery of electrons to electrodes in BES [29, 30]. This low selectivity goes hand-in-hand with low affinity, with dissociation constants with downstream redox partners in the range of hundreds of micromolar [31, 32, 33]. This ensures that the protein is predominantly free to donate electrons to different downstream partners.
The periplasmic space of S. oneidensis MR-1 is filled with cytochromes, with the small tetraheme cytochrome (STC) and FccA being the most abundant MC that participate in EET. The STC is a tetraheme cytochrome c of 12 kDa where the four hemes present bis-histidine axial coordination. It is one of the major players in the conduction of electrons across the periplasmic space to reach redox complexes at the outer-membrane, responsible for the reduction of extracellular terminal acceptors. Despite its small size, STC displays specificity in the way that interacts with CymA upstream and with different downstream partners [31, 32]. This shows that this protein engages in specific recognition and docking for electron transfer across the periplasmic space. The reduction potentials of the individual hemes were discriminated and surprisingly, the STC in different organisms show different order of reduction of the hemes [33, 34]. However, when STC from different organisms are expressed in trans in S. oneidensis MR-1 deletion mutants, the reduction of extracellular electron acceptors is restored to the same level observed for the wild-type protein [33]. This reveals that although this protein is essential for EET, the perturbation on the rates of electron uptake or donation by this MC caused by the replacement of homologs from other organisms is insufficient to modify EET. However, overexpression of this protein was demonstrated to enhance EET processes [35, 36]. FccA is the sole fumarate reductase in Shewanella, and is unique as a soluble periplasmic tetraheme flavocytochrome c. This protein with approximately 64 kDa is organized in three domains: a heme domain, a flavin domain and a clamp domain that links the previous two [37, 38]. Like STC, FccA binds its upstream partner CymA and its downstream partners specifically via one heme (heme II, numbered according to the position of the respective CXXCH binding motif in the polypeptide chain), again showing that it engages in specific recognition and docking [31, 32]. FccA is a moonlighting enzyme, taking the role of both fumarate reductase and redox shuttle across the periplasmic space [39]. As in the case of STC, the redox properties of the hemes in proteins from different organisms are different [40]. However, a common pattern of modulation of the rates of fumarate reduction according to the degree of reduction of the hemes is observed, showing how switching between EET and fumarate reduction is achieved [39]. Surprisingly, STC and FccA do not interact with each other, establishing electron transfer pathways that co-exist in the periplasmic space but that do not mix. This appears to be a consequence of their surface electrostatic properties which are predominantly negatively charged [31].
On the outer-membrane, several porin-cytochrome super complexes can be present.
The known genomes of Shewanella species show different numbers of these
complexes. By far the most prevalent is the complex encoded by the
mtrCAB-omcA operon, which is also the most abundant in S.
oneidensis MR-1. MtrB is a porin beta-barrel of 28 strands where the MC
MtrA and MtrC are inserted from the periplasmic and cell surface side,
respectively, forming an electrically conductive complex across the
outer-membrane [41]. MtrA is a 40 kDa decaheme cytochrome c
that is obtained from the periplasmic and membrane fractions of
Shewanella, showing that its attachment to the MtrB pore is transient
[42]. Surprisingly, the site of interaction with STC and FccA is the same, as
these proteins were shown to compete for binding to MtrA [32]. MtrC is a
decaheme cytochrome c of approximately 75 kDa with a lipidated cysteine
in the N-terminus to provide anchoring to the external surface of the
outer-membrane. In addition to the 10 hemes, MtrC also contains a CX
The MtrDEF porin cytochrome complex, homologous to MtrCAB complex, was also shown to be important for EET [48]. In this complex, only the MtrF decaheme cytochrome c has been structurally characterized, showing the same staggered cross arrangement of the 10 hemes found in MtrC and OmcA [49]. The affinity of this protein to soluble redox shuttles is much lower than that found for MtrC and OmcA in agreement with slower electron transfer kinetics measured in vitro [46]. Another porin-cytochrome complex found in Shewanella appears to have gone under extra specialization towards the extracellular respiration of DMSO [50]. The proteins DmsE and DmsF show homology with MtrA and MtrB, respectively. DmsE is capable of interacting with STC but the mode of interaction is different from that of MtrA, and it does not interact with FccA [32]. DmsA is a molybdenum containing protein responsible for DMSO reduction likely localised on the cell surface. The genome of S. oneidensis MR-1 also shows the presence of a gene cluster that codes for a fourth complex with genes SO4359 and SO4360 that are highly homologous to MtrAB. This complex can rescue a MtrAB deletion mutant during EET [51]. The question of the functional redundancy of the proteins in these complexes was explored using ferric citrate assays of deletion mutants complemented by expression of diverse combinations of proteins from the various complexes [48, 52]. Although these studies revealed that the modular arrangement of the porin cytochrome complexes allow the functional replacement of proteins from one of the complexes by proteins of the other, not all combinations were able to reduce iron citrate at similar rates, and the causes for the differences remain to be identified.
Geobacter species, side by side with Shewanella, have been on the focus for the development of EET based technologies. Geobacter are usually the predominant iron reducers in soils and sediments where this is an important geochemical process, and they can be found in aquatic sediments, contaminated ditches and wetlands [53]. Several microbial processes were observed for the first time with Geobacter, as the capacity to use iron and manganese oxides as electron acceptors, reduction of uranium, or the ability to use electrodes as electron donors or acceptors [53]. However, its electron transfer pathways are less understood, since the number of putative components involved is much higher (Fig. 3). In fact, the genome of G. sulfurreducens, the model strain, codes for more than one hundred proteins containing c-type heme(s), from which at least 78 are multiheme [54, 55]. The clarification of the role of the proteins involved in the reduction of terminal extracellular electron acceptors has been the target of different studies using diverse strategies, including proteomic studies in specific growth conditions and the evaluation of the capability of individual knock-out mutants to grow on each electron acceptor [56, 57, 58, 59, 60, 61].
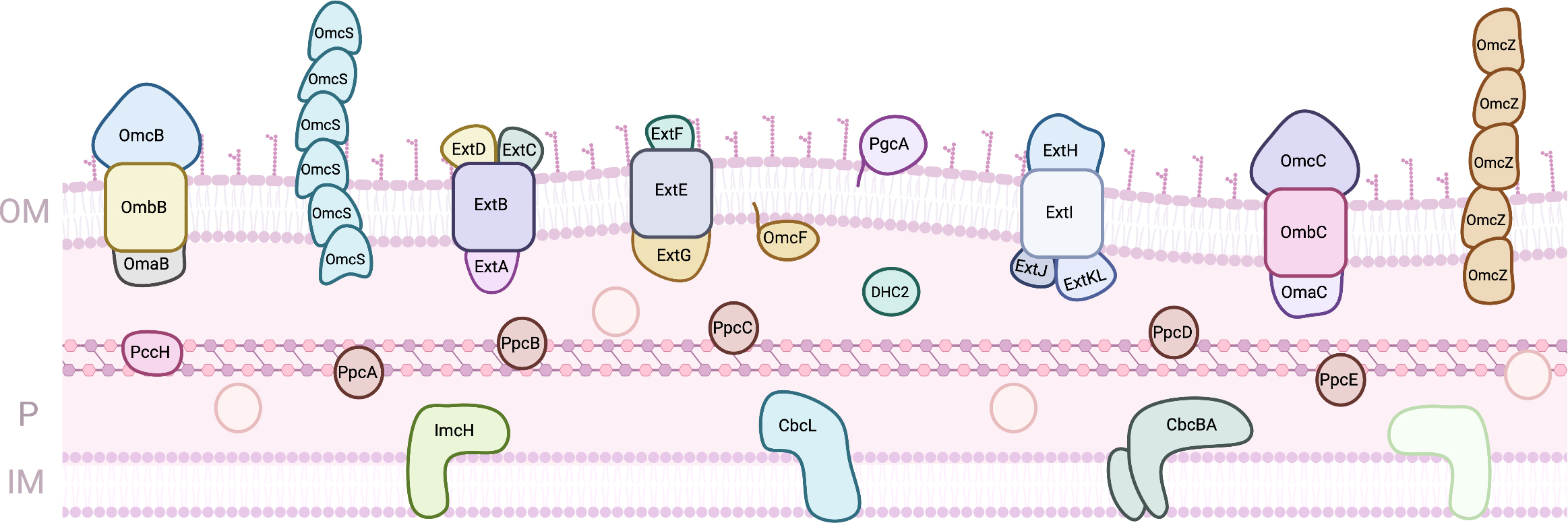 Fig. 3.
Fig. 3.Proteins involved in EET processes of G. sulfurreducens. (OM, outer-membrane; IM, Inner-membrane; P, periplasm).
In the inner-membrane, six putative quinone oxidoreductases are predicted to transfer electrons from the menaquinone pool to the periplasm: four Cbc complexes that contain b- and c-type heme groups (Cbc3/CbcVWX, Cbc4/CbcSTU, Cbc5/CbcEDCBA, and Cbc6/CbcMNOPQR), one Cbc single protein (CbcL) and one c-type cytochrome (ImcH) [55, 62]. From these, up to now, only three have been studied by gene knock-out experiments: ImcH, CbcL and CbcBA. ImcH is an heptaheme cytochrome that is anchored to the inner-membrane by three transmembrane helices. Mutant strains lacking the imcH gene were not able to reduce terminal electron acceptors with a redox potential above –100 mV (vs SHE) as iron chelates, Mn(IV) oxides and electrodes poised above this potential [61]. In its place, CbcL, a protein with a periplasmic domain with nine c-type heme groups attached to an inner-membrane associated domain with six transmembrane helices and two b-type heme groups, was shown to be essential for the reduction of Fe(III) oxides and electrodes below –100 mV (vs SHE) [60]. More recently, studies on CbcBA revealed that this complex formed by a di-heme b-type cytochrome with four transmembrane helices (CbcB) and an heptaheme c-type cytochrome (CbcA) is essential for extracellular metal and electrode reduction below –210 mV (vs SHE) [62]. Despite this knowledge, neither of these proteins has been biochemically characterized. MacA is also an inner-membrane associated diheme cytochrome c peroxidase, whose deletion mutant had impaired growth on soluble and insoluble iron, and it was proposed to be a regulator for the expression of OmcB [57, 63]. Although MacA was shown to interact and transfer electrons to the periplasmic MC PpcA [64, 65], its physiological function is still to be defined.
From the inner-membrane, electrons are transferred to the periplasm, where several cytochromes co-exist. The best characterized are the homologous cytochromes PpcA, PpcB, PpcC, PpcD and PpcE, known as the PpcA family. These are triheme cytochromes with approximately 10 kDa and all the hemes coordinated by two histidine residues and negative apparent midpoint reduction potentials with apparent midpoint values between –143 and –117 mV (vs SHE) [66]. Despite the high sequence and structural similarity, their detailed thermodynamic characterization revealed distinct properties: PpcA and PpcD can couple electron transfer with proton transfer, while PpcB and PpcE cannot at physiological pH [67]. This feature was proposed to be fundamental for an additional contribution to the proton gradient across the cytoplasmic membrane for ATP synthesis [68]. PpcA was shown to be necessary and sufficient for EET across the periplasm [69], and remarkably, there is at least one PpcA homolog in all Geobacter strains, which demonstrates that PpcA is a key component for the respiratory pathways of these bacteria [54]. G. sulfurreducens is also able to grow using electrodes as electron donors, and the monoheme cytochrome PccH was shown be required for this process [70]. For a cytochrome with an His-Met coordinated heme, PccH present a low reduction potential (–24 mV vs SHE), which can be explained by the high heme surface exposure when compared to other monoheme cytochromes [71, 72]. DHC2 is a diheme cytochrome found in the periplasm of G. sulfurreducens. Its gene its upregulated in cells grown on Fe(III) and Mn(IV) oxides compared with growth on Fe(III) citrate [57]. Both hemes are bis-His and have reduction potentials of –289 and –135 mV (vs SHE), and the protein’s structure revealed that the hemes are packed in a parallel motif [73].
Other MC have been identified in G. sulfurreducens’ periplasm, including the dodecaheme cytochrome GSU1996, for which a physiological role has not been clearly determined. The protein is arranged in four triheme domains with similar structure, each containing two His-His hemes and one His-Met heme [74]. The high number of heme groups led to proposal that this cytochrome could act as an electron capacitor, since it was observed that Geobacter biofilms can grow even when terminal electron acceptors are not available [75, 76]. Indeed, the detailed thermodynamic characterization of domain CD from GSU1996 has shown that this protein could function as a nanowire in the periplasmic space of Geobacter [77]. OmcF is a monoheme cytochrome present in the periplasm of G. sulfurreducens, and it is not clear if it is directly involved in EET or if it is only necessary for the expression of other cytochromes [78, 79]. The heme is coordinated by an histidine and a methionine and has a reduction potential of +180 mV (vs SHE) [80].
As observed for Shewanella, the outer-membrane of G.
sulfurreducens contains different porin-cytochrome complexes [81]. However, in
contrast with Shewanella where the complexes are paralogs, in
Geobacter the complexes have low homology in size, content or heme
number. A thorough study where the genes coding for each and/or combinations of
these complexes were deleted in G. sulfurreducens showed that different
complexes are involved in different EET pathways, namely the reduction of Fe(III)
citrate, Fe(III) and Mn(IV) oxides, and graphite electrodes [59]. OmabcB
and OmabcC were the first identified complexes, and are the ones that
are more similar to the Mtr complexes of Shewanella [82]. These two
complexes are part of two tandem gene clusters that code for an octaheme
periplasmic cytochrome (OmaB and OmaC), a transmembrane porin (OmbB and OmbC) and
an outer-membrane cytochrome with 12 hemes (OmcB and OmcC). Interestingly, the
porins and the outer-membrane components share the same sequence, and are
probably the result of a gene duplication [83]. These complexes were shown to be
essential for the reduction of iron compounds [59]. The complex ExtABCD,
formed by a porin (ExtB), a periplasmic dodecaheme cytochrome (ExtA) and
two outer-membrane lipoprotein cytochromes with 5 and 12 heme binding sites
respectively (ExtC and ExtD), was shown to be fundamental for growth with
electrodes [59, 84]. Another complex, ExtEFG, composed by the porin
ExtE, the periplasmic multiheme cytochrome ExtG, that has thirteen CXXCH typical
binding motifs plus some putative unusual heme binding motifs CX
PgcA is a cytochrome that is loosely bound to the outer-membrane of G. sulfurreducens in its extracellular side. Besides its three c-type heme groups, the protein is composed by proline and threonine tandem repeats. Deletion mutants were unable to transfer electrons to metal oxides but maintained the ability to reduce Fe(III) citrate and electrodes [85]. OmcS and OmcZ are multiheme cytochromes that arrange in polymeric assemblies forming extracellular conductive filaments and allow long-range (micrometer) electron transfer [86, 87, 88]. OmcS monomer contains six hemes that are hexacoordinated with two histidines and its filaments are involved in Fe(III) oxides reduction [87, 89]. OmcZ monomer contains eight hemes that are also hexacoordinated with two histidines and is essential for electron transfer to electrodes [88, 90]. The high number of heme groups spans the proteins’ redox working windows, and isolated monomer forms of OmcS and OmcZ have midpoint reduction potentials of –212 and –220 mV (vs SHE) respectively [89, 90]. In addition to these groups of cytochromes, electrically conductive filaments were also reported and implicated in the electron transfer to extracellular metals and electrodes [91].
Thermophilic iron-reducing bacterium belonging to Thermincola spp. were demonstrated to be able to perform EET to electrodes either by direct or indirect electron transfer [92, 93]. As for Gram-negative bacteria, these bacteria also rely on MC to perform EET [14]. The EET pathway proposed for bacteria belonging to Thermincola spp. is composed by four MC that form a pathway from the inner-membrane across the cell wall composed by peptidoglycan, towards outside of the cell (Fig. 4). ImdcA is a decaheme cytochrome that resides at the unique membrane of this Gram-positive bacteria. It receives electrons from the quinone pool and transfer them to the periplasmic space. This protein contains nine low-spin hemes, with axial ligands that are parallel and perpendicular to each other, and one high spin heme [94] that is probably involved on the interaction with the quinones from the quinone pool. At the periplasmic space, PdcA mediates electron transfer between ImdcA and CwcA, a MC that is present at the peptidoglycan layer [95]. The decaheme cytochrome PdcA contains nine hemes that are hexacoordinated with two histidines, and one heme coordinated with an histidine and a methionine [94]. It was proposed that several monomers of the hexaheme cytochrome CwcA can be stacked on top of each other forming a conductive nanowire embedded in the peptidoglycan, allowing electrons to be transferred across the cell wall of these Gram-positive bacteria [94]. The structure arrangement of this nanowire enables the hemes to be close to each other, allowing fast intra and intermolecular electron transfer, enabling electrons to be transferred across the peptidoglycan, to OcwA or extracellular electron acceptors. The nonaheme cytochrome OcwA was proposed to be the terminal metal reductase of T. potens strain JR, as isolated and identified by trypsin shaving [95]. This protein was shown to contain hemes with three distinct coordination environments: hexacoordinated with two histidines (hemes I, III, IV, VI, VII and VIII); hexacoordinated with an histidine and a methionine (heme IX), and pentacoordinated hemes (heme II and V) with an histidine serving as proximal iron ligand [96]. These structural features were different from other terminal metal reductases identified and studied so far, and are a novelty within MC. OcwA, besides being able to exchange electrons with an electrode and reduce iron oxides, it can also reduce soluble electron shuttles, and nitrogen compounds, including nitrite and hydroxylamine. This enzymatic capacity may provide an advantage for cell survival in different anaerobic environments, where OcwA may work as a detoxifying enzyme when encounter these compounds.
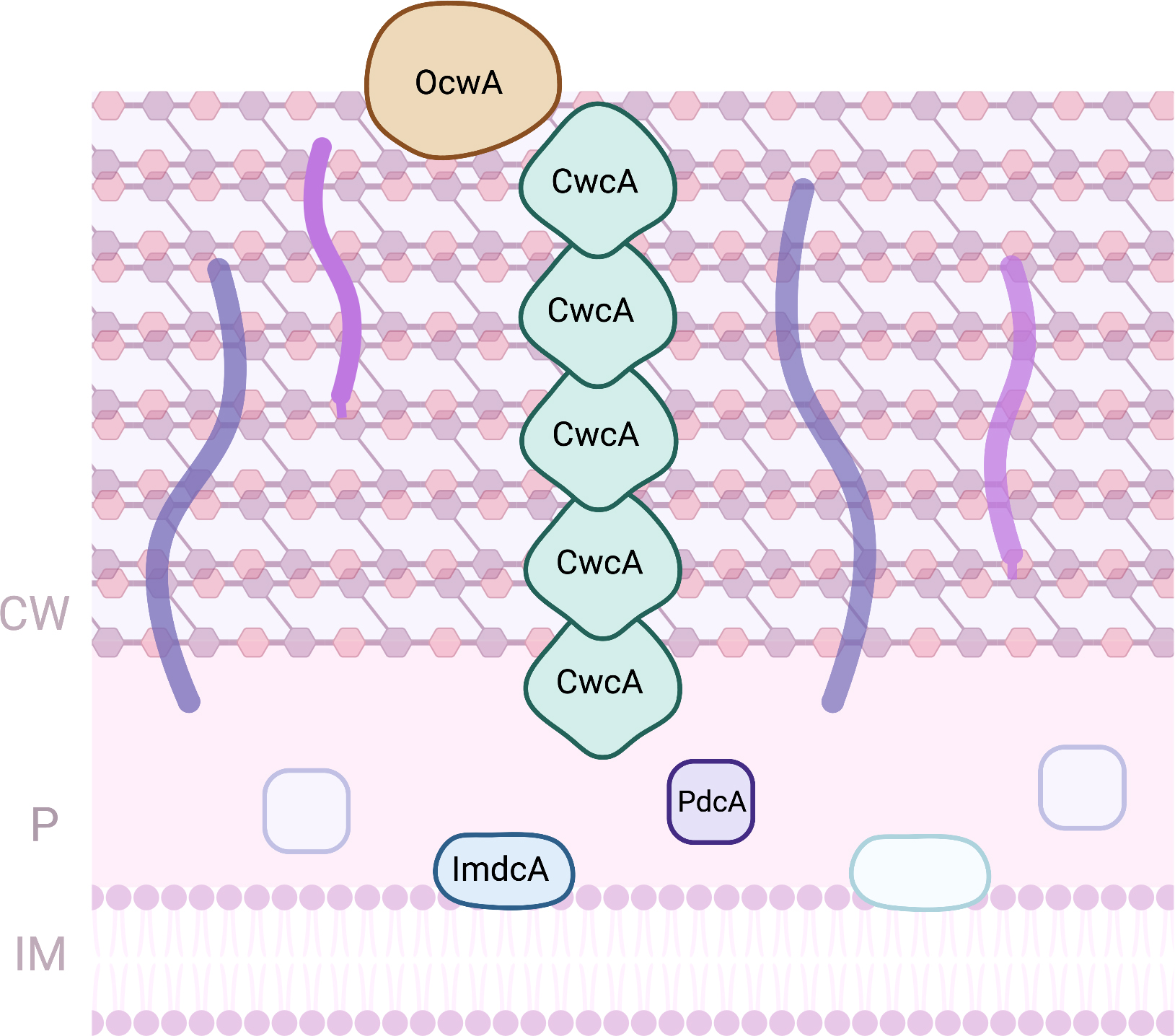 Fig. 4.
Fig. 4.Proteins involved in EET processes of Thermincola sp. (CW, Cell-wall; IM, Inner-membrane; P, periplasm).
ImdcA, PdcA and OcwA are redox active within a similar range of electrochemical potential (between –300 and +100 mV vs SHE), which were shown to provide the necessary electrochemical window to establish a connection between the Thermincola cell metabolism and extracellular acceptors of higher reduction potentials, such as iron minerals or electrodes in BES [94, 96].
Rhodopseudomonas palustris TIE-1 is a Gram-negative bacterium with a
broad metabolic versatility that includes the capacity to grow autotrophically
using iron minerals instead of water as electron source [97]. This
photoferrotrophic metabolism is underpinned by the operation of proteins coded by
the pio operon (Fig. 5) [98]. This operon codes for three proteins:
PioA, PioB and PioC. PioA is a decaheme cytochrome with homology with
MtrA from Shewanela spp. It is proposed to be inserted in the cavity
formed by PioB that forms a 28-strand beta-barrel located on the
outer-membrane. PioA is proposed to be the iron oxidase and to allow conduction
of the electrons across the outer-membrane towards the periplasmic space. The
macroscopic redox characterization of PioA shows that it titrates in a range of
potentials from –400 to +250 mV (vs SHE) [99]. PioC is a high potential
iron sulfur protein and is the only protein of this operon for which the
structure has been obtained [100]. It is proposed to deliver the electrons
collected by PioA to the photosynthetic reaction center for light activation. The
electron can be used in non-cyclic photosynthesis or recycled from the cytochrome
bc
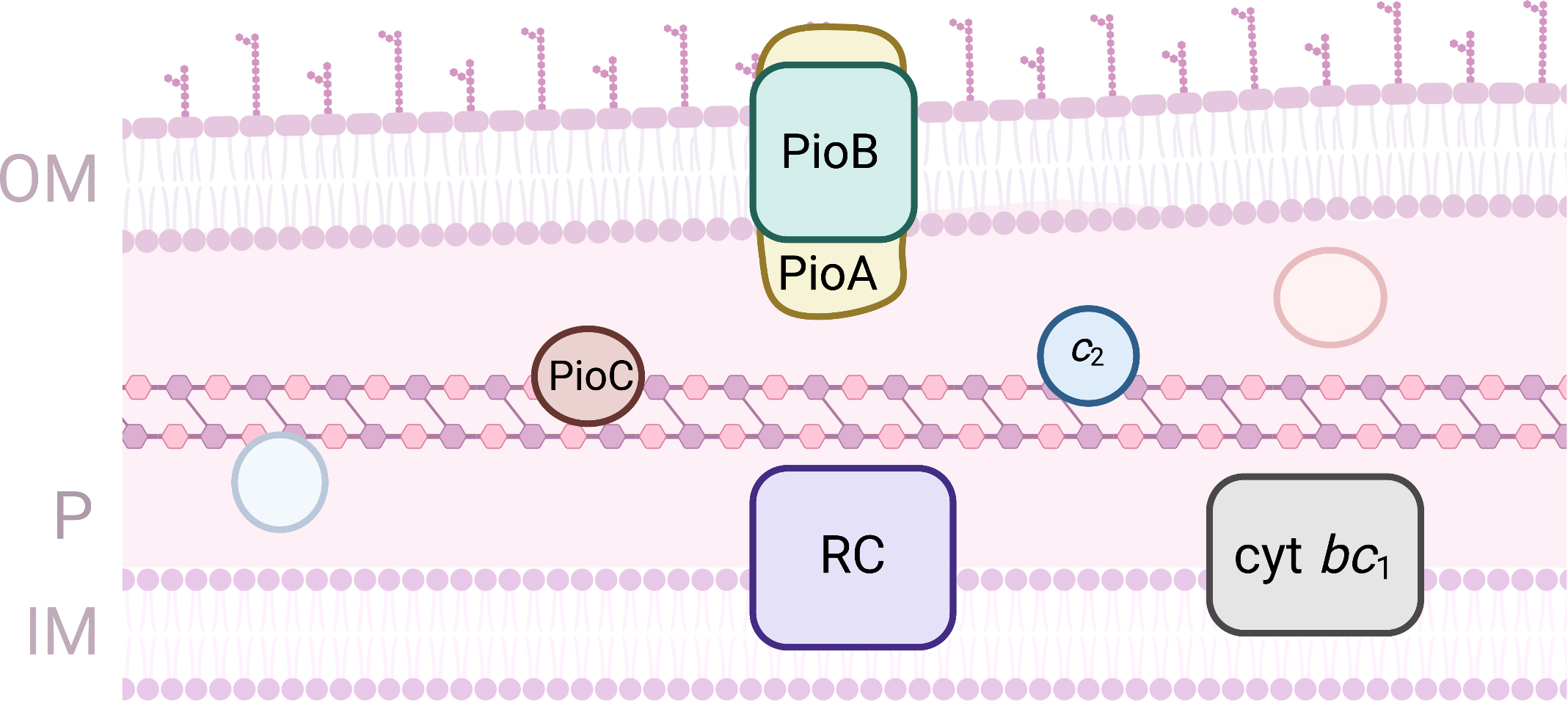 Fig. 5.
Fig. 5.Proteins involved in EET processes of Rhodopseudomonas palustris. (OM, outer-membrane; IM, Inner-membrane; P, periplasm).
The freshwater chemolithoautotrophic Gram-negative bacterium S.
lithotrophicus ES-1 is an Fe(II) oxidizing organism that grows on FeCO
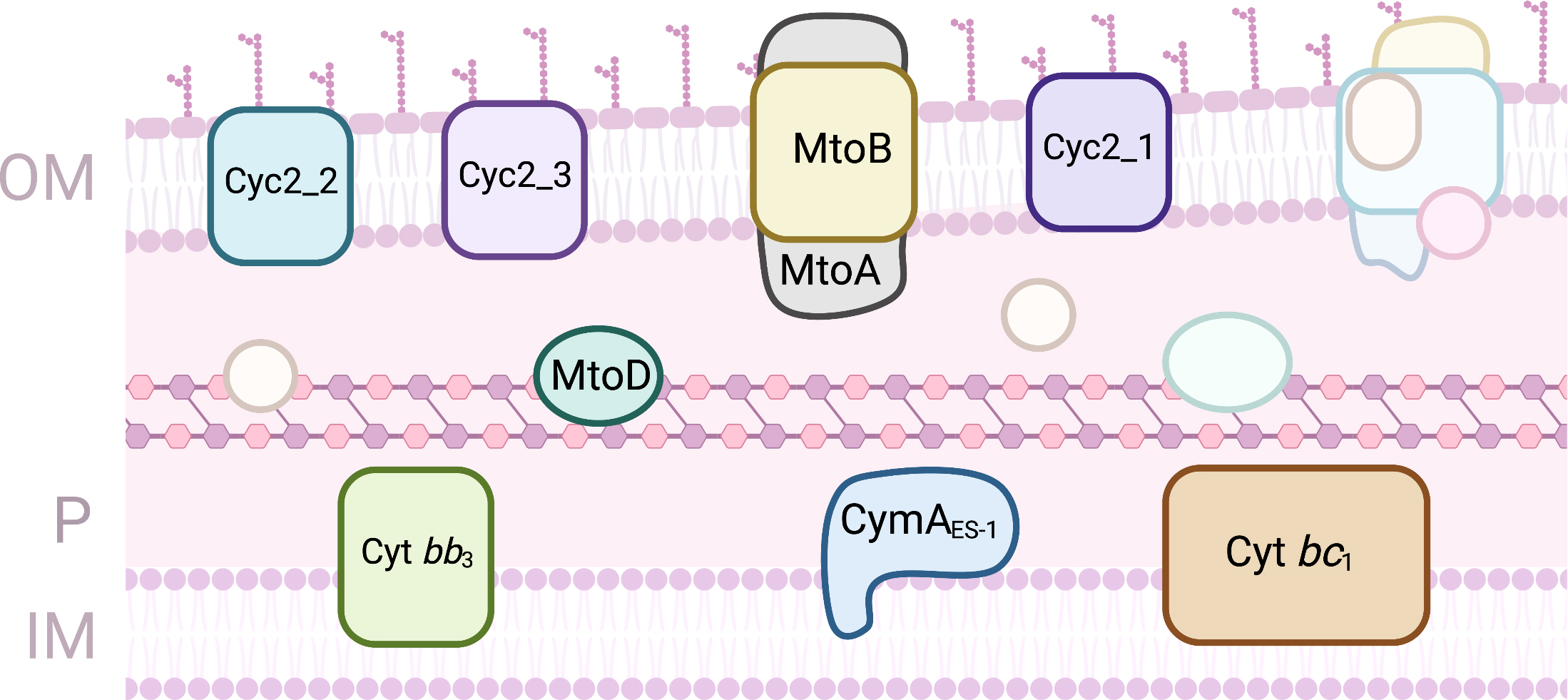 Fig. 6.
Fig. 6.Proteins involved in EET processes of Sideroxydans lithotrophicus ES-1. (OM, outer-membrane; IM, Inner-membrane; P, periplasm).
The evaluation of the electron transfer processes performed by the MC involved in EET pathways requires the production of the proteins in sufficient amounts for their characterization, that typically includes the 3D-structure determination and elucidation of their thermodynamic and kinetics properties [21, 22]. Although this is not sufficient to elucidate the role of the protein in the EET process, that commonly requires knock-out approaches and in vivo studies [19], it enables to unravel their electron transfer processes and understand their mode of action. Indeed, the thermodynamic properties of these proteins are one of the most important parameters to reveal the function of the protein, providing knowledge on the electrochemical window in which the protein is active [108]. This is fundamental to envisage possible electron donors and acceptors. For most MC involved in EET only a macroscopic thermodynamic characterization has been achieved (Table 1), with no detailed information about the individual hemes.
| MC | Organism | Localization | Number of hemes | Reduction window range | Reference |
| CymA | S. oneidensis MR-1 | IM | 4 | –300 mV – +100 mV | [109] |
| MtrA | S. oneidensis MR-1 | P | 10 | –400 mV – –100 mV | [110] |
| MtrC | S. oneidensis MR-1 | OM | 10 | –500 mV – +100 mV | [111] |
| OmcA | S. oneidensis MR-1 | OM | 10 | –330 mV – +0 mV | [112] |
| MtrF | S. oneidensis MR-1 | OM | 10 | –400 mV – +50 mV | [49] |
| MacA | G. sulfurreducens | IM | 2 | –300 mV – 0 mV | [64] |
| OmcS | G. sulfurreducens | OM | 6 | –350 mV – +50 mV | [89] |
| OmcZ | G. sulfurreducens | OM | 8 | –420 mV – –60 mV | [90] |
| ImdcA | T. ferriacetica | IM | 10 | –300 mV – +100 mV | [94] |
| PdcA | T. ferriacetica | P | 10 | –300 mV – +100 mV | [94] |
| OcwA | T. potens strain JR | CS | 9 | –350 mV – –80 mV | [96] |
| PioA | R. palustris TIE-1 | OM | 10 | –400 mV – +250 mV | [99] |
| MtoA | S. lithotrophicus ES-1 | OM | 10 | –350 – +30 mV | [104] |
| IM, Inner-membrane; CS, Cell-surface; P, Periplasm; OM, Outer-membrane. | |||||
While for a redox protein with one heme detailed information about the redox center can be achieved by the direct application of the Nernst equation, in the case of MC the situation is much more complex due to the presence of several closely spaced hemes [113, 114]. In this case, the redox properties of the protein are defined by the microscopic reduction potential of each individual heme, the interaction between the hemes and the interaction between the hemes and protonable(s) center(s) [108, 114]. The determination of these parameters depends on the capacity to discriminate experimentally each individual heme and follows the respective signals as the cytochrome transitions from the fully reduced to fully oxidized state. This can only be achieved by spectroscopic techniques, including nuclear magnetic resonance [115]. Up to date, this type of analysis has only been performed for few cytochromes involved in EET processes (Table 2). Of these, the largest is FccA with 64 kDa, whereas the most complex is domain CD of GSU1996 with six hemes [40, 77].
| MC | Organism | Localization | Number of hemes | Individual heme reduction potential (mV vs SHE) | Reference |
| STC | S. oneidensis MR-1 | P | 4 | –243 (I); –222 (II); –189 (III); –171 (IV) | [34] |
| FccA | S. oneidensis MR-1 | P | 4 | –145 (I); –286 (II); –247 (III); –160 (IV) | [40] |
| PpcA | G. sulfurreducens | P | 3 | –154 (I); –138 (III); –125 (IV) | [67] |
| PpcB | G. sulfurreducens | P | 3 | –150 (I); –166 (III); –125 (IV) | [67] |
| PpcD | G. sulfurreducens | P | 3 | –156 (I); –139 (III); –149 (IV) | [67] |
| PpcE | G. sulfurreducens | P | 3 | –167 (I); –175 (III); –116 (IV) | [67] |
| PccH | G. sulfurreducens | OM | 1 | –24 | [71] |
| OmcF | G. sulfurreducens | P | 1 | +180 | [80] |
| Domain CD (GSU1996) | G. sulfurreducens | P | 6 (12) | –106 (IC); –136 (IIIC); –125 (IVC);–155 (ID); –178 (IIID); –113(IVD) | [77] |
| DHC2 | G. sulfurreducens | P | 2 | –239; –135 | [73] |
| MtoD | S. lithotrophicus ES-1 | P | 1 | +155 | [105] |
| P, Periplasm; OM, Outer-membrane. | |||||
The microscopic redox characterization of diverse MC has shown that the close proximity between the hemes, that ensures very fast intramolecular electron transfer [116], gives rise to substantial electrostatic interactions as the protein gains or loses electrons. These interactions can be described by a simple model of Coulomb decay enhanced by a Debye-Huckel shielding [117]. Microscopic thermodynamic characterization demonstrated that the redox state of a MC can establish preferential electron transfer pathways. For example, in the case of FccA, thermodynamic and kinetic data showed that at low electron flux, FccA does not become fully reduced, and transfer electrons to the outer-membrane reductases for the reduction of solid phase acceptors. As the electron flux increases, FccA then becomes increasingly reduced, switching to an efficient catalyst for fumarate reduction. This switching mechanism enables FccA to function as a moonlighting protein, and assist Shewanella to switch back and forth between reducing soluble and insoluble electron acceptors, circumventing transcriptional events [39]. Another example is the case of domain CD from GSU1996, that is predicted to conduct long range electron transfer within the periplasmic space of Geobacter. The detailed redox characterization of domain CD with six hemes demonstrated that GSU1996 could function as a capacitor receiving electrons from the heme that is at one edge of the protein, and transferring them to the other hemes within the protein [77].
For larger and more complex proteins, this detailed analysis is much more difficult due to the complexity of the system, preventing the discrimination of the hemes [108, 114]. For some of these cases, other approaches, including computational methods have been used to predict individual redox properties of the hemes [118, 119]. However, in this case, the reduction potential is only obtained for a particular state of the protein, preventing the elucidation of the electron transfer mechanisms within the protein during the reduction or oxidation process.
Different MC present distinct electrochemical behavior. The large potential
range of these proteins is attributed to the wide range of reduction potentials
for the hemes present in the protein. Interestingly, most of them are active in
the same electrochemical window (Fig. 7), enabling electrons to be transferred
from the electron donor (e.g., NADH/NADPH or iron oxides) to the electron
acceptor (e.g., iron hydroxides, H
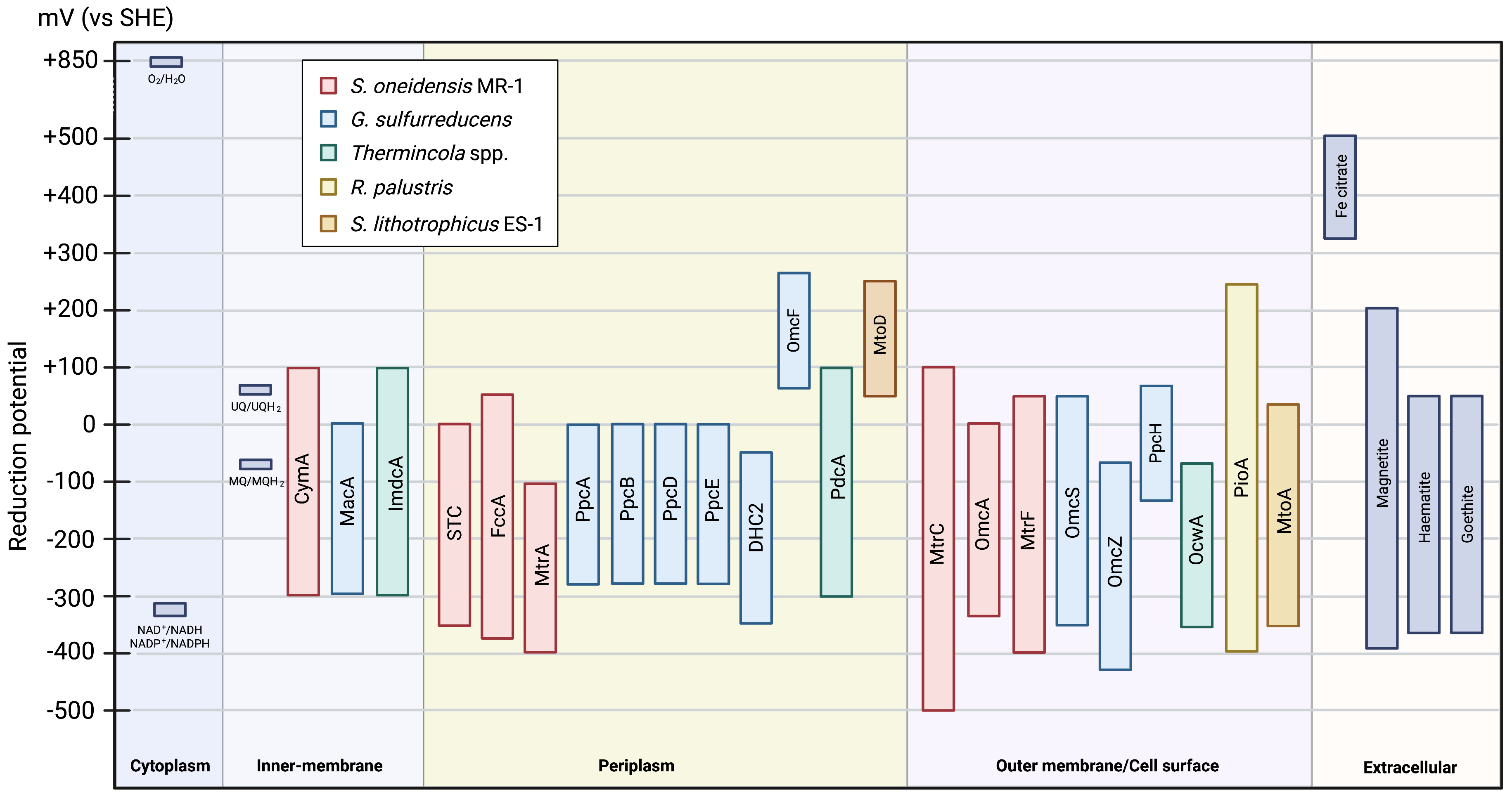 Fig. 7.
Fig. 7.Scheme of the electrochemical active window of the MC involved in EET pathways (values taken from Table 1 and Table 2). The reduction potentials of the other compounds were taken from [5].
MC can perform both reduction and oxidation processes. Indeed, it has been shown that EET can happen both ways [120], indicating that it is the electron donor and acceptor that dictates the electron transfer direction. Although this opens the door to manipulate the EET pathways toward desired functionalities, an interaction between the two redox partners also must occur so that the electron transfer event can actually happen.
Although the redox properties of MC are critical to predict possible physiological partners, electron transfer between them may not be possible due to the slow electron transfer rate or the lack of interaction given by structural constrains [31]. Electron transfer between two proteins typically depends on the formation of an encounter complex, before the actual electron transfer occurs [114, 121]. This is usually governed by random collisions, where electrostatic forces contribute to the formation of the complex, placing the hemes close to each other and with a correct orientation for the electron transfer event to occur. This is fundamental to prevent the loss of electrons and the formation of reactive oxygen species that would damage the cell. In the case of MC, NMR spectroscopy has been used to study interactions between physiological partners [31, 122]. This technique is very powerful to study interactions since the chemical shifts of the nuclei are very sensitive to changes in their chemical environment, and therefore if the spectra of one MC is affected after the addition of a redox partner, this indicates that an interaction occurs. A full assignment of the NMR signals to each of the protein’s residues is desirable to enable the identification of the interacting region and determination of the dissociation constant [19]. However, considering that the electron exchange must occur between the protein redox centers, in most case the assignment of the NMR signals of the heme substituents is sufficient. This approach has been successfully used for several MC involved in the EET of S. oneidensis MR-1 and G. sulfurreducens (Table 3).
| MC | Ligand | Organism | Dissociation Constant ( |
Reference |
| FccA | CymA | S. oneidensis MR-1 | 398 (5) | [31] |
| FccA | MtrA | S. oneidensis MR-1 | 35 (14) | [31] |
| STC | CymA | S. oneidensis MR-1 | 250 (17) | [31] |
| STC | MtrA | S. oneidensis MR-1 | 572 (5) | [31] |
| STC | OTR | S. oneidensis MR-1 | 1600 (400) | [32] |
| STC | DmsE | S. oneidensis MR-1 | 783 (227) | [32] |
| PpcA | MacA | G. sulfurreducens | 223 (1) | [65] |
| PpcA | GSU1996 | G. sulfurreducens | 1170 (100) | [127] |
| PpcA | AQDS | G. sulfurreducens | 17 800 (500) (n = 1) | [123] |
| OmcA | FMN | S. oneidensis MR-1 | 29 (11) (n = 2) | [46] |
| MtrC | FMN | S. oneidensis MR-1 | 255 (126) (n = 1) | [46] |
In the case of S. oneidensis MR-1 this approach has been used to identify the redox partners involved in bridging the periplasmic gap between CymA and the outer-membrane complex MtrCAB [31]. In this work, NMR was used to evaluate the interaction between STC and FccA, which are the most abundant proteins found in the periplasmic space of Shewanella, with different redox partners, including CymA and MtrA. It was shown that these proteins form weak complexes between them (i.e., CymA with STC or FccA, and MtrA with STC or FccA), which is crucial to prevent the blockage access to CymA by the various redox partners. Furthermore, using this technique it was demonstrated that both STC and FccA interact with CymA and MtrA with the same heme (heme II in the case of FccA and heme IV in the case of STC), indicating that a complex between the three proteins (e.g., CymA-STC-MtrA or CymA-FccA-MtrA) cannot occur, and that both STC and FccA move back and forth in the periplasmic space to receive and donate electrons during EET [31]. In another study using the same approach, it was demonstrated that STC is the major player in connecting CymA with DmsE responsible for electron transfer to DMSO reductase at the outer-membrane, and with OTR in the periplasmic space [32]. However, it is not involved in the interactions with ccNir or MtrD. These studies clearly show that STC functions like an electronic cul-de-sac where electrons enter and leave the cytochrome by the same heme, which enables the transfer of electrons in a controlled and efficient manner to different redox partners within the periplasmic space of Shewanella, minimizing the risk of diverting the electrons to side redox pathways or to the production of radical species [32]. NMR was also a powerful tool to explore the interactions between the outer-membrane cytochromes of S. oneidensis MR-1 and different electron shuttles [46]. FMN, riboflavin, anthraquinone-2,6-disulfonate (AQDS) and phenazines provide distinct electrostatic environments for interaction with the MC. These led to observation of perturbations of different signals in the NMR spectra of MC upon binding of the various shuttles, and different binding sites predicted by molecular simulation. Electron transfer with the MC is dominated by thermodynamics with the extent of electron extraction from the cytochromes set by the redox potential of the mediators. The results obtained in this work showed that despite the structural similarities between the outer-membrane cytochromes, the detailed characteristics of their interaction with soluble electron shuttles are quite distinct. The values of the apparent dissociation constants obtained for MtrC and OmcA with FMN indicate that these proteins have weak affinity for this electron shuttle, which is in agreement with an electron shuttling mechanism. The stoichiometry of binding of FMN with these proteins matches the number of redox active disulfide bridges found in these cytochromes with 1:1 binding for MtrC and 2:1 binding for OmcA [46].
In G. sulfurreducens, the NMR-probed interaction studies have been
conducted with the well-characterized PpcA family of triheme cytochromes.
Interactions between these proteins and several terminal acceptors, namely
anthra(hydro)quinone-2,6-disulfonate (A(H
The increased interest in EET processes for understanding the biogeochemical cycle of elements and the development of biotechnological processes has led to the identification and characterization of key proteins involved in these pathways. Most of these proteins are MC that form conductive conduits, enabling electrons to be transferred across long distances, including from the inner-membrane to the exterior of the cell, or vice-versa. These proteins are crucial to define the electron transfer route within the electroactive organism. Although most of the electroactive organisms are known to oxidize or reduce solid electron donors or acceptors, respectively, several of them have been shown to perform both processes. Indeed, most of the MC involved in EET processes titrate at the same electrochemical range, which enables the electron transfer to occur both ways. This suggests that it is the electron donor and acceptor that predict the direction of the electron transfer process, and it is the thermodynamic driving force that defines the pathway. Additionally, an interaction between physiological partners must occur, and this has to happen in a way that enables the formation of a transient complex that is required for a fast and efficient electron transfer event. Electroactive organisms contain different proteins that perform similar function, revealing that distinct electron transfer pathways coexist in a cell, and by which the microorganism have to have a way to control the electron transfer to different donors or acceptors. Although significant knowledge on the redox properties and interaction processes already exist for several MC, there are still numerous questions to be answered, including what controls the electron transfer process in a cell. Only with a detailed characterization of the electron transfer processes performed by all the redox proteins involved in these pathways, it will be possible to completely understand what is happening in a living organism during EET.
CMP draft the manuscript; CMP, LM, CS and ROL wrote and revised and the manuscript. All authors read and approved the final manuscript.
Not applicable.
This work was supported by national funds from Fundac¸aõ para a Cien̂cia e a Tecnologia (FCT) in the scope of Project MOSTMICRO-ITQB with refs UIDB/04612/2020 and UIDP/04612/2020, projects UIDP/04378/2020 and UIDB/04378/2020 of the Research Unit on Applied Molecular Biosciences – UCIBIO, project LA/P/0087/2020 of the Associate laboratory Life Sciences for a Sustainable and Healthy Future and the project LA/P/0140/2020 of the Associate Laboratory Institute for Health and Bioeconomy - i4HB. FCT is also acknowledged for funding through the following grants: SFRH/BPD/114848/2016 (LM), PTDC/BIA-BQM/31981/2017 (CAS), PTDC/BIA-BQM/4967/2020 (CAS), PTDC/BIA-BQM/4143/2021 (CMP) and EXPL/BIA-BQM/0770/2021 (LM). This project was also supported by the European EC Horizon2020 TIMB3 (Project 810856).
This research received no external funding.
The authors declare no conflict of interest. CMP is serving as the guest editor and editorial board member of this journal. ROL is serving as the guest editor of this journal. We declare that CMP and ROL had no involvement in the peer review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to RA.







