Academic Editor: Graham Pawelec
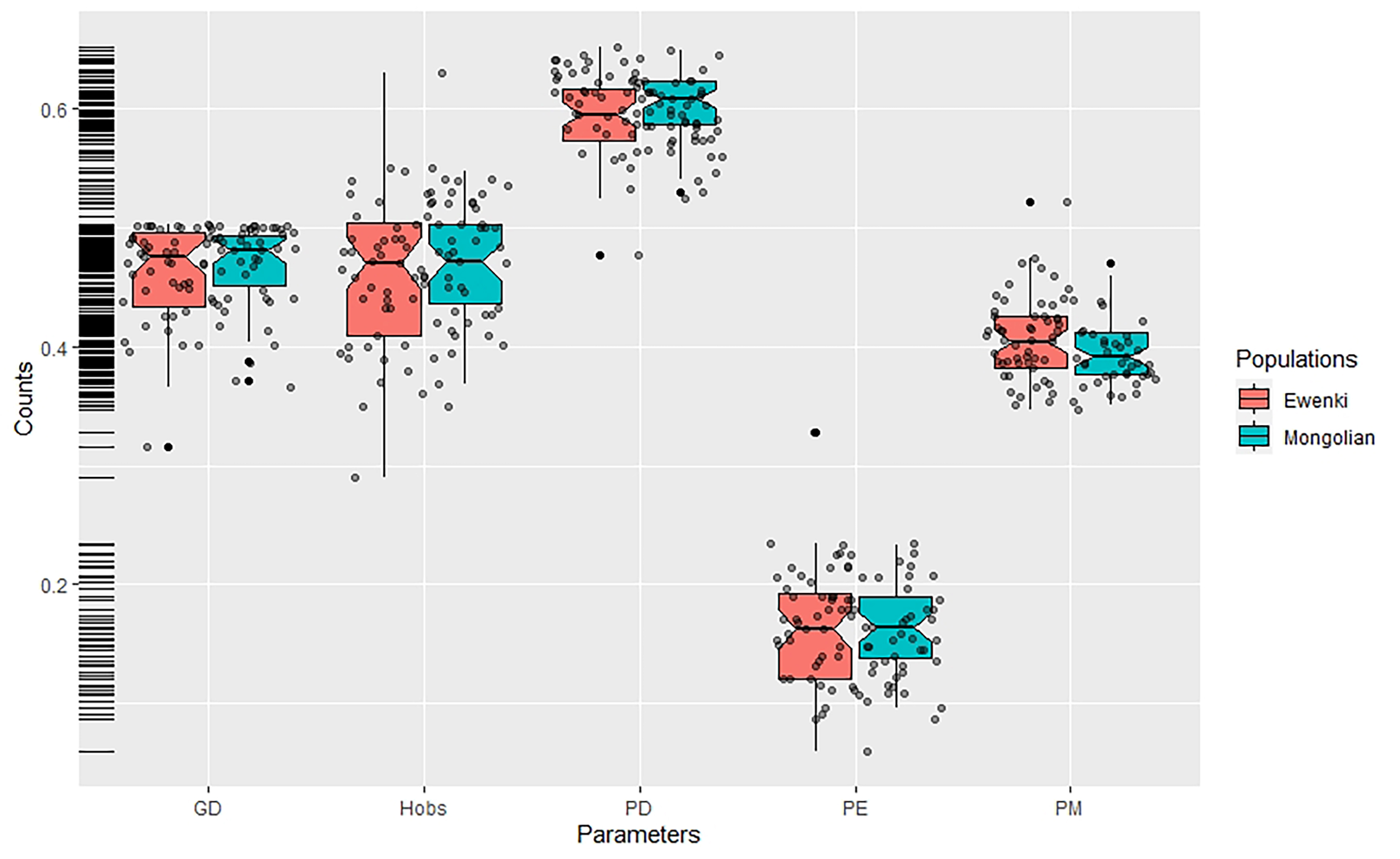
Background: InDel polymorphisms show great potential for use with challenging DNA samples in forensic practice due to having similar advantages to STRs and SNPs. Large-scale InDel genotype data are becoming available world wide populations, thus providing an alternative for investigating genetic architectures in rarely studied populations from a genome perspective. Methods: Here, we genotyped 47 highly polymorphic InDel variations in 157 Mongolian and 100 Ewenki individuals from the Inner Mongolia Autonomous Region of China in order to evaluate their utility for forensic purposes. Results: The CDPs of the 47 InDels for these groups were calculated to be 0.999999999999999999874 and 0.999999999999999999677, respectively, while the CPEs were 0.99981 and 0.99975, respectively. The 47 InDel variations were therefore an efficient tool for forensic personal identification in the Mongolian and Ewenki ethnic groups. Comparison of results from the present study with datasets from previously published literatures and from the 1000 Genomes Project revealed a prominent East Asian ancestry component in the gene pool of both ethnic groups, hinting at the close genetic relationships between Mongolian, Ewenki and most East Asian populations. Furthermore, Han populations from northern China showed even closer genetic affinities with the Mongolian and Ewenki groups. Conclusions: The data presented here would facilitate the forensic application of InDels for Mongolian and Ewenki ethnic minorities and expand our knowledge regarding the genetic diversities of these populations.