1 Department of Experimental and Clinical Biomedical Sciences, University of Florence, 50134 Florence, Italy
2 Research Centre for Agriculture and Environment, Council for Agricultural Research and Economics, 50125 Firenze, Italy
3 Institute of Agricultural Biology and Biotechnology, National Research Council, 56124 Pisa, Italy
4 Department of Biology, University of Pisa, 56126 Pisa, Italy
5 Department of Experimental and Clinical Medicine, University of Florence, 50134 Florence, Italy
6 Laboratory of Microbial and Molecular Evolution, Department of Biology, University of Florence, 50019 Sesto Fiorentino (Firenze), Italy
Academic Editor: Ru Chen
Abstract
Background: Azurin, a bacterial cupredoxin firstly isolated from the bacterium Pseudomonas aeruginosa, is considered a potential alternative therapeutic tool against different types of cancer. Aims: In this work we have explored the relationship possibly existing between azurin and colorectal cancer (CRC), in light of the evidence that microbial imbalance can lead to CRC progression. Methodology/Results: To this aim, the presence of azurin coding gene in the DNA extracted from saliva, stool, and biopsy samples of 10 CRC patients and 10 healthy controls was evaluated by real-time PCR using primers specifically designed to target the azurin coding gene from different bacterial groups. The correlation of the previously obtained microbiota data with real-time PCR results evidenced a “preferential” enrichment of seven bacterial groups in some samples than in others, even though no statistical significance was detected between controls and CRC. The subset of azurin gene-harbouring bacterial groups was representative of the entire community. Conclusions: Despite the lack of statistical significance between healthy and diseased patients, HTS data analysis highlighted a kind of “preferential” enrichment of seven bacterial groups harbouring the azurin gene in some samples than in others.
Keywords
- azurin
- Pseudomonas aeruginosa
- colorectal cancer
- real-time PCR
- microbiota
At the end of the 19th century bacteria and their metabolites were tested for the first time as anticancer agents [1, 2]. This interest in the development of alternative therapies based on the use of bacteria and/or their secondary metabolites has renewed and increased in the last years. Indeed, some bacterial (secondary) metabolites have been found to specifically affect the tumor cells survival or, otherwise, interfere with signalling pathways that allow cancer progression [3, 4, 5, 6, 7].
Among these, cupredoxins are metalloproteins involved in bacterial electron
transport chain [8, 9] showing different activities including antimalarial [10]
and anticancer ones [5, 11, 12, 13, 14, 15]. The cupredoxin family includes azurins,
rusticyanins, pseudoazurins, auracyanins, amicyanins, and halocyanins [16, 17].
For several decades, the interest in anticancer activity has mainly focused on
azurin [5, 14, 18]. Even though Azurin was purified in 1958 [19], it was
discovered for the first time in Pseudomonas aeruginosa, where it is
secreted as periplasmatic protein [5]. Azurin belongs to the copper proteins type
1, and it is involved in the electron transfer chain within the denitrification
process in P. aeruginosa [3, 20, 21]. This protein has a
tertiary
Besides, different in vivo studies demonstrated that azurin and p28 inhibit tumor growth in 4T1 breast model, B16 melanoma and Dalton’s lymphoma ascites model [30, 32, 33].
More in-depth, regarding in vitro evidence, azurin/p28 can induce inhibition of cancer cells proliferation via p53 pathway, by complexing and stabilizing p53, inducing overexpression of pro-apoptotic genes such as BAX, and, consequently, decreasing BCL-2 level that leads to the release of mitochondrial cytochrome c in the cytosol, thus promoting downstream activation of apoptotic machinery [30, 34, 35, 36, 37]. Despite this activity has been demonstrated in vitro, mainly in breast, colon, melanoma, glioblastoma, and prostate cancer cell lines [24, 30, 31, 35, 38, 39, 40, 41] the proliferation inhibition was observed also in other cancer cell lines such as ovarian cancer, fibrosarcoma, osteosarcoma, pancreatic cancer, and neuroblastoma [24, 31, 42].
In addition, azurin improves anticancer response also through modulation of cell
membrane properties, impacting for instance on
Considering these promising evidence, two recent clinical trials (phase I) have been completed [44, 45], demonstrating the efficacy of a safety and optimal dose of p28 on (i) 15 patients bearing metastatic solid tumors - NCT00914914; (ii) 18 younger patients with recurrent or progressive central nervous system tumors - NCT01975116.
Recent studies have highlighted how azurin is able to impact on cellular processes such as adhesion and invasion of bacterial pathogens (e.g., S. aureus, Salmonella sp.) in Colorectal Cancer (CRC) lines CaCo-2 [20]. The azurin involvement in bacterial-cancer cell interactions was demonstrated for CRC through experiments of exclusion, competition, and replacement. These results paved the way to innovative ideas for potential adjuvant therapies. Moreover, regarding CRC pathogenesis, an involvement of the gut microbiota in CRC development and therapy response is documented [46]. Accordingly, increasing evidences from metagenomic analyses highlighted a microbial imbalance (dysbiosis) typical of the CRC patient gut, that supports a pathological state [47]. Recently, the analysis of exopolysaccharides (EPSs) activity from Pseudomonas sp. strains against HT-29 CRC cell line, confirmed that microbial molecules are able to carry out anti-tumor activity in CRC through apoptosis mechanism [48].
Surprisingly, there is a considerable lack of investigation about azurin gene and protein in biological samples of human origin, especially related to the occurrence of cancer. An enrichment in azurin gene content of the microbiota in the cancer context, could suggest that the azurin is able to confer a selective advantage to such microorganisms. This evidence, in turn, could be leveraged to induce enrichment in azurin-harbouring microorganisms, potentially leading to an in-situ production of azurin able to have a pharmacological value for the host.
In this work, samples collected from CRC patients and healthy controls [49] were analysed for the presence of specific bacterial groups related to the production of azurin. The analysis was conducted on High-throughput sequencing (HTS) data previous collected for the FAS project “MICpROBIMM” [49], and via quantitative amplification protocol by real-time PCR. Six primer pairs specifically designed to target the azurin gene from different bacterial groups (mainly different Pseudomonas species) were designed and validated through end-point PCR. Four of the validated primer pairs were used for quantitative real-time PCR in samples collected from 10 colorectal cancer patients (saliva, stool and biopsy samples), in comparison with samples collected from 10 healthy subjects (in saliva and stool samples). Our aim was therefore to evaluate the presence of azurin gene in the available samples, to evaluate any differences in relation to the sample type and also in relation to the bacterial groups producing azurin.
The primer sets for azurin amplification were designed based on the conserved regions obtained from the alignment of the azurin nucleotide sequences using the BioEdit software version 7.2.6 [50]. For the primer design the following parameters were considered: 18–25 base pairs length, a similar denaturation temperature of the primer forward with the primer reverse in each couple (Tm) and the nucleotide composition of the primer sequences, whose GC content was established as being about 60% [51]. A total of 6 primer pairs were designed (Table 1).
| PRIMER PAIRS | ||||||
| Pseudomonas aeruginosa | Pseudomonas group 12 | Pseudomonas group 1 | Pseudomonas group 4 | Aeromonas | Pseudomonas group 5 | |
| Primer forward (F) | 5′-GGCTGCCGAGTGCTCGG -3′ | 5′-AAGAGCTGCAAGACCTT -3′ | 5′-ATGAYTCGCAAGCTTGTA -3′ | 5′-ATCGACAAGAGCTGCAAG -3′ | 5′-CATGGGYCACAACTGGG -3′ | 5′-ATGTTTGCCAAACTCGT -3′ |
| Primer reverse (R) | 5′-CCCTGCATGTCGGCGGC -3′ | 5′-GGGAAGGAGCAGAAGA -3′ | 5′-GCGCCTTTCATCATGCT -3′ | 5′-CCTTTCATCATCGAGATRT -3′ | 5′-CCGGGGAAGGAGCAGAA -3′ | 5′-GTTRTGVCCCATSACGTT -3′ |
| Tm primer F | 62 °C | 50 °C | 50 °C | 54 °C | 56 °C | 48 °C |
| Tm primer R | 62 °C | 50 °C | 52 °C | 52 °C | 57 °C | 50 °C |
| Annealing temperature in PCR | 62 °C | 50 °C | 50 °C | 52 °C | 56 °C | 48 °C |
| Target | P. aeruginosa | P. fluorescens, P. cichorii, P. coronafaciens, P. delhiensis, P. pseudoalcaligenes, P. mendocina | P. syringae, P. savastanoi, P. amygdali, P. cannabina | P. fluorescens, P. marginalis, P. putida, P. simiae, P. trivalis, P. costantinii | A. aquatica, A. caviae, A. fluvialis, A. hydrophila, A. rivuli, A. salmonicida. | P. fluorescens, P. sp., P. cedrina, P. chlororaphis, P. protegens |
| N. of target sequences | 2290 | 98 | 268 | 221 | 195 | 143 |
| Fragment length (bp) | 177 | 275 | 431 | 305 | 216 | 201 |
The 6 primer pairs were validated through end-point PCR on a panel of bacterial
strains that were present in the laboratory collections of the Dept. of Biology,
University of Florence. The strains used are listed in Table 2 (Ref. [52, 53, 54]).
The amplification with the designed primer pairs were performed in 20-
| Strain | Source | Taxonomy | Reference |
| 3.33 | Acquarossa river, red epilithon | Pseudomonas sp. | [52] |
| 5.14 | Acquarossa river, black epilithon | Pseudomonas chloraphis | [52] |
| 11.7 | Infernaccio waterfalls, red epilithon | Pseudomonas fluorescens/P. protegens | [52] |
| 2.13 | Acquarossa river, red epilithon | Pseudomonas fluorescens/P. protegens | [52] |
| 14.6 | Infernaccio waterfalls, red epilithon | Pseudomonas moraviensis | [52] |
| 3.18 | Acquarossa river, red epilithon | Aeromonas sp. | [52] |
| 11.45 | Infernaccio waterfalls, red epilithon | Aeromonas sp. | [52] |
| 11.47 | Infernaccio waterfalls, red epilithon | Aeromonas sp. | [52] |
| Ep_R1 | Echinacea purpurea roots | Pseudomonas sp. | [53, 54] |
| ATCC 9027 | Department of Health Sciences, University of Florence, Laboratory of Prof. A. Lo Nostro. | Pseudomonas aeruginosa | |
| 4739 | Department of Health Sciences, University of Florence, Laboratory of Prof. A. Lo Nostro. | Pseudomonas aeruginosa | |
| 5254 | Department of Health Sciences, University of Florence, Laboratory of Prof. A. Lo Nostro. | Pseudomonas aeruginosa |
Real-time PCR reactions were performed on a panel of samples collected from CRC, and healthy subjects (HC, Table 3, Ref. [49]), in a QuantStudioTM 7 apparatus (Applied Biosystems, Waltham, MA, USA), using a SYBR Green mix (Thermo Fisher Scientific), and according to the protocol described in Checcucci et al. [55]. Four out of the six primers pairs were chosen for real-time PCR on samples listed in Table 3, namely: “Pseudomonas group 4”, “Pseudomonas group 5”, “Pseudomonas group 12” and “Pseudomonas aeruginosa”. The choice was made on the basis of the end-point PCR results. For each primers pair, a standard curve was constructed starting from the corresponding azurin amplicon, that was previously quantified. Amplicon quantifications were performed with GelQuant.NET software provided by biochemlabsolutions.com (http://biochemlabsolutions.com/GelQuantNET.html). The copy number of azurin amplicon in each point of the standard curve and in samples giving a positive signal was calculated using the free software “dsDNA copy number calculator” (https://cels.uri.edu/gsc/cndna.html).
| Sample name | CRC patient/ healthy subject | Sample type |
| CM7_S | CRC patient | Saliva |
| CM7_F | CRC patient | Stool |
| CM7_B | CRC patient | Biopsy |
| CM8_S | CRC patient | Saliva |
| CM8_F | CRC patient | Stool |
| CM8_B | CRC patient | Biopsy |
| CM10_S | CRC patient | Saliva |
| CM10_F | CRC patient | Stool |
| CM10_B | CRC patient | Biopsy |
| CM11_S | CRC patient | Saliva |
| CM11_F | CRC patient | Stool |
| CM11_B | CRC patient | Biopsy |
| CM18_S | CRC patient | Saliva |
| CM18_F | CRC patient | Stool |
| CM18_B | CRC patient | Biopsy |
| CM19_S | CRC patient | Saliva |
| CM19_F | CRC patient | Stool |
| CM19_B | CRC patient | Biopsy |
| CM20_S | CRC patient | Saliva |
| CM20_F | CRC patient | Stool |
| CM20_B | CRC patient | Biopsy |
| CM22_S | CRC patient | Saliva |
| CM22_F | CRC patient | Stool |
| CM22_B | CRC patient | Biopsy |
| CM23_S | CRC patient | Saliva |
| CM23_F | CRC patient | Stool |
| CM23_B | CRC patient | Biopsy |
| CM24_S | CRC patient | Saliva |
| CM24_F | CRC patient | Stool |
| CM24_B | CRC patient | Biopsy |
| CFP1_S | healthy subject | Saliva |
| CFP1_F | healthy subject | Stool |
| CFP2_S | healthy subject | Saliva |
| CFP2_F | healthy subject | Stool |
| CFP3_S | healthy subject | Saliva |
| CFP3_F | healthy subject | Stool |
| CFP4_S | healthy subject | Saliva |
| CFP4_F | healthy subject | Stool |
| CFP6_S | healthy subject | Saliva |
| CFP6_F | healthy subject | Stool |
| CFP7_S | healthy subject | Saliva |
| CFP7_F | healthy subject | Stool |
| CFP8_S | healthy subject | Saliva |
| CFP8_F | healthy subject | Stool |
| CFP9_S | healthy subject | Saliva |
| CFP9_F | healthy subject | Stool |
| CFP10_S | healthy subject | Saliva |
| CFP10_F | healthy subject | Stool |
| CFP11_S | healthy subject | Saliva |
Raw sequencing data obtained in Russo et al. [49] were downloaded from the SRA database using fastq-dump command in the SRA-Toolkit and using the accession numbers (SRR) associated to the single samples included in the BioProject at accession PRJNA356414 (SRA accession SRP094636). According to Vitali et al. [56], the obtained sequence fastq files were pre-processed as follows: (i) sequencing primer was removed with CUTADAPT [57], while the low-quality ends at 5’ was removed using Sickle with a quality cutoff of 18. (ii) MICCA v1.7.2 [58] was then used for all analysis steps from pre-treated sequences to final Amplicon Sequence Variance (ASV) picking. The latter was performed using the UNOISE3 algorithm [59] and taxonomy was assigned using the VSEARCH [60] algorithm and the SILVA 132 database [61].
According to the presence of azurin gene in the different bacterial groups [62], the following bacterial taxa were searched and quantified in each sample from both healthy and CRC patients: Pseudomonas sp., (Phylum Proteobacteria), Akkermansiaceae, Opitutaceae (Phylum PVC), Holophagales, Vicinamibacteria (Phylum Acidobacteria), and the phyla Bacteroidetes, Chloroflexi and Actinobacteria.
R software version 4.2 was used to analyse these bacterial group subsets (i.e., their distribution among samples) and the whole HTS dataset, using vegan [63] and ggplot2 [64] for diversity analysis. Data of the azurin-subset community were normalized with z-score transformation prior to distance matrix calculation using the Euclidean dissimilarity index. This transformation was necessary to account for the large difference in relative abundance between the selected variables, some at the species-genus level while others at the level of entire phyla. Data of the whole HTS dataset were normalized with relative abundance transformation prior to distance matrix calculation using the Bray-Curtis dissimilarity index. In both cases, distance matrixes were used for diversity evaluation with non-metric Multidimensional Scaling (nMDS), as well their correlation was measured with Mantel test in the vegan package (9999 permutations, Pearson correlation).
End-point amplifications with the six designed primer sets on the panel of bacterial strains listed in Table 2, gave different results. Fig. 1 and Table 4 summarizes the outcome of the amplifications.
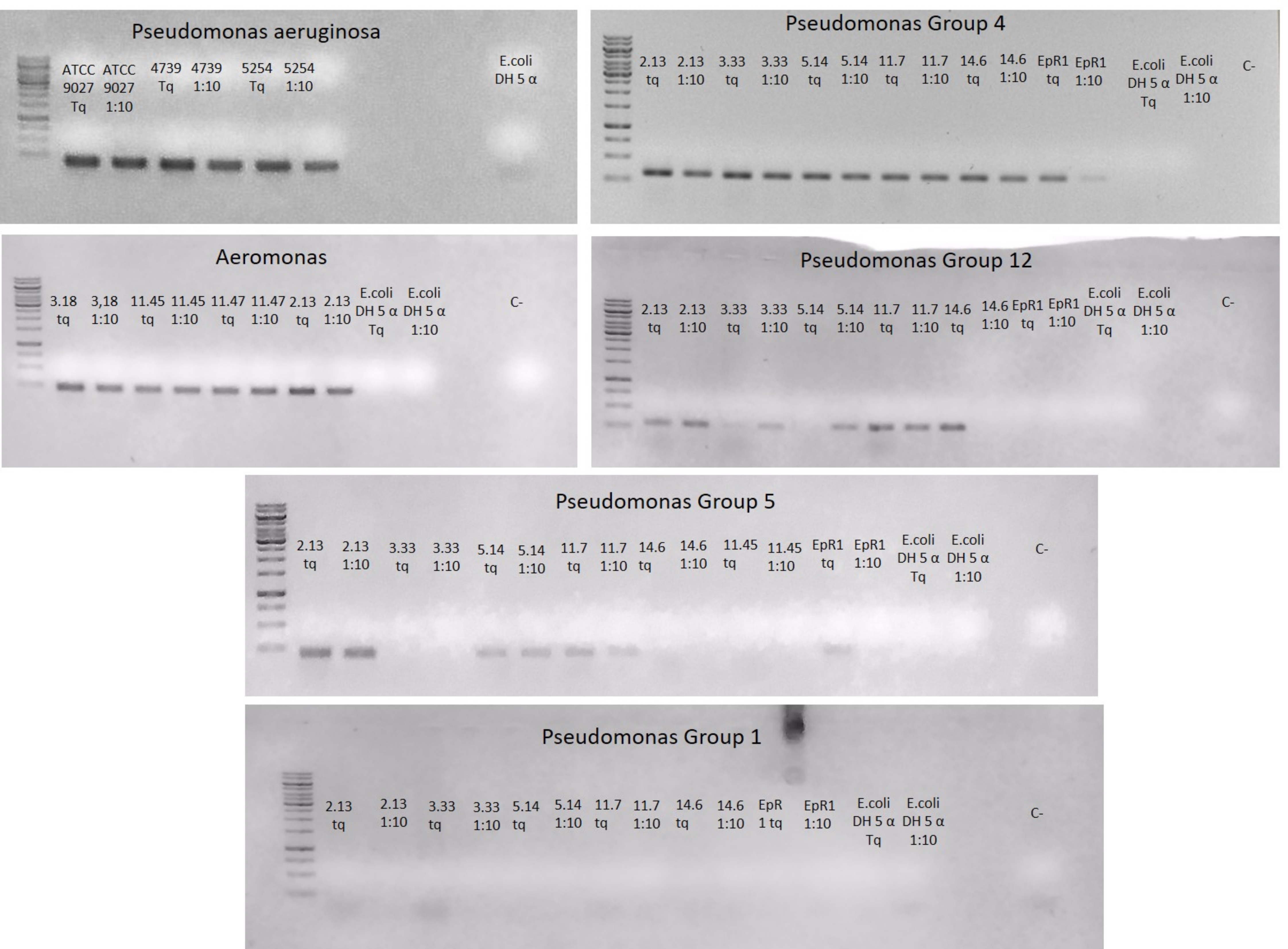 Fig. 1.
Fig. 1.End-point PCR results using the six primer pairs designed in this work. DNA were used both not diluted (Tq) and diluted (1:10). Negative controls indicated as “C-”.
| Pseudomonas aeruginosa | Pseudomonas group 12 | Pseudomonas group 1 | Pseudomonas group 4 | Aeromonas | Pseudomonas group 5 | |
| 3.33 | - | YES | NO | YES | - | NO |
| 5.14 | - | YES | NO | YES | - | YES |
| 11.7 | - | YES | NO | YES | - | YES |
| 2.13 | - | YES | NO | YES | YES | YES |
| 14.6 | - | YES | NO | YES | - | NO |
| 3.18 | - | - | - | - | YES | - |
| 11.45 | - | - | - | - | YES | - |
| 11.47 | - | - | - | - | YES | - |
| Ep_R1 | - | NO | NO | YES | - | - |
| ATCC 9027 | YES | - | - | - | - | - |
| 4739 | YES | - | - | - | - | - |
| 5254 | YES | - | - | - | - | - |
| DH5 |
NO | NO | NO | NO | NO | NO |
An amplicon of the expected size was detected in all samples except for the primer pair “Pseudomonas group 1” (Fig. 1). For this reason, this primer pair was excluded from the further analyses. Each primer pair specifically amplified the azurin gene fragment of the bacterial strains belonging to its specific group, with the exception of primer pairs “Aeromonas”, which gave amplicons not only from Aeromonas sp. strains 3.18, 11.45, and 11.47, but also from strains 2.13 affiliated to the genus Pseudomonas (Fig. 1). For this reason, also this primer pair was removed from the subsequent analysis.
The nucleotide sequence of seven randomly chosen end-point amplicon was determined and submitted to Blastx online databases for comparison with the available deposited sequences, in order to verify whether not only the fragment length, but also the amplified sequence corresponded to the azurin encoding gene. All the sequences retrieved at the lowest t-value the azurin gene, thus confirming the efficacy/specificity of the designed primer pairs and are available in the supplementary material.
Once the azurin amplicons from each primers pair were quantified, a standard curve was constructed for each couple. Table 5 (Ref. [52, 53, 54]) shows the bacterial strains from which DNA was used for azurin amplification and standard curve construction, and also the azurin copy number calculation for the standard curves prepared for each primers pair selected for real-time PCR.
| Primer pairs | Bacterial strain | Reference | Amplicon concentration | No. copies of amplicon in 1 µL |
| Pseudomonas aeruginosa | P. aeruginosa ATCC 9027 | 40 ng/µL | 5.26 | |
| Pseudomonas group 12 | 2.13 | [52] | 1 ng/µL | 3.37 |
| Pseudomonas group 5 | 5.14 | [52] | 0.2 ng/µL | 9 |
| Pseudomonas group 4 | Ep_R1 | [53, 54] | 0.2 ng/µL | 6.2 |
Real-time reactions revealed the presence of the azurin gene in both CRC and healthy patients as shown in Table 6. The primer pairs “Pseudomonas group 4” and “Pseudomonas aeruginosa” were not effective in detecting the azurin gene in the samples analysed. Primer pair “Pseudomonas group 5” gave positive results for the azurin gene in 7 CRC samples (4 samples of stool and 3 sample of biopsy) and in 2 healthy subjects (stool samples). Finally, primer pair “Pseudomonas group 12” successfully amplified the azurin coding gene from 8 CRC patients (7 stool samples and 1 biopsy) and 5 stool samples collected from healthy subjects (Table 6).
| n. row | Chloroflexi | Opitutaceae | Akkermansia | Pseudomonas | Bacteroidetes | Actinobacteria | Acidobacteria. Holophagales | Pseudomonas group 4 | Pseudomonas aeruginosa | Pseudomonas group 12 | Pseudomonas group 5 | ||
| 1 | CM7_S | Saliva | 0.00 | 0.00 | 0.00 | 0.00 | 20.93 | 10.97 | 0.00 | ||||
| 2 | CM7_F | Stool | 0.00 | 0.00 | 0.02 | 0.00 | 62.00 | 0.08 | 0.00 | 300 | 9 | ||
| 3 | CM7_B | Biopsy | 0.00 | 0.00 | 0.00 | 0.00 | 16.08 | 5.74 | 0.00 | 9 | |||
| 4 | CM8_S | Saliva | 0.00 | 0.00 | 0.00 | 0.00 | 22.73 | 4.94 | 0.00 | ||||
| 5 | CM8_F | Stool | 0.00 | 0.00 | 0.17 | 0.00 | 68.17 | 0.35 | 0.00 | 300 | 90 | ||
| 6 | CM8_B | Biopsy | 0.00 | 0.00 | 0.42 | 0.05 | 37.84 | 1.17 | 0.00 | 30 | |||
| 7 | CM10_S | Saliva | 0.01 | 0.00 | 0.00 | 0.00 | 28.77 | 4.62 | 0.00 | ||||
| 8 | CM10_F | Stool | 0.00 | 0.00 | 0.01 | 0.00 | 32.94 | 0.58 | 0.00 | 9 | |||
| 9 | CM10_B | Biopsy | 0.00 | 0.00 | 0.00 | 0.00 | 18.80 | 2.72 | 0.00 | ||||
| 10 | CM11_S | Saliva | 0.00 | 0.00 | 0.00 | 0.00 | 26.73 | 6.57 | 0.00 | ||||
| 11 | CM11_F | Stool | 0.00 | 0.00 | 1.42 | 0.00 | 69.22 | 0.28 | 0.00 | 300 | 90 | ||
| 12 | CM11_B | Biopsy | 0.00 | 0.00 | 0.99 | 0.05 | 24.64 | 9.83 | 0.00 | ||||
| 13 | CM18_S | Saliva | 0.00 | 0.00 | 0.00 | 0.00 | 19.51 | 1.67 | 0.00 | ||||
| 14 | CM18_F | Stool | 0.01 | 0.00 | 3.73 | 0.00 | 50.36 | 1.25 | 0.00 | ||||
| 15 | CM18_B | Biopsy | 8.22 | 0.00 | 0.02 | 2.63 | 10.17 | 11.03 | 0.00 | ||||
| 16 | CM19_S | Saliva | 0.00 | 0.00 | 0.00 | 0.00 | 16.45 | 18.82 | 0.00 | ||||
| 17 | CM19_F | Stool | 0.00 | 0.00 | 4.09 | 0.00 | 80.82 | 0.16 | 0.00 | 30 | |||
| 18 | CM19_B | Biopsy | 7.43 | 0.08 | 0.15 | 2.15 | 10.39 | 8.51 | 0.15 | 9 | |||
| 19 | CM20_S | Saliva | 0.01 | 0.00 | 0.01 | 0.00 | 27.25 | 2.71 | 0.00 | ||||
| 20 | CM20_F | Stool | 0.01 | 0.00 | 5.80 | 0.00 | 70.98 | 0.79 | 0.00 | 3 | |||
| 21 | CM20_B | Biopsy | 6.83 | 0.00 | 0.00 | 2.05 | 7.59 | 8.71 | 0.49 | 9 | |||
| 22 | CM22_S | Saliva | 0.00 | 0.00 | 0.00 | 0.00 | 28.20 | 3.41 | 0.00 | ||||
| 23 | CM22_F | Stool | 0.00 | 0.00 | 0.00 | 0.00 | 63.68 | 0.38 | 0.00 | ||||
| 24 | CM22_B | Biopsy | 1.61 | 0.05 | 0.00 | 0.16 | 35.47 | 1.76 | 0.04 | ||||
| 25 | CM23_S | Saliva | 0.00 | 0.00 | 0.00 | 0.00 | 22.90 | 7.39 | 0.00 | ||||
| 26 | CM23_F | Stool | 0.00 | 0.00 | 5.26 | 0.00 | 46.63 | 0.12 | 0.00 | 30 | |||
| 27 | CM23_B | Biopsy | 4.50 | 0.00 | 0.03 | 0.79 | 21.52 | 5.29 | 0.10 | ||||
| 28 | CM24_S | Saliva | 0.01 | 0.00 | 0.01 | 0.00 | 11.53 | 11.04 | 0.00 | ||||
| 29 | CM24_F | Stool | 0.01 | 0.00 | 0.00 | 0.00 | 52.03 | 0.35 | 0.00 | 30 | |||
| 30 | CM24_B | Biopsy | 3.57 | 0.00 | 0.00 | 0.62 | 44.20 | 4.54 | 0.00 | ||||
| 31 | CFP1_S | Saliva | 0.00 | 0.00 | 0.02 | 0.00 | 27.14 | 5.06 | 0.00 | ||||
| 32 | CFP1_F | Stool | 0.00 | 0.00 | 4.70 | 0.00 | 68.68 | 0.28 | 0.00 | ||||
| 33 | CFP2_S | Saliva | 0.00 | 0.00 | 0.00 | 0.00 | 39.19 | 3.83 | 0.00 | ||||
| 34 | CFP2_F | Stool | 0.00 | 0.00 | 0.02 | 0.00 | 70.82 | 0.44 | 0.00 | 300 | 90 | ||
| 35 | CFP3_S | Saliva | 0.00 | 0.00 | 0.00 | 0.00 | 38.22 | 6.30 | 0.00 | ||||
| 36 | CFP3_F | Stool | 0.00 | 0.00 | 0.02 | 0.00 | 44.35 | 0.25 | 0.00 | 30 | |||
| 37 | CFP4_S | Saliva | 0.00 | 0.00 | 0.00 | 0.00 | 33.96 | 0.99 | 0.00 | ||||
| 38 | CFP4_F | Stool | 0.00 | 0.00 | 0.00 | 0.00 | 25.00 | 0.00 | 0.00 | 300 | |||
| 39 | CFP6_S | Saliva | 0.00 | 0.00 | 0.01 | 0.00 | 18.92 | 11.43 | 0.00 | ||||
| 40 | CFP6_F | Stool | 0.00 | 0.00 | 0.00 | 0.00 | 21.01 | 0.50 | 0.00 | ||||
| 41 | CFP7_S | Saliva | 0.01 | 0.00 | 0.01 | 0.00 | 23.51 | 3.26 | 0.00 | ||||
| 42 | CFP7_F | Stool | 0.00 | 0.00 | 4.30 | 0.00 | 20.70 | 1.20 | 0.00 | 30 | |||
| 43 | CFP8_S | Saliva | 0.00 | 0.00 | 0.00 | 0.00 | 16.99 | 1.97 | 0.00 | ||||
| 44 | CFP8_F | Stool | 0.01 | 0.00 | 0.01 | 0.25 | 64.33 | 0.39 | 0.00 | ||||
| 45 | CFP9_S | Saliva | 0.00 | 0.00 | 0.00 | 0.00 | 31.37 | 2.80 | 0.00 | ||||
| 46 | CFP9_F | Stool | 0.00 | 0.00 | 0.14 | 0.04 | 69.50 | 0.13 | 0.00 | 300 | |||
| 47 | CFP10_S | Saliva | 0.00 | 0.00 | 0.00 | 0.00 | 23.12 | 5.62 | 0.00 | ||||
| 48 | CFP10_F | Stool | 0.00 | 0.00 | 1.10 | 0.00 | 21.48 | 0.72 | 0.00 | 9 | |||
| 49 | CFP11_S | Saliva | 0.01 | 0.00 | 0.00 | 0.00 | 8.62 | 11.00 | 0.00 | ||||
| 50 | CFP11_F | Stool | 0.00 | 0.00 | 1.03 | 0.00 | 20.40 | 0.11 | 0.00 |
The relative abundances of Pseudomonas sp., (Phylum Proteobacteria), Akkermansiaceae, Opitutaceae (Phylum PVC), Holophagales, Vicinamibacteria (Phylum Acidobacteria), and the phyla Bacteroidetes, Chloroflexi and Actinobacteria, were evaluated in each sample through the analysis of the HTS data reported in Russo et al. [49]. Data obtained are resumed in Fig. 2 and Fig. 3 (Ref. [49]). It was worth of noticing that Holophagales, Opitutaceae, and Chloroflexi were detected only in samples of biopsy from CRC patients, and not in stool and saliva samples from either healthy or CRC subjects (Fig. 2).
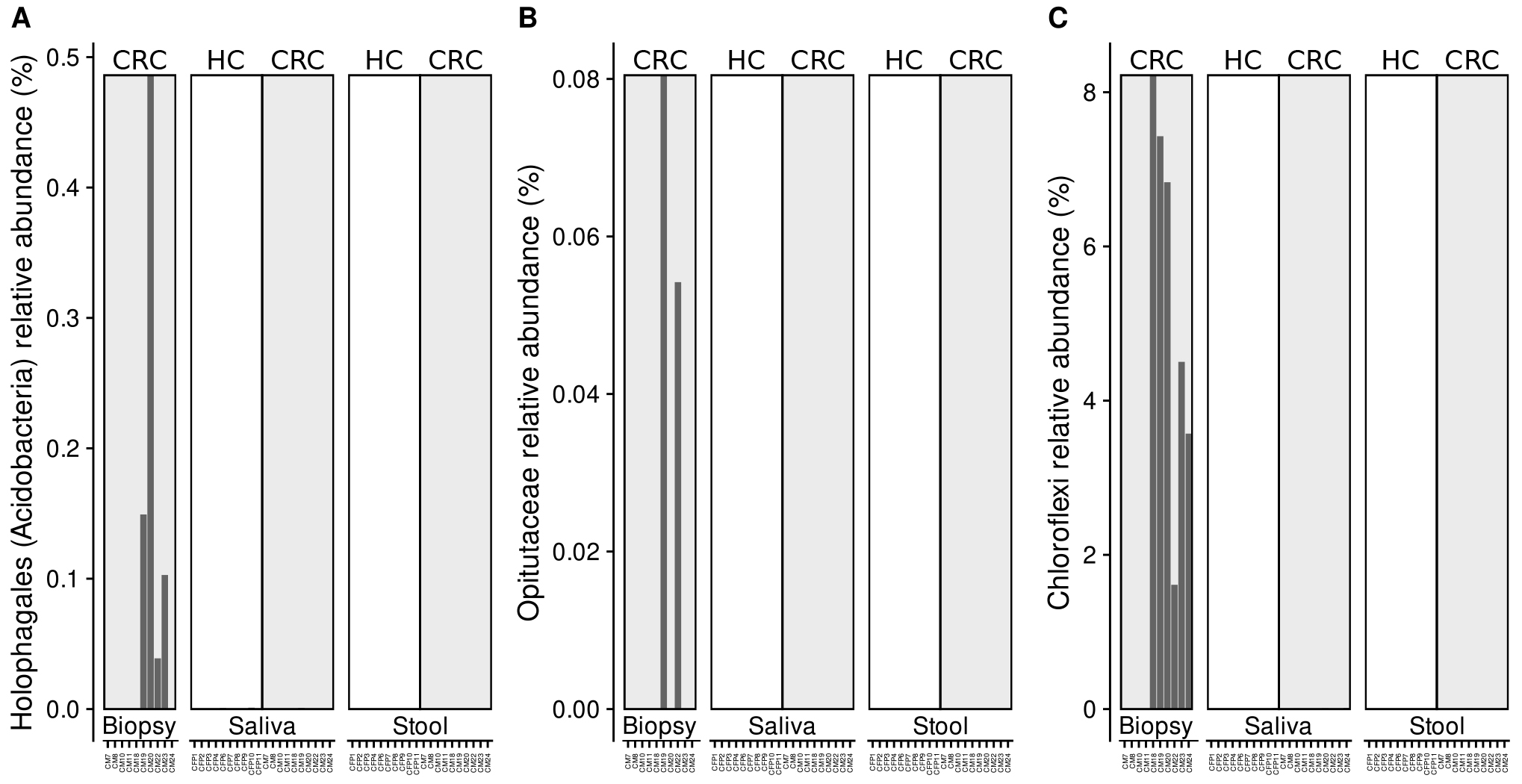 Fig. 2.
Fig. 2.Relative abundance of Holophagales (A), Opitutaceae (B) and Chloroflexi (C) reads detected in the samples analysed through HTS technique.
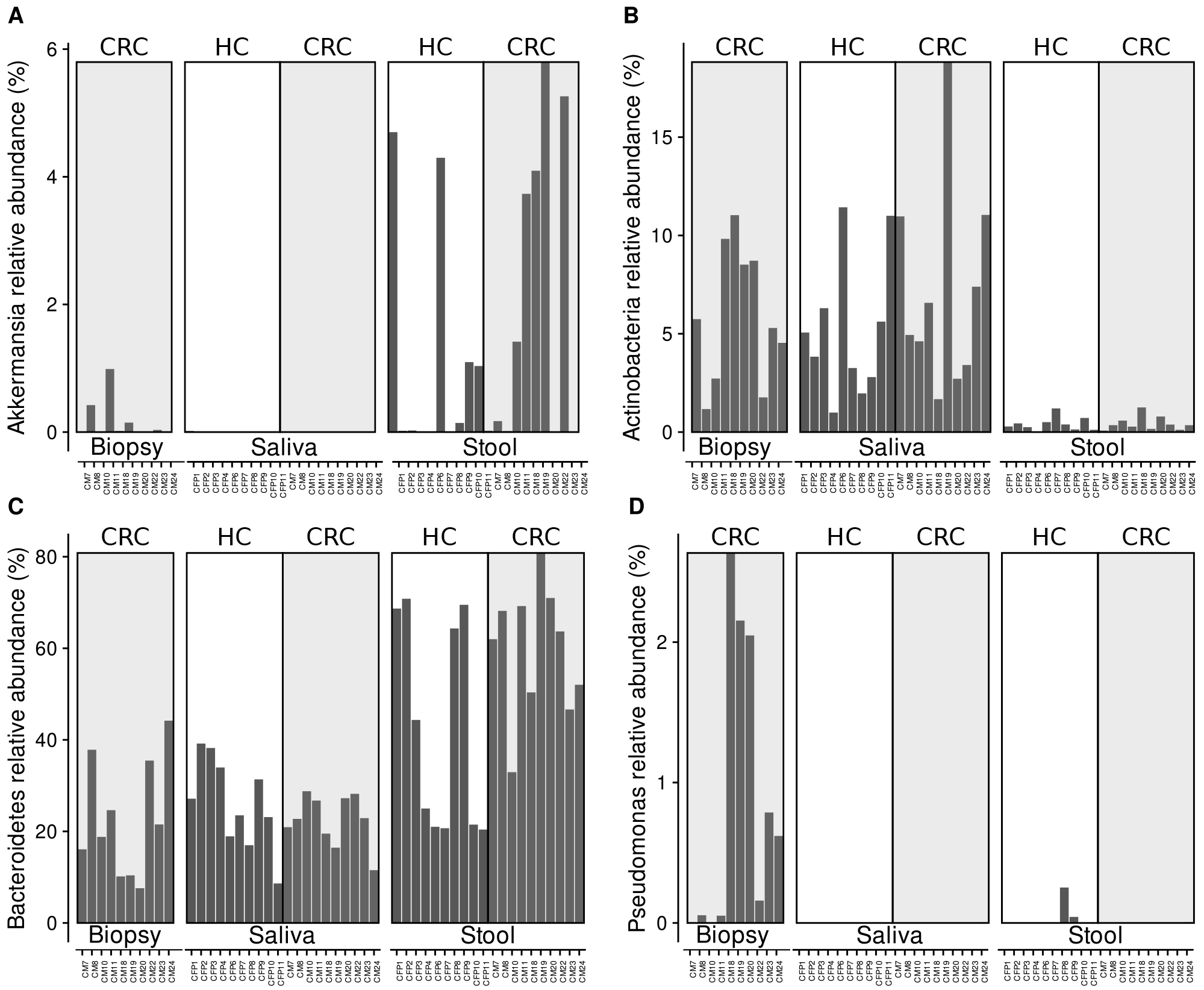 Fig. 3.
Fig. 3.Relative abundance of Akkermansia (A), Actinobacteria (B), Bacteroidetes (C), and Pseudomonas (D) reads detected in the samples analysed through HTS technique [49].
Also, the relative abundance of Pseudomonas sp. (Fig. 3D) suggest that this bacterial genus is highly represented in the biopsy samples of CRC patients, while it is about completely lacking in all the other samples, except for two stool samples from healthy controls (samples CFP 8 and 9), showing a low Pseudomonas sp. abundance. Interestingly, while Akkermansia sp. is highly represented in stool samples, and slightly more abundant in CRC patients respect to healthy controls (Fig. 3A—difference not statistically significant with p-value = 0.4057), the relative abundance of Actinobacteria (Fig. 3B) revealed that this bacterial group is highly represented in saliva samples of both CRC and healthy subjects (no statistically significant difference of the Actinobacteria relative abundance between CRC and healthy subjects in Kruskal-Wallis test), as well as in CRC biopsy samples.
Finally, Bacteroidetes relative abundance revealed that this group is generally more represented in stool than in saliva and suggested a higher abundance in CRC patients’ stool than in healthy subject, even though the difference failed to reach a statistically significance with a p value = 0.13.
Data obtained were further inspected with ordination analysis, using nMDS on the
subset of bacterial groups harbouring the azurin gene (Fig. 4A) and on the whole
set of HTS data (Fig. 4B). The aim of this analysis was investigating the extent
to which the taxa subset harbouring azurin gene was related to the whole HTS
dataset. To this aim, we firstly investigated community diversity using
ordination analysis. In both cases, the ordination, provided a good
representation of the real distances among samples as indicated by a stress value
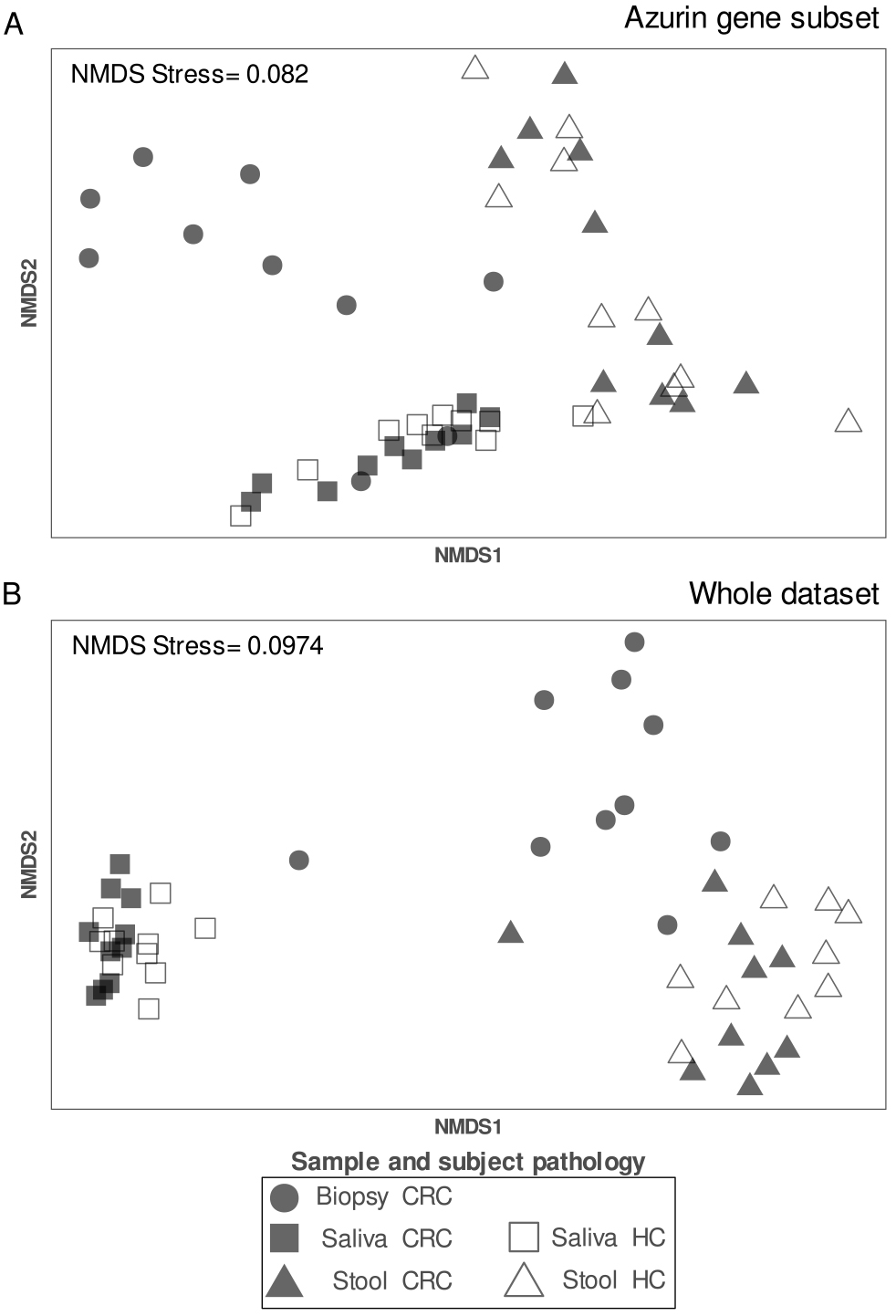 Fig. 4.
Fig. 4.Ordination plot resulting from nMDS based on Bray-Curtis dissimilarity on the (A) subset of bacterial groups harbouring the azurin gene and (B) the whole HTS dataset. Plot is annotated with stress values (i.e., a measure of the similarity of distances between samples in the reduced data of the ordination, respect to the distances between samples of the complete dataset).
Finally, we asked the degree of correlation of the diversity between the two
datasets using Mantel test (9999 permutations, Pearson correlation measure) to
obtain the overall correlation between the Euclidean distance matrix calculated
on the “azurin-related” subset, and the Bray-Curtis distance matrix calculated
on the whole HTS dataset. Mantel test showed a moderate but highly significant
correlation between the two matrices (Mantel Pearson r = 0.3554, p-value
From decades, the research of new anticancer molecules has raised great interest within the scientific community. In particular, microbial secondary metabolites are under the spotlight as promising candidates of innovative anticancer approaches. Interestingly, some of them are successfully applied as therapy for the cancer treatment as well (recently reviewed by Mohammadi et al. [20] resuming bacterial secondary metabolites which have been clinically used or are undergoing clinical trial). Among them, azurin synthesized from P. aeruginosa MTCC 2453, was demonstrated to induce tumor regression. In vivo experiments using Dalton’s lymphoma mice model demonstrated that azurin is able to induce apoptosis [32]. Likewise, anticancer activity of azurin was demonstrated in vitro in gastrointestinal cancers cell lines [65]. These evidences brought to the investigation on the possible optimization of azurin formulation to improve its efficacy [13, 66, 67], such as the drug delivery into chitosan nanoparticles [65].
In the field of CRC, given its known environmental pathogenesis (i.e., related to unhealthy diet or lifestyle) and given the known abundance and complexity of the microbial community in its target organ, the use of bacterial and bacterial-derived (secondary) metabolites is particularly promising for innovative treatment options [68].
Stemming from those premises, in this work we have evaluated the presence of azurin genes in different biological samples (i.e., saliva, stool, and biopsy) from CRC patients and healthy controls, investigating on the possible relationship existing between the disease occurrence and the different microbiota body compartments associated. Moreover, as the presence of azurin genes has been correlated to specific bacterial groups of environmental and gut origin [62], it might help in a future translation to clinical practice.
In this scenario, the construction of a valid amplification protocol for both bacterial pure cultures and biological samples is mandatory. Indeed, primer pairs are not easily available in scientific literature, and little is known about the amplification protocols of the azurin coding gene. To this purpose, our study was at first aimed at creating valid primer sets for a successful amplification and/or quantification of the gene encoding azurin in different samples. Interestingly, in a recent work [20] the amplification of azurin gene was applied to detect the differences occurring among bacterial isolates obtained from burn patients with P. aeruginosa infection. The authors demonstrated that the expression of azurin gene was significantly higher in the P. aeruginosa strains isolated from the blood of patients with systemic infections, hypothesizing an azurin role in the increased pathogenicity of the strains isolated in those patients, as well as in their ability to escape the host immune system and disseminate into the bloodstream [20].
As a further step, we analyzed HTS results obtained from previous research [49] in the light of the evidences from Gammuto et al. [62], this last specifically evaluating the phylogenetic distribution of azurin gene. These results allowed us to focus on a defined set of taxa known for harbouring azurin genes, rather than the entire community. This analysis revealed that bacterial taxa Holophagales, Opitutaceae, and Chloroflexi were only retrieved in CRC biopsy samples (Fig. 2). Although no evidence in scientific literature correlates the presence of Holophagales and Opitutaceae with tumor conditions, thus avoiding any speculation on the possible relationship of this pathology with these bacterial groups, the phylum Chloroflexi seems to be involved in the microbiota dysbiosis, which plays a documented key role in carcinogenesis [69].
In agreement with these observations, it has been demonstrated that the species Chloroflexus aurantiacus encodes for four auracyanins [70], which belong to the type 1 (T1) blue copper protein family, also known as cupredoxins; these auracyanins play a role as periplasmatic electron carriers [71]. Interestingly, it has been also highlighted that the Chloroflexi was one of the bacterial phyla that were significantly enriched in tissues of subsolid nodules from patients with lung adenocarcinoma [69]. A similar trend was also found in other tumor types; accordingly, microbiota alterations are associated with the different histological stages of gastric cancer [72], and higher Chloroflexi abundance was found in CRC biopsy samples [73]. Therefore, our observations on the Chloroflexi presence in CRC biopsies (Fig. 2) could direct future studies in the investigation of the possible correlations between this bacterial phylum and the azurin presence, and also in the putative relationship with cancer progression.
Another interesting result is that of the presence of Pseudomonas sp. bacteria only in biopsy samples, with an almost total lack in saliva and stool samples of CRC patients and healthy controls (Fig. 3). These results may be partially in agreement with the scientific literature. Indeed, despite Pseudomonas aeruginosa is a common colonizer of the human intestine in cancer patients facing particular clinical conditions (i.e., hospitalization, immunosuppression, antibiotic treatment, surgery, severe trauma), this bacterium increases in the number and also become more virulent and damaging to the intestinal epithelium upon surgery, injury, and severe stress [74]. Additionally, P. aeruginosa isolated from humans can induce intestinal pathology and cancer-related epithelial phenotypes in genetically predisposed model hosts [74]. On the other side, results from real time PCR revealed the azurin presence from Pseudomonas sp. not only in biopsies but also in stool samples from both CRC and healthy patients. This incongruence might be related to the different resolution of the used molecular techniques. Indeed, HTS is aimed at a taxonomical analysis on 16S rRNA genes, while real time primers were designed on the sequence of the azurin genes from different Pseudomonas sp. groups. Thus, we cannot exclude a priori the possibility that our azurin primers might have amplified the gene from other taxonomical groups.
Recently, Yu and co-authors [75] observed a higher relative abundance of Actinobacteria, Acidobacteria, and Chloroflexi in the gut microbiota of myeloid leukemia compared to healthy controls; this observation confirms an alteration of the microbiota in tumor conditions. This enrichment is in line with our experimental findings, not only about Chloroflexi, but also regarding the presence of Actinobacteria in CRC biopsies (Fig. 3). Moreover, our analysis supports higher relative abundances of Actinobacteria in saliva samples of both healthy and CRC patients respect to stool samples. These data partially agree with results obtained analyzing other diseases; to this purpose, Actinobacteria, together with other phyla, were more abundant in samples from patients with periodontitis instead of in healthy controls [76].
Finally, we constructed a subset of azurin gene harbouring bacteria and tested in beta diversity analysis. The subset was intrinsically very variable in term of taxonomic level of the variables (i.e., some represented phyla, some genera) and consequently in terms of numeric count. Nevertheless, we found that this subset was able to fairly depict the entire community diversity as seen with HTS. Although this evidence is necessarily preliminary and observational due to the low number of samples involved and the strong taxonomic variability of the created subset, it still represents interesting evidence worthy of further investigation.
Overall, despite the study limitations such as the low number of recruited patients, in this work for the first time, six primer pairs targeting the bacterial azurin gene were specifically designed, tested, and validated via end-point and real-time PCR. Two out of the six designed primers were efficient in detecting the azurin gene in real-time PCR on human samples from CRC and healthy patients. Despite the lack of statistically significant differences between healthy and diseased patients, HTS data analysis on seven bacterial groups harbouring azurin gene in their genome highlighted a kind of “preferential” enrichment of these bacterial groups in some samples than in others (i.e., Pseudomonas sp., Holophagales, Opitutaceae and Chloroflexi in CRC biopsy, Akkermansia sp. in stool samples, Actinobacteria in saliva samples). Moreover, the subset of azurin gene-harbouring bacterial groups was representative of the entire community diversity, despite this observation needs further investigations and confirmation, on a wider number of samples.
The partial 16S rRNA gene sequence data analysed in this study were previously generated and published (Russo et al., 2018), and are available in the GenBank Sequence Read Archive with the accession number PRJNA356414.
Conceptualization—RF MI, CC; methodology—CC, MI; software—FV; validation—CC, MI; formal analysis—CC, FV, MI; investigation—RF, AA, CC, LG, MI, FV; resources—RF, AA; data curation—CC, FV, MI; writing - original draft preparation—CC, MI, FV; writing - review and editing—RF, CC, LG, MI, FV, AA, AT; supervision and project administration—RF. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
The study has received the local ethics (Toscana - area vasta centro) committee approval (CE: 11166_spe), and informed written consent has been obtained from each participant.
Not applicable.
The research was founded with a grant from the Regione Toscana, the Programma Attuativo Regionale (Toscana) funded by FAS (now FSC), that supported the project “MICpROBIMM”.
The authors declare no conflict of interest. AA is serving as one of the Editorial Board members and Guest editors of this journal. We declare that AA had no involvement in the peer review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to RC.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/j.fbl2711305.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.




