1 Department of Vascular Surgery, The First Hospital of China Medical University, 110001 Shenyang, Liaoning, China
2 School of Life and Pharmaceutical Sciences, Dalian University of Technology, 116001 Dalian, Liaoning, China
† These authors contributed equally.
Abstract
Background: It remains largely unclear about the function of 5-methylcytosine (m5C) RNA modification in the context of abdominal aortic aneurysm (AAA). In this regard, the present work focused on investigating m5C RNA methylation and related modulator expression levels in AAA. Materials and methods: To this end, we quantified the m5C methylation levels in AAA tissues (n = 32) and normal aortic tissues (n = 12) to examine the mRNA m5C status and m5C modulator expression at mRNA and protein levels. Meanwhile, modulator localization within AAA tissue samples was detected by immunohistochemistry (IHC). Moreover, RNA immunoprecipitation-sequencing (RIP-seq) was also used to analyze the lncRNAs and mRNA binding to Aly/REF, as an m5C reader. Results: m5C expression markedly elevated in AAA in comparison with normal aortic samples in the AAA cases. The major 5-methylcytosine modulators, including NSUN2, NSUN5, and Aly/REF, which represented the major parameters related to the abnormal m5C modification level, were observed up-regulating in AAA tissues at both protein and mRNA levels. In addition, NSUN2 mRNA level remarkably related to Aly/REF expression, and they were co-expressed in the same cells in AAA group. Regarding the cellular location, Aly/REF was associated with inflammatory (CD45+, CD3+) infiltrates. Simultaneously, after screening for reads in AAA tissue compare with anti-Aly/REF group relative to IgG as control, we obtained totally 477 differentially expressed Aly/REF-binding lncRNAs and 369 differentially expressed Aly/REF-binding mRNAs in AAA tissue. The functions of Aly/REF-interacting lncRNA were involved in immune system process and macrophages infiltration. Through regulatory network (lncRNA-mRNA) analysis, our findings predicted the potential mechanism of Aly/REF-induced lncBCL2L1 and Aly/REF-lncFHL1 axis in AAA and inspire the understanding of m5C and lncRNA in AAA. Conclusions: This study is the first to examine m5A modification within human AAA samples. Our results indicate that m5C modulators, namely, Aly/REF and NUSN2, play vital parts in the human AAA pathogenic mechanism, which shed new lights on the function of m5C modification within AAA. Taken together, findings in this work offer a possible RNA methylation modification mechanism within clinical AAA.
Keywords
- Abdominal aortic aneurysm
- 5-methylcytosine (m5C) RNA modification
- RNA methylation
- Aly/REF
- NUSN2
Abdominal aortic aneurysm (AAA) accounts for a primary cause leading to cardiovascular event among the old male population [1, 2]. AAA is featured by the local while persistent abdominal aortic weakening and expansion [3], and it may be symptomatic, asymptomatic, or may present as rupture. The open retroperitoneal or transperitoneal selection operation has been the frequently adopted repair method [4]. Nonetheless, it is now suggested that the placement of the endoluminal stent graft in aneurysm may be applied in replacement of open intervention [5, 6]. There is no effective pharmacological treatment capable of limiting AAA progression or avoiding AAAs rupture. So far, pharmacological intervention is unavailable, while monitoring aneurysm size prior to operation is the only choice [7].
The pathogenic mechanism of AAA has been suggested to be complicated and multifactorial. In previous studies, AAA is suggested to be related to the deficient adventitial/medial arterial layers, like fibroblasts and smooth muscle cells (SMCs). Recent research with either human tissue or animal models has led to a shift regarding AAA, and AAA development is now considered to be part of a significant and dynamic remodeling process in the vessel [8]. Moreover, the critical pathological features are vascular inflammation [9], oxidative stress (OS) [10], aortic extracellular matrix (ECM) destruction [11], and aortic wall thinning due to vascular smooth muscle cell (VSMC) losses [12].
Epigenetic alterations, such as DNA methylation [13], histone modification [14] or RNA modification [15], has been found to exert vital parts in AAA due to their strong impacts on regulating gene levels. N6-methyladenosine (m6A) has been well investigated and most frequently observed in mRNA, which is found to be related to pre-mRNA translation, processing, mRNA decay and miRNA biogenesis. m6A modifications are reversible and dynamic among mammalian cells, and they are suggested to be the other epigenetic regulating layer associated with histone and DNA modifications [16, 17]. Our recent study has reported that aberrant RNA epigenetic modifications, such as m6A RNA methylation, are present in AAA [18]. The above results can shed novel lights on the AAA pathogenic mechanism.
Compared with m6A modification, 5-methylcytosine (m5C) is rarely investigated. m5C may be discovered in mRNA, tRNAs, rRNAs and recently poly(A)RNAs. It represents the abnormal RNA modification existing in various RNA species, such as numerous non-coding RNAs (ncRNAs), mitochondrial and cytoplasmic ribosomal RNAs (rRNAs), enhancer RNAs (eRNAs), transfer RNAs (tRNAs) and messenger RNAs (mRNAs) [19, 20]. m5C is a vital factor to regulate gene expression, like ribosomal assembly, RNA export, RNA stability and translation [21, 22, 23].
Hence in this study, we focused on the function of m5C mRNA modification involved in human AAA tissues, and detected m5C mRNA methylation level as well as related modulator expression. In addition, this study examined the relationship of m5C modification with the clinical data of patients. Moreover, RNA immunoprecipitation-sequencing (RIP-seq) was also used to analyze the lncRNAs and mRNA binding to Aly/REF, and to investigate the role of m5C RNA methylation in progression of AAA.
This study obtained human AAA tissue and relevant peripheral blood samples from altogether 147 consecutive cases at the Department of Vascular Surgery, The First Hospital of China Medical University (CMU) following the open operation for aneurysm according to previous description from January 2009 to December 2020 [18, 24]. In addition, each case provided the informed consent for participation. All human AAA samples were collected in line with guidelines from the World Medical Association Declaration of Helsinki. The Ethics Committees of the First Hospital of CMU approved our study protocol (ethical approval number: 2019-97-2).
We collected aneurysm tissues from 147 Chinese AAA cases at the time of emergency or elective open surgical repair. In addition, we obtained the clinical and history data from all patients, like medication history, rupture history, peripheral/coronary artery disease history, or risk factors like hypertension, smoking, hyperlipidemia and diabetes mellitus (DM). Meanwhile, patients with concurrent Marfan syndrome, Ehlers-Danlos syndrome, or additional identified connective tissue or vascular diseases were excluded from this study. Within 147 AAAs, for present study, 32 AAA tissues (28 male patients and 4 female patients) were available for further analyses. For inclusive 32 AAA patients, AAA was analyzed by computed tomography angiography (CTA). For all AAA cases, their diameter of infra-renal abdominal aorta was over 30 mm or 1.5–2 folds as high as the abdominal aorta diameter in corresponding normal segment [25] under CTA diagnosis. Furthermore, the included patients had no evidence or medical history of any cancer disease.
Over the same period, abdominal aorta samples from 12 heart-beating brain-dead organ donors (10 males and 2 females) were used as the controls. For controls, those with concurrent drug history, cancer, infection and additional immune-related disorders potentially affecting this work were excluded.
TRIzol reagent (TaKaRa Bio, Shiga, Japan) was utilized to extract total mRNA from aortic tissues according to the standard protocol as described previously. The extracted total mRNA was used to directly detect the m5c RNA methylation level by adopting the fluorometricm m5C RNA methylation ELISA kit (Epigentek, Farmingdale, NY, USA). Briefly, a pipette was utilized to add mRNA (200 ng) in the detection wells. Later, an m5C detection complex solution with a specific m5c antibody was added to the wells. After washing, the wells were added with fluorescence development solution for incubation under ambient temperature away from direct light. The fluorescence development solution will turn pink in the presence of sufficient m5c products. The fluorescence was read on a fluorescence microplate reader within 2 to 10 min at 530EX/590EM nm. Then, the mRNA m5C methylation level was assessed through the m5c-modified mRNA proportion in overall mRNA (m5C%) following the specific instructions.
For the high-quality RNAs, their A260/A280 ratio was greater than 1.8. The
PrimeScript RT Master Mix (Q711-02/03, Vazyme Biotech, Nanjing,
China) was used to synthesize cDNA from total mRNA, and the Universal SYBR qPCR
Master Mix (Q711-02/03, Vazyme Biotech, Nanjing, China) was
utilized to conduct quantitative real-time PCR (qRT-PCR) in
LightCycler 480 system (Roche Molecular
Systems, Indianapolis, IN, USA) in accordance with the amplification protocols
after modification. The PCR conditions were as follows, 30 s of initial
denaturation under 95
| Gene | Forward primer | Reverse primer |
| NSUN1 | AAGGGTGCCGAGACAGAACT | GAGCACGACTAGACAGCCTC |
| NSUN2 | CAAGCTGTTCGAGCACTACTAC | CTCCCTGAGAGCGTCCATGA |
| NSUN5 | CGCTACCATGAGGTCCACTAC | GCATCTCGCACCACGTCTT |
| NSUN6 | TCTCAGCCCTTCATTTGACAGT | TCCAGTGCTATAACTTCTCCCTG |
| Dnmt1 | TGCCAAGACGATTGAAGGCAT | GCAGGGAGGGCTCATTAAAAT |
| Aly/REF | TATGATCGCTCTGGTCGCAG | AGAGGGACGCCGTTGTACT |
| GAPDH | ACAACTTTGGTATCGTGGAAGG | GCCATCACGCCACAGTTTC |
The RIPA-based reagents were utilized to extract proteins from fresh frozen tissues in line with specific instructions; later, the concentration of each sample was measured by a Pierce BCA Protein Assay kit. The samples were isolated through SDS-PAGE, followed by transfer onto PVDF membranes. Afterwards, membranes were blocked using skimmed milk and incubated using suitable primary as well as secondary antibodies. Later, the ECL detection system was employed for membrane developing in line with specific instructions. GAPDH was used for normalization. In the present work, the following primary antibodies were utilized, anti-Aly/REF (ab202894; Abcam, Cambridge, UK; dilution 1:1000) and anti-NSUN2 (Cat. No. 20854-1-AP; Proteintech, Wuhan, China; dilution 1:1000) and anti-GAPDH (dilution 1:2000; Zsbio, Beijing, China). The densitometry was performed with Image J software (Java 1.8.0_172, National Institutes of Health, Bethesda, Rockville, MD, USA) and normalized to the signal intensity of GAPDH for equal protein loading control of each sample in each experiment.
The typical 2–3-
In IHC assay, antigen retrieval was performed through boiling tissue sections in
sodium citrate solution (pH = 6.0), after washing, suitable antibodies were used
to treat the sections, then they were deparaffinized and hydrated. To analyze
cells within AAA wall, endothelial cells (ECs) were analyzed by anti-CD34 (1:100;
Proteintech, Wuhan, China), SMCs were measured through anti-
The Nikon ScanScope 90i system (Nikon, Melville, NY, USA) was
used to obtain the digital images of the stained tissues (slides). To be
specific, the settings for digital image capturing were 20
To conduct standard and traditional staining, the extent of calcification and cellularity of vascular wall were characterized for histological classification. Thereafter, basic cell count and related cell intensity were used to evaluate the grade, such as new vessel formation, infiltrates and SMCs.
To determine the expression of biomarkers in single cells in aneurysms, we made successive slides of every sample and incubated them using appropriate antibodies. In all cases, one slide was stained with an antibody to detect a certain cell type; thereafter, the anti-biomarker antibodies were used to stain the successive slides.
In brief, typical 2–3-
First of all, the Magna RIP™ RNA-binding Protein
Immunoprecipitation Kit (Millipore, Billerica,
MA, USA) was utilized for RIP-seq in line with specific instructions. In brief,
after coating with 10
With regard to mRNA and lncRNA, we used hisat2 software to align high-quality reads to human reference genome (UCSC hg19). Thereafter, we adopted Cuffdiff (version 2.1.0: http://cole-trapnell-lab.github.io/cufflinks/) for obtaining the fragments per kilobase of transcript per million mapped reads (FPKM) under the guidance of Ensembl gtf gene expression profiles, which were the mRNA and lncRNA expression profiles [27]. Further, we determined p-values and fold changes (FCs) according to FPKM, and discovered the differentially expressed mRNAs and lncRNAs. The target genes of lncRNAs were estimated according to their locations relative to adjacent genes. Additionally, the predicted target genes were subjected to GO (www.geneontology.org) and KEGG (www.genome.jp/kegg) enrichment to determine the major functions and related pathways involved in differentially expressed mRNAs and lncRNAs on the basis of differentially expressed mRNAs.
Since the expression of lncRNAs shows significant relation with the surrounding protein encoding genes, numerous lncRNAs play roles of the cis regulators [28]. LncRNA and its adjacent mRNAs were integrated and differentially expressed in the biome to explore the function of lncRNA. LncExpDB (https://ngdc.cncb.ac.cn/lncexpdb) estimates lncRNA genes’ expression reliability and capacities, used for the lncRNA-mRNA interaction network analysis.
PPI networks can offer precious data about cell functions or the signal transduction pathways. In this study, we searched the interactions of DEGs-encoded proteins through the Search Tool for the Retrieval of Interacting Genes/Proteins (http://string-db.org/) online database. Thereafter, the PPI network was built using 5 calculation algorithms (EPC, Degree, EcCentricity, MNC and MCC) and visualized using Cytoscape (http://cytoscape.org/). At last, those overlapping genes obtained by the above 5 algorithms encoded core proteins that had vital biological regulatory activities.
Statistical analysis was completed using SPSS22.0 (SPSS Inc., Chicago, IL, USA).
Differences among the categorical variables were compared by chi-square test
between groups. In addition, the one-sample Kolmogorov-Smirnov test was used to
examine the distribution of data. Later, the non-parametric Mann–Whitney U test
or parametric t-test for unpaired samples was used for analysis
according to variable distribution. Thereafter, partial correlation analysis was
conducted to examine the associations among continuous variables after adjusting
for smoking, age and sex. Correlations between continuous variables were
quantified by using Spearman’s rank correlation coefficient. A difference of
p
Table 2 presents the clinical and demographic data of AAA cases and normal
subjects. For AAA cases, their age ranged from 43 to 86 years, and the average
AAA maximum diameter was found to be 69.25
| Characteristics | AAA group (N = 32) | Control group (N = 12) | p | |||
| Baseline | n | mean |
n | mean |
||
| Age, Year | 32/32 | 61.94 |
12/12 | 58.67 |
0.279 | |
| female | 4/32 | 12.50% | 2/12 | 16.67% | 0.529 | |
| BMI | 31/32 | 25.75 |
11/12 | 24.41 |
0.344 | |
| maximum AAA diameter, mm | 32/32 | 74.23 |
NA | NA | NA | |
| Comorbidity | ||||||
| Hypertension | 17/32 | 53.13% | 5/12 | 41.67% | 0.368 | |
| Smoking | 18/31 | 58.06% | 5/12 | 41.67% | 0.265 | |
| Hyperlipidemia | 5/32 | 15.63% | 1/12 | 8.33% | 0.471 | |
| Diabetes mellitus | 1/32 | 3.13% | 0/12 | 0% | 0.727 | |
| Cardiac disease | 4/31 | 12.90% | 1/12 | 8.33% | 0.569 | |
| Renal disease | 2/31 | 6.45% | 0/12 | 0% | 0.515 | |
| Medication use | 10/32 | 31.25% | 1/12 | 0% | 0.118 | |
| Note, AAA, abdominal aortic aneurysm; SD, standard deviation; BMI, body mass index; NA, not available. | ||||||
The symptoms of the AAA patients are summarized in Table 2. Altogether 22% AAA cases had aneurysmal rupture, whereas 5 out of the 32 cases had iliac aneurysms. Among AAA cases, each AAA sample was semi-quantitatively and histologically characterized for evaluating each histopathological characteristic degree within AAA wall as previously [14]. In Table 3, IHC was used to differentiate between the four main cell types in AAAs, i.e., endothelial cells, lymphocytes, macrophages, and smooth muscle cells to assess the extension of the individual histopathological features in AAA wall.
| No. | Abdominal pain | Pulsating sensations in the abdomen | Comorbidity-Iliac artery aneurysms | Ruptured AAA | Cellularity (HE) | Infiltrates (CD45) | Macrophages (CD68) | SMCs ( |
Neovessel (CD34) |
| A1 | √ | + | +/++ | + (+/++) | ++ | + (+/++) | |||
| A2 | √ | +/– | + | + (+/–) | +/++ | +/++ | |||
| A3 | √ | + | + | –/+ | ++/+++ | +/– | |||
| A4 | √ | +/++ | ++++ | + (+/–) | +/++ | +++ | |||
| A5 | ++ | +/– | ++ | +/– | –/+ | ||||
| A6 | √ | +/++ | +/++ | +/– | +++ | ||||
| A7 | √ | +/++ | ++++ | ++ | ++ (+/++) | +/++ | |||
| A8 | √ | +/– | +/++ | –/+ | + | + | |||
| A9 | √ | + | ++ | – | +/– | + (+/–) | |||
| A10 | √ | +/– | +/– | + | +/– | –/+ | |||
| A11 | √ | +/++ | +++ | + | + | ++/+++ | |||
| A12 | √ | √ | √ | + | +/++ | –/+ | +/– | +/++ | |
| A13 | √ | ++ | + | + (+/–) | +/++ | –/+ | |||
| A14 | √ | √ | + | ++++ | +/– | +/– | +/– | ||
| A15 | √ | +/++ | ++++ | + (+/–) | + | +/++ | |||
| A16 | √ | + | +/++ | +/– | +/++ | +/– | |||
| A17 | √ | √ | + | ++ (+/++) | + (+/–) | +/++ | +/++ | ||
| A18 | √ | √ | +/– | +++ | + | + (+/–) | + | ||
| A19 | √ | √ | ++ | +/++ | – | + | +++ | ||
| A20 | + | + | +/– | ++ | ++ | ||||
| A21 | √ | √ | + | +/– | + | + (+/–) | –/+ | ||
| A22 | √ | ++ | +++ | –/+ | + | ++ | |||
| A24 | √ | +/++ | ++ | + | +/++ | + | |||
| A25 | √ | √ | √ | √ | +/– | +++ | –/+ | +/++ | +/– |
| A26 | √ | √ | + | + | +/++ | + | +/++ | ||
| A27 | √ | √ | + | +++ | +/++ | + | –/+ | ||
| A29 | √ | +/– | + | ++ | +/++ | + (+/–) | |||
| A30 | + | +/– | +/++ | +/++ | + | ||||
| A31 | +/– | +++ | ++ | +/– | + (+/–) | ||||
| A32 | √ | √ | + | ++ (+/++) | + | +/– | –/+ | ||
| A33 | √ | √ | √ | +/++ | + | –/+ | + | +++ |
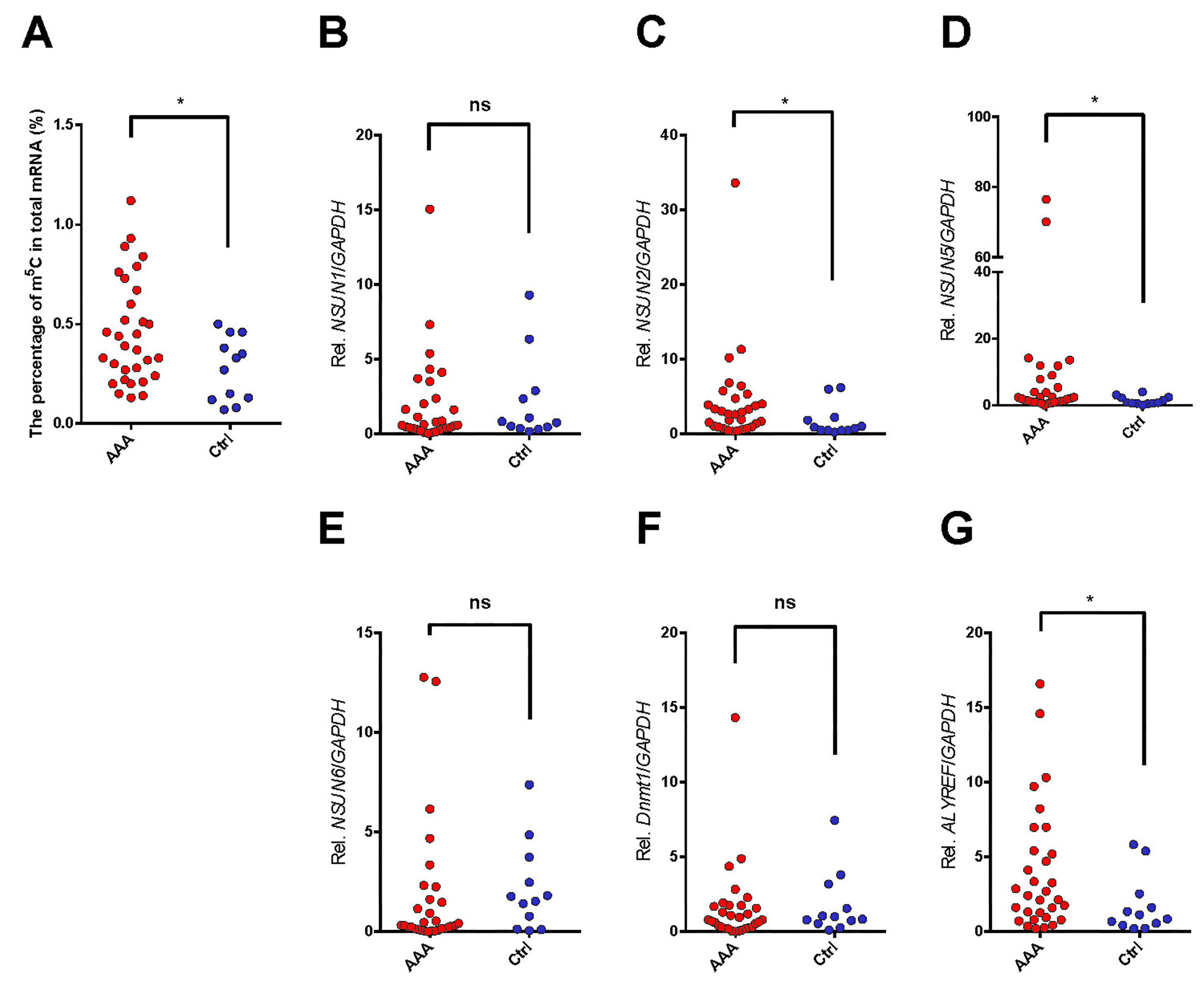
According to our analysis, relative m5c mRNA methylation level within AAA tissues showed significant correlation with an increased m5c proportion in the overall mRNA relative to controls (more than 1.5-fold; p = 0.040; Fig. 1A).
 Fig. 1.
Fig. 1.Expression of m5C RNA methylation status (A) and m5C methylation
modulators (B, C, D, E, F, and G) at the mRNA level in AAA tissue samples
compared with the healthy control aortas analyzed by qRT-PCR.
The non-parametric Mann-Whitney U test was all applied to analyze the m5C
methylation ratio (A); the m5C “writers” family, NSUN1 (B),
NSUN2 (C), NSUN5 (D), NSUN6 (E), Dnmt2 (F);
and “readers” family Aly/REF (G). The expression of individual m5C RNA
methylation modulators related to the expression of GAPDH set as 1 (100%
expression).
*, p
Thereafter, certain typical molecules that might be related to m5C mRNA
modification were analyzed for their expression at mRNA level, including
NSUN1, NSUN2, NSUN5, NSUN6, Dnmt2 and Aly/REF (Fig. 1B–G). In
the current study, the mRNA expressions of NSUN2, NSUN5 and
Aly/REF were observed up-regulating in the AAA tissue samples relative
to matched normal tissues (
According to our results, the m5C% in total mRNA was significantly correlated
with NSUN2, NSUN5 and Aly/REF expression levels (R = 0.316, 0.320, and
0.312 respectively, and p = 0.039, p = 0.037 and p =
0.042, respectively; Table 4). Afterwards, the associations of mRNA expression
were examined. As a result, NSUN2 mRNA expression showed significant
relationship with the expressions of NSUN5 and Aly/REF (R =
0.538, p
| m5c | NSUN1 | NSUN2 | NSUN5 | NSUN6 | Dnmt1 | Aly/REF | |
| m5c | - | ||||||
| - | |||||||
| NSUN1 | –0.264 | - | |||||
| 0.087 | - | ||||||
| NSUN2 | 0.316* | 0.209 | - | ||||
| 0.039 | 0.18 | - | |||||
| NSUN5 | 0.320* | 0.218 | 0.538** | - | |||
| 0.037 | 0.16 | - | |||||
| NSUN6 | –0.145 | 0.451** | 0.287 | 0.410** | - | ||
| 0.352 | 0.002 | 0.062 | 0.006 | - | |||
| Dnmt1 | –0.099 | 0.470** | 0.143 | 0.349* | 0.673** | - | |
| 0.527 | 0.001 | 0.361 | 0.022 | - | |||
| Aly/REF | 0.312* | 0.313* | 0.464** | 0.642** | 0.214 | 0.339* | - |
| 0.042 | 0.041 | 0.002 | 0.169 | 0.026 | - | ||
| Note, significant correlation: *p | |||||||
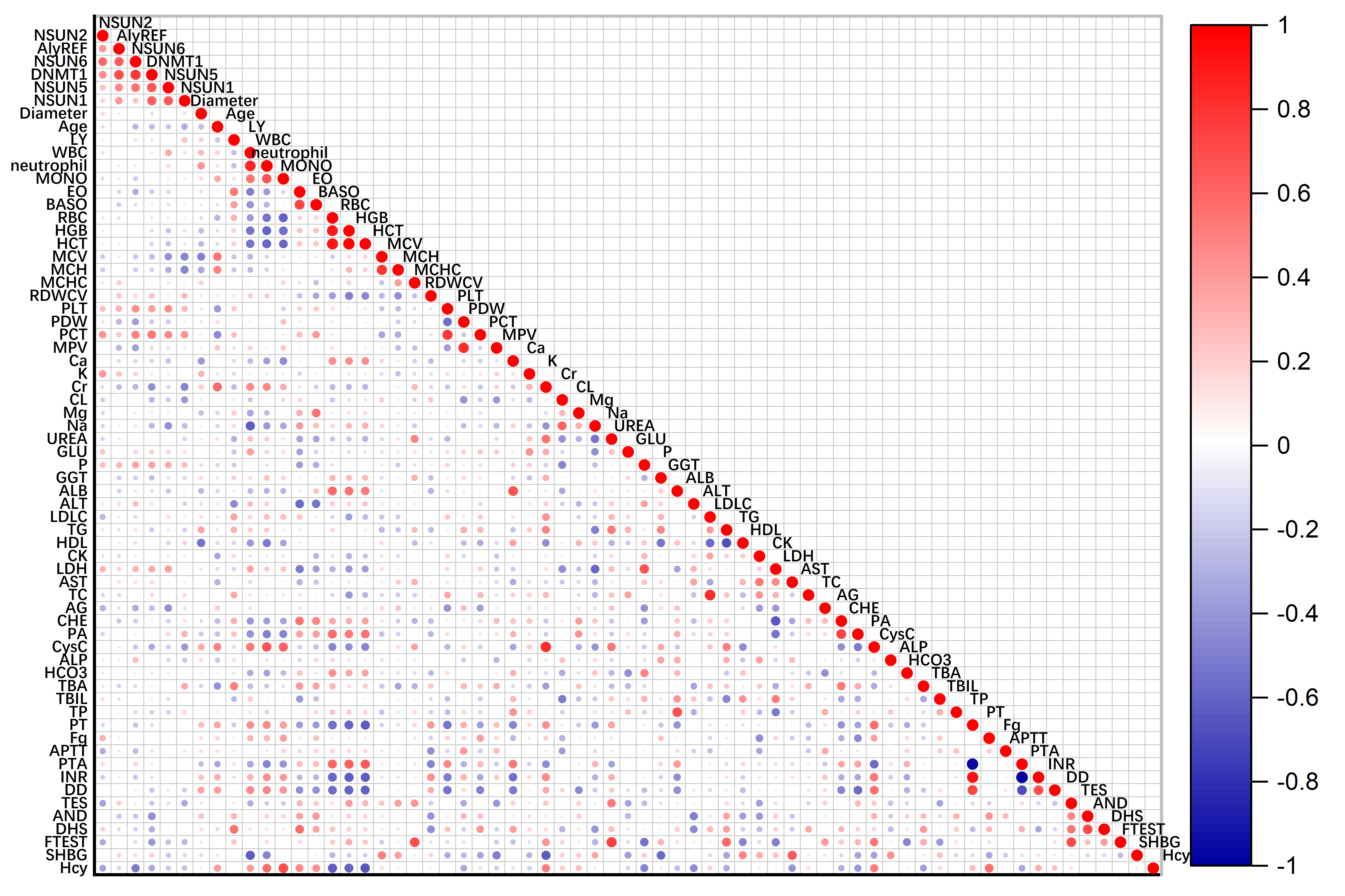
Thereafter, the associations of m5C modulator expression levels with clinical features were analyzed. Similarly, the expression of NSUN1, NSUN2, NUSN5, NSUN6 and Dnmt1 positively correlated to platelet hematocrit (PCT) (R = 0.429, 0.429, 0.462, 0.494, and 0.537; p = 0.016, 0.016, 0.009, 0.005, and 0.002, respectively).
NSUN5, NSUN6, and Dnmt1 expression significantly correlated to platelets count (PLT) (R = 0.443, 0.456, and 0.382; p = 0.013, 0.010, and 0.034, respectively). The expression of NSUN1 and NSUN5 at mRNA level were negative associated with mean corpuscular volume (MCV), the correlation coefficient was –0.470 and –0.419 (p = 0.008 and 0.019 respectively) in Fig. 2.
 Fig. 2.
Fig. 2.Correlation among the mRNA expressions of m5C modulators and clinical parameters. Partial correlation analysis was adjusted with age, gender, BMI, and smoking. BMI, body mass index; LY, lymphocyte; WBC, white blood cell; MONO, monocyte; EO, eosinophils; BASO, basophil; RBC, red blood cell; HGB, hemoglobin; HCT, hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; RDWCV, red blood cell volume distribution width; MCHC, mean corpuscular hemoglobin concentration; PLT, platelets; PDW, platelet distribution width; PCT, platelet hematocrit; MPV, mean platelet volume; Ca, serum calcium; K, serum kalium; Cr, creatinine; Cl, serum chlorine; Mg, serum magnesium; Na, serum sodium; GLU, fasting plasma glucose; P, serum phosphate; GGT, gamma glutamyl transpeptidase; ALB, albumin; ALT, alanine aminotransferase; LDL-C, low-density lipoprotein-cholesterol; TG, triglycerides; HDL-C, high-density lipoprotein-cholesterol; CK, creatine kinase; LDH, lactate dehydrogenase; AST, aspartate aminotransferase; TC, total cholesterol; AG, anion gap; CHE, cholinesterase; PA, pre-albumin; CysC, cystatin C; ALP, alkaline phosphatase; TBA, total bile acid; TBIL, total bilirubin; TP, total protein; PT, prothrombin time; Fg, fibrinogen; APTT, activated partial thromboplastin time; PTA, prothrombin activity; INR, international normalized ratio; DD, D-dimer; TES, testosterone; AND, androgen; DHS, dehydroepiandrosterone; F-TEST, free testosterone; SHBG, sex hormone-binding globulin; Hcy, Homocysteine.
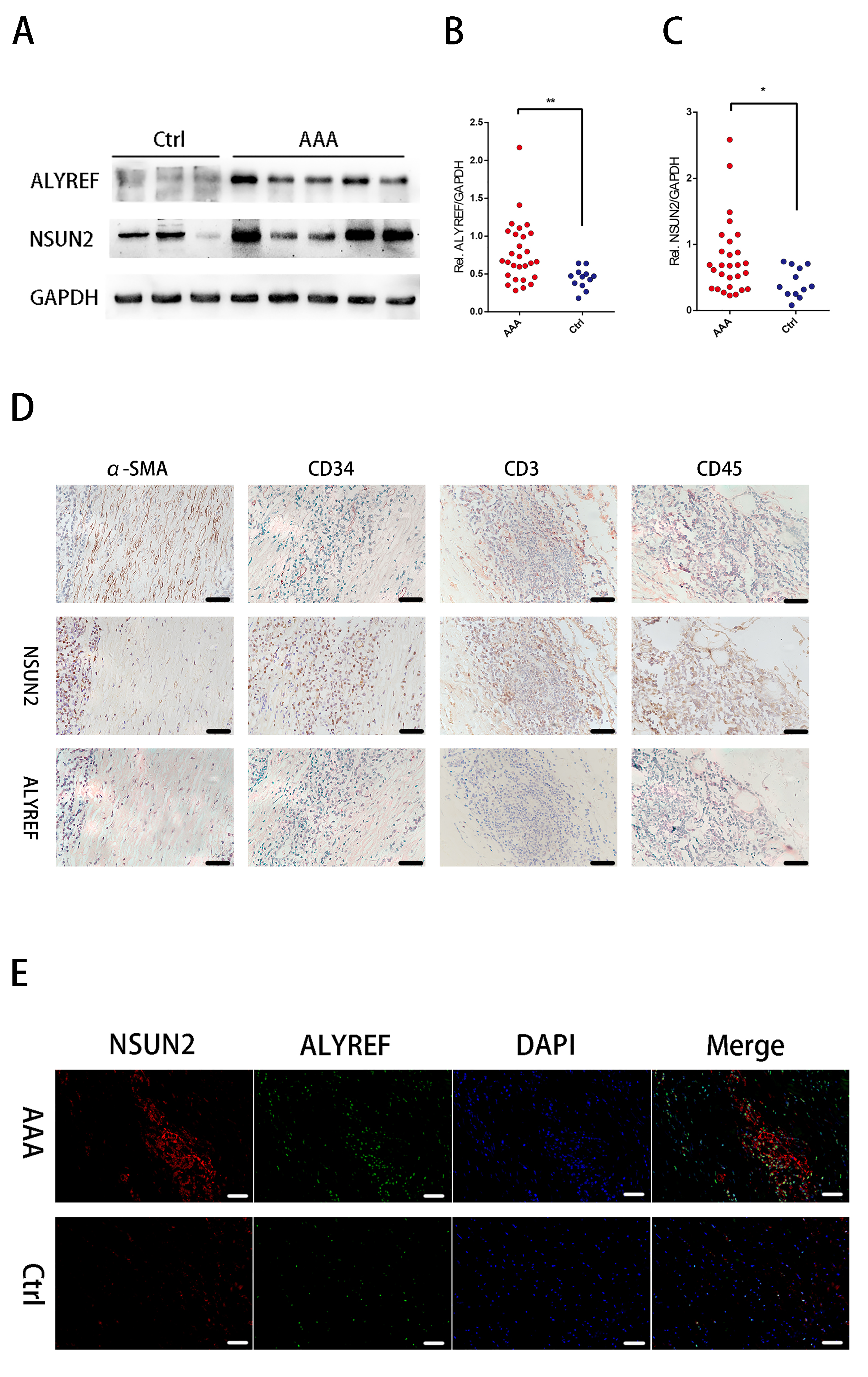
We selectively performed western blot analysis for NSUN2 and Aly/REF, because their roles on the 5-mC modification in mRNA, their expressions at mRNA levels and their interrelatedness with mRNA m5C status in human AAA tissue samples. Fig. 3A displays the representative images of NSUN2 and Aly/REF expression levels measured through Western blotting (Supplementary Fig. 1). Aly/REF and NSUN2 protein expression markedly increased within AAA tissues relative to normal controls (more than 1.5-fold for both comparison; p = 0.039, and p = 0.003, respectively; Fig. 3B,C).
 Fig. 3.
Fig. 3.Protein level of Aly/REF and NUSN2 are highly expressed in human
abdominal aortic aneurysm (AAA).
The representative images of blots between
AAA and healthy aortas at the protein level (A); The density of the protein
signals for Aly/REF (B) and NUSN2 (C) through the non-parametric Mann-Whitney U
test quantification of the band intensities relative to the expression of GAPDH.
*, p
Similarly, significantly increased expressions of NSUN2 and Aly/REF were also observed in AAA tissue samples, both were observed in the healthy aorta tissue through IHC analysis, date was not shown. Additionally, we examined the colocalization of NSUN2 and Aly/REF within the successive sections by IHC. As a result, NSUN2 staining showed weak colocalization with CD34+ ECs, and strongly with CD45+ leukocytes and CD3+ T lymphocytes. Aly/REF also showed strong colocalization with CD3+ T lymphocytes and CD45+ leukocytes (Fig. 3D).
Later, the AAA wall and normal aortic wall co-localization of Aly/REF with NSUN2 was examined using IF analysis (Fig. 3E). Here, we also observed that the expressions of NSUN2 and Aly/REF were higher in AAA sections compared with the controls. The result is similar to the results of western blot analysis and IHC analysis. Furthermore, for the co-localization analysis, NSUN2 and Aly/REF were co-expressed in the one cell in AAA group (Fig. 3E).
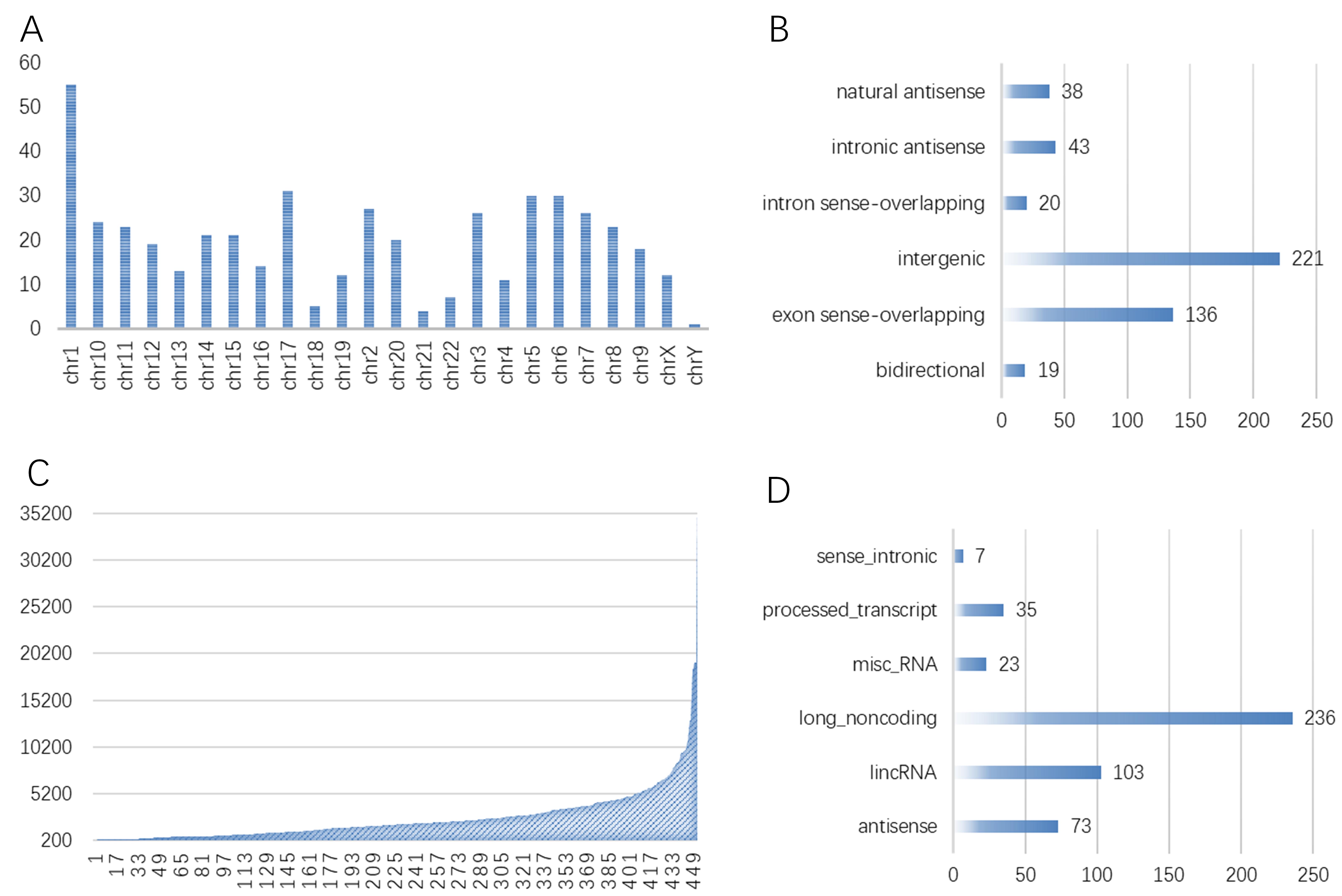
The possible Aly/REF target genes were predicted by RIP-seq assay. lncRNAs have
the length of over 200 nucleotides (nt). In addition, this study analyzed those
known lncRNAs for their length distribution. As a result, there were markedly
more lncRNAs with the length of 500–1000 and
 Fig. 4.
Fig. 4.The results demonstrated that 477 differentially expressed genes Aly/REF-binding lncRNAs in AAA tissue. The locus, genomic locus, chromosome of Aly/REF binding lncRNAs (A); thebiotype of Aly/REF binding lncRNAs (B); the lncRNA length of Aly/REF binding lncRNAs (C); and relationship of Aly/REF binding lncRNAs (D) in abdominal aortic aneurysm tissue. Note, exon sense-overlapping: the LncRNA’s exon is overlapping a coding transcript exon on the same genomic strand; intron sense-overlapping: the LncRNA is overlapping the intron of a coding transcript on the same genomic strand; intronic antisense: the LncRNA is overlapping the intron of a coding transcript on the antisense strand; natural antisense: the LncRNA is transcribed from the antisense strand and overlapping with a coding transcript; bidirectional: the LncRNA is oriented head-to-head to a coding transcript within 1000 bp; intergenic: there are no overlapping or bidirectional coding transcripts nearby the LncRNA.
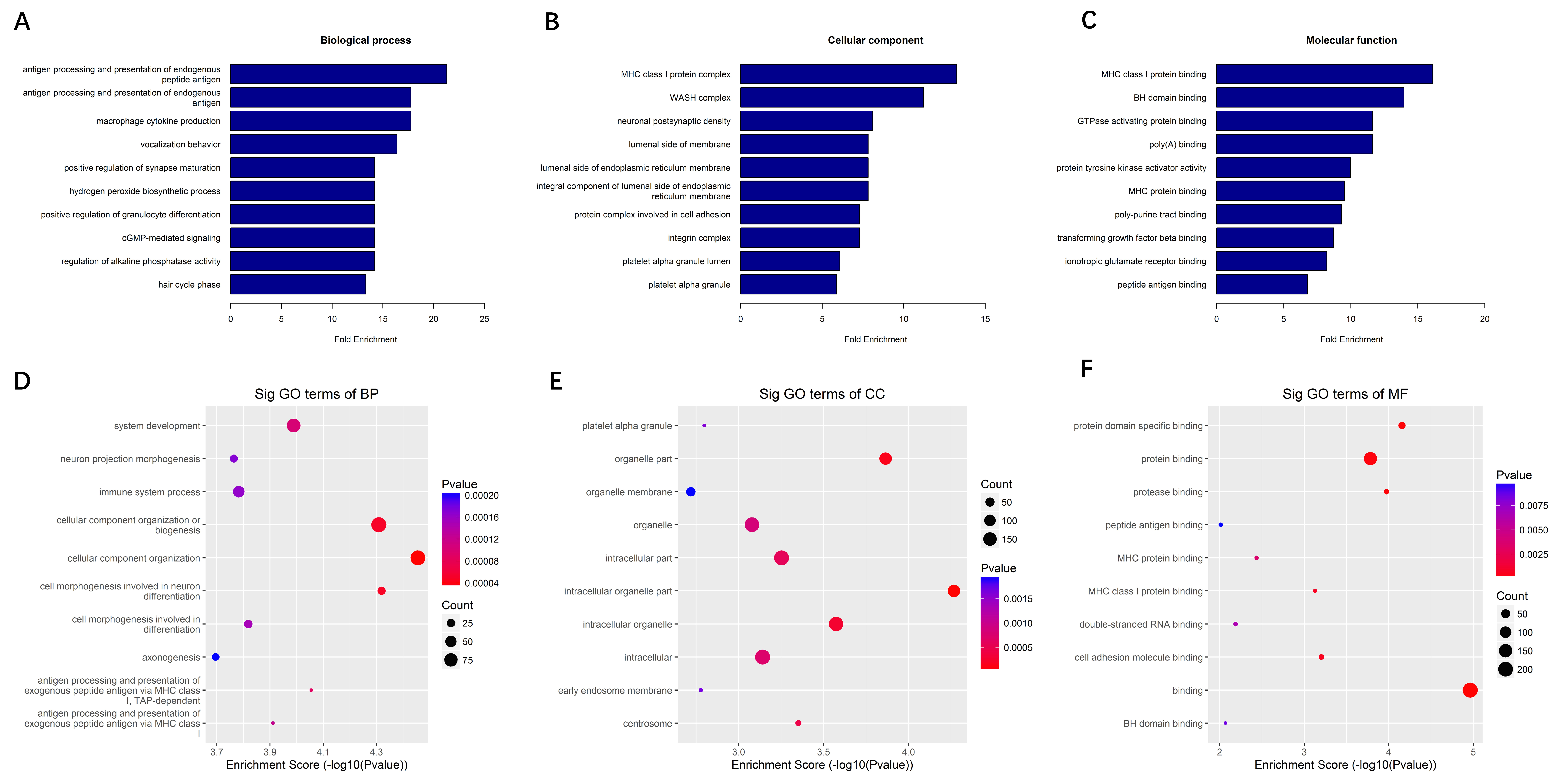
GO analysis indicated that the functions of Aly/REF-interacting lncRNA were involved in a variety of biological processes, including immune system process and macrophages infiltration, i.e., macrophage cytokine production (GO:0010934), immune system process (GO:0002376) for Biological Process; MHC class I protein complex (GO:0042612), platelet alpha granule (GO:0031091) for Cellular Component; and MHC class I protein binding (GO:0042288), MHC protein binding (GO:0042287) for Molecular Function (Fig. 5).
 Fig. 5.
Fig. 5.Go analysis of 477 differentially expressed genes Aly/REF-binding lncRNAs in AAA tissue. Fold enrichment in biological process (A), cellular component (B), and molecular function (C); The enrichment score in biological process (D), cellular component (E), and molecular function (F).
KEGG pathway analysis indicated the up-regulation of 26 pathways, mainly including ECM-receptor interaction (KEGG: hsa04512), PPAR (KEGG: hsa03320) and phagosome (KEGG: hsa04145) signal transduction pathways, suggested that dysregulated lncRNA pathways in Aly/REF were closely associated with signal transduction in AAA tissue (Supplementary Fig. 2). Based on the FC, the top 20 dysregulated lncRNAs are summarized in Table 5. Among these dysregulated lncRNAs, distributions among the human chromosomes were also illustrated.
| No. | Transcript ID | Biotype | Strand | Gene ID | Chromosome | LncRNA Source | LncRNA length | Relationship | Association gene name |
| 1 | ENST00000416191 | antisense | + | ENSG00000258486 | chr14 | Ensembl | 297 | bidirectional | RPS29 |
| 2 | ENST00000598952 | misc RNA | − | ENSG00000265150 | chr14 | Ensembl | 297 | intergenic | |
| 3 | uc003ine.3 | lincRNA | − | ENSG00000260402 | chr16 | Ensembl | 2484 | intergenic | |
| 4 | ENST00000473958 | misc RNA | − | ENSG00000200488 | chr2 | Ensembl | 332 | intergenic | |
| 5 | TCONS_l2_00025918 | misc RNA | + | ENSG00000265735 | chr9 | Ensembl | 288 | intronic antisense | PTPRD |
| 6 | uc009ykc.1 | lincRNA | + | ENSG00000224934 | chr10 | Ensembl | 2998 | bidirectional | GOT1 |
| 7 | ENST00000455068 | antisense | + | ENSG00000269296 | chr19 | Ensembl | 3653 | natural antisense | ZNF780A |
| 8 | ENST00000493013 | long noncoding | − | BC039551 | chr4 | UCSC knowngene | 1124 | intronic antisense | FHDC1 |
| 9 | TCONS_00004186 | antisense | + | ENSG00000244300 | chr3 | Ensembl | 722 | intronic antisense | GATA2 |
| 10 | ENST00000565978 | long noncoding | − | SNHG10 | chr14 | RefSeq | 1980 | bidirectional | GLRX5 |
| 11 | ENST00000510986 | antisense | − | ENSG00000225733 | chr3 | Ensembl | 1874 | intronic antisense | NR2C2 |
| 12 | uc002xuq.1 | lincRNA | − | ENSG00000267427 | chr19 | Ensembl | 2103 | exon sense-overlapping | MLLT1 |
| 13 | ENST00000516507 | antisense | − | ENSG00000249042 | chr5 | Ensembl | 1151 | bidirectional | FAM151B |
| 14 | ENST00000586625 | antisense | + | ENSG00000259985 | chr18 | Ensembl | 1169 | natural antisense | B4GALT6 |
| 15 | TCONS_l2_00005763 | lincRNA | + | ENSG00000254339 | chr8 | Ensembl | 697 | intergenic | |
| 16 | NR104639 | long noncoding | − | CENPJ | chr13 | RefSeq | 5246 | exon sense-overlapping | CENPJ |
| 17 | ENST00000332012 | lincRNA | + | ENSG00000248587 | chr5 | Ensembl | 1941 | exon sense-overlapping | GDNF-AS1 |
| 18 | ENST00000556819 | long noncoding | − | AK055386 | chr20 | UCSC knowngene | 3404 | intergenic | |
| 19 | ENST00000602575 | antisense | + | ENSG00000227061 | chr2 | Ensembl | 4775 | intergenic | |
| 20 | ENST00000434707 | processed transcript | + | ENSG00000012171 | chr13 | Ensembl | 1765 | exon sense-overlapping | SEMA3B |
After RPKM value distribution was analyzed, we analyzed the sample gene
expression profiles on the whole. When IP showed significant enrichment relative
to input group, the combined expression for each gene of IP group increased
relative to input group. Through RIP-seq, mapping data revealed that FMRP had
3060 potential target genes, which showed extensive activities in cell
physiological processes, moreover, there were 369 mRNAs showing differential
expression (p
To reduce the mRNAs binding to Aly/REF to better investigate and enrich mRNAs
that might be related to AAA, this study screened the significantly
differentially expressed mRNAs (FC
| No. | Gene symbol | Gene ID | Strand | Chromosome | Map location | Gene name |
| 1 | MYCBP2 | ENSG00000005810 | − | 13 | 13q22 | MYC binding protein 2, E3 ubiquitin protein ligase |
| 2 | LUC7L | ENSG00000007392 | − | 16 | 16p13.3 | LUC7-like |
| 3 | TYMP | ENSG00000025708 | − | 22 | 22q13.33 | thymidine phosphorylase |
| 4 | CYFIP2 | ENSG00000055163 | + | 5 | 5q33.3 | cytoplasmic FMR1 interacting protein 2 |
| 5 | MYO9A | ENSG00000066933 | − | 15 | 15q22-q23 | myosin IXA |
| 6 | JMJD6 | ENSG00000070495 | − | 17 | 17q25 | jumonji domain containing 6 |
| 7 | FGFR1 | ENSG00000077782 | − | 8 | 8p11.23-p11.22 | fibroblast growth factor receptor 1 |
| 8 | SDF4 | ENSG00000078808 | − | 1 | 1p36.33 | stromal cell derived factor 4 |
| 9 | CREM | ENSG00000095794 | + | 10 | 10p11.21 | cAMP responsive element modulator |
| 10 | MED15 | ENSG00000099917 | + | 22 | 22q11.2 | mediator complex subunit 15 |
| 11 | ZC3H14 | ENSG00000100722 | + | 14 | 14q31.3 | zinc finger CCCH-type containing 14 |
| 12 | DCAF11 | ENSG00000100897 | + | 14 | 14q11.2 | DDB1 and CUL4 associated factor 11 |
| 13 | ZMYND8 | ENSG00000101040 | − | 20 | 20q13.12 | zinc finger MYND-type containing 8 |
| 14 | SIRPB1 | ENSG00000101307 | − | 20 | 20p13 | signal regulatory protein beta 1 |
| 15 | JAG1 | ENSG00000101384 | − | 20 | 20p12.1-p11.23 | jagged 1 |
| 16 | MTMR9 | ENSG00000104643 | + | 8 | 8p23-p22 | myotubularin related protein 9 |
| 17 | VRK3 | ENSG00000105053 | − | 19 | 19q13 | vaccinia related kinase 3 |
| 18 | PALD1 | ENSG00000107719 | + | 10 | 10q22.1 | phosphatase domain containing, paladin 1 |
| 19 | XPNPEP1 | ENSG00000108039 | − | 10 | 10q25.3 | X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| 20 | ZMIZ1 | ENSG00000108175 | + | 10 | 10q22.3 | zinc finger MIZ-type containing 1 |
| 21 | CDK5RAP3 | ENSG00000108465 | + | 17 | 17q21.32 | CDK5 regulatory subunit associated protein 3 |
| 22 | PPP6R3 | ENSG00000110075 | + | 11 | 11q13 | protein phosphatase 6 regulatory subunit 3 |
| 23 | ELMOD3 | ENSG00000115459 | + | 2 | 2p11.2 | ELMO/CED-12 domain containing 3 |
| 24 | SERPINC1 | ENSG00000117601 | − | 1 | 1q25.1 | serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| 25 | MTRR | ENSG00000124275 | + | 5 | 5p15.31 | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| 26 | ERMARD | ENSG00000130023 | + | 6 | 6q27 | ER membrane-associated RNA degradation |
| 27 | YIPF2 | ENSG00000130733 | − | 19 | 19p13.2 | Yip1 domain family member 2 |
| 28 | DNMT1 | ENSG00000130816 | − | 19 | 19p13.2 | DNA (cytosine-5-)-methyltransferase 1 |
| 29 | SCLY | ENSG00000132330 | + | 2 | 2q37.3 | selenocysteine lyase |
| 30 | HSD17B4 | ENSG00000133835 | + | 5 | 5q21 | hydroxysteroid (17-beta) dehydrogenase 4 |
| 31 | NUMB | ENSG00000133961 | − | 14 | 14q24.3 | numb homolog (Drosophila) |
| 32 | ZCCHC11 | ENSG00000134744 | − | 1 | 1p32.3 | zinc finger CCHC-type containing 11 |
| 33 | ELP2 | ENSG00000134759 | + | 18 | 18q12.2 | elongator acetyltransferase complex subunit 2 |
| 34 | LMO7 | ENSG00000136153 | + | 13 | 13q22.2 | LIM domain 7 |
| 35 | CALCOCO2 | ENSG00000136436 | + | 17 | 17q21.32 | calcium binding and coiled-coil domain 2 |
| 36 | FPGS | ENSG00000136877 | + | 9 | 9q34.1 | folylpolyglutamate synthase |
| 37 | PML | ENSG00000140464 | + | 15 | 15q22 | promyelocytic leukemia |
| 38 | RHOT2 | ENSG00000140983 | + | 16 | 16p13.3 | ras homolog family member T2 |
| 39 | DEF8 | ENSG00000140995 | + | 16 | 16q24.3 | differentially expressed in FDCP 8 homolog (mouse) |
| 40 | ANAPC11 | ENSG00000141552 | + | 17 | 17q25.3 | anaphase promoting complex subunit 11 |
GO analysis indicated that the functions of Aly/REF-interacting mRNA were related to various biological processes, such as UDP-N-acetylglucosamine metabolic process and endocytic recycling; and cellular component, including eukaryotic 48S preinitiation complex, eukaryotic 43S preinitiation complex; and platelet-derived growth factor biding, histone methyltansferases activity (Supplementary Fig. 3).
KEGG Moreover, regulated pathways were enriched in protein processing in endoplasmic reticulum (KEGG: hsa04141), lysine degradation (KEGG: hsa00310), and Focal adhesion (KEGG: hsa04510) (Supplementary Fig. 4).
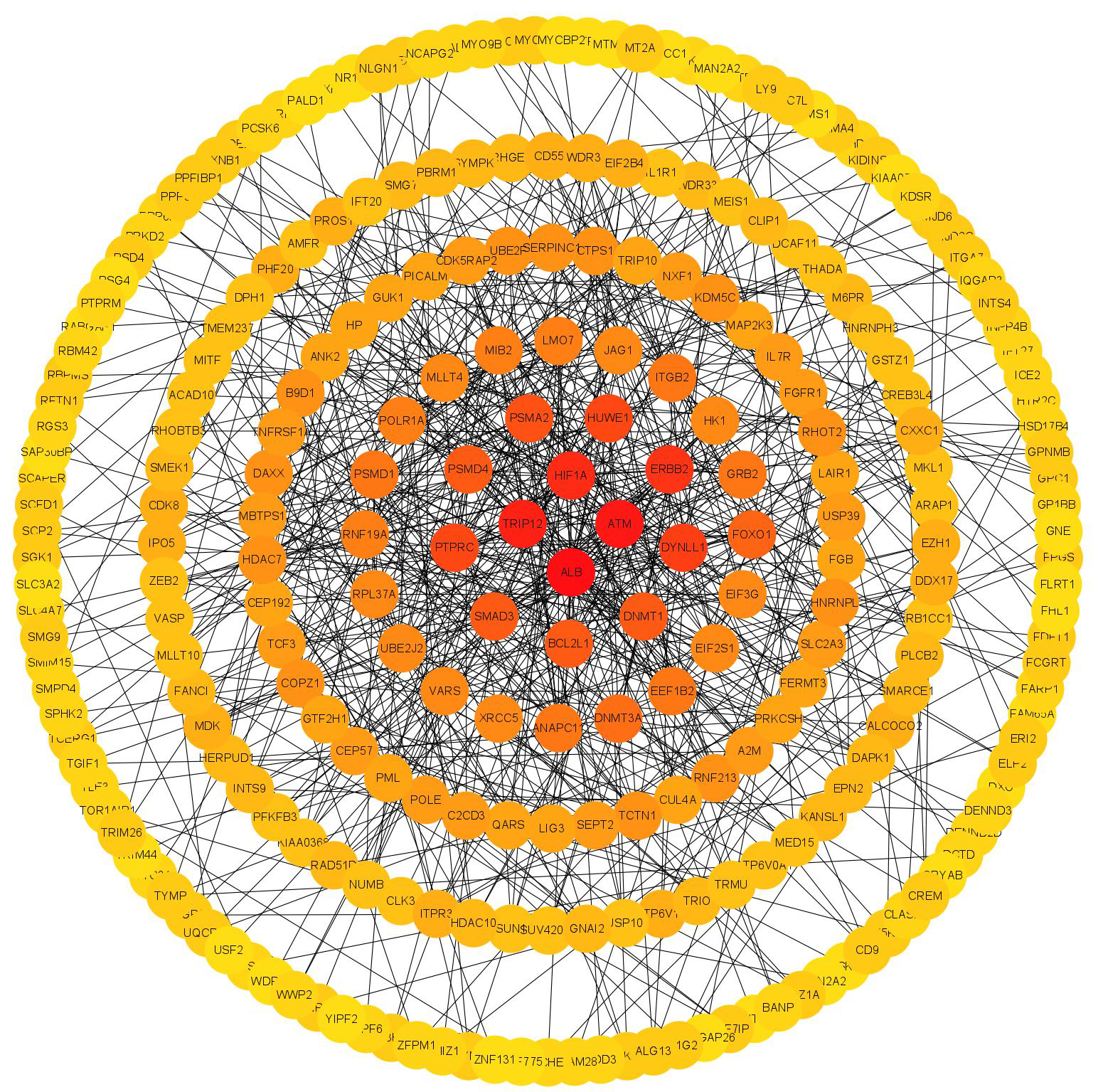
Altogether 369 DEGs were analyzed against the STRING database. At the same time,
Cytoscape was used to construct a PPI network through neighborhood,
co-expression, settings experiments, text-mining and database. Afterwards, the
separated genes were removed (those that did not interact with the remaining
genes), and the DEGs regulated by Aly/REF were exhibited with 745 edges and 372
nodes (Fig. 6). Based on every gene degree, 4 hub genes that had a
 Fig. 6.
Fig. 6.Construction of PPI network, analysis of 369 differentially expressed, and identification of hub genes.
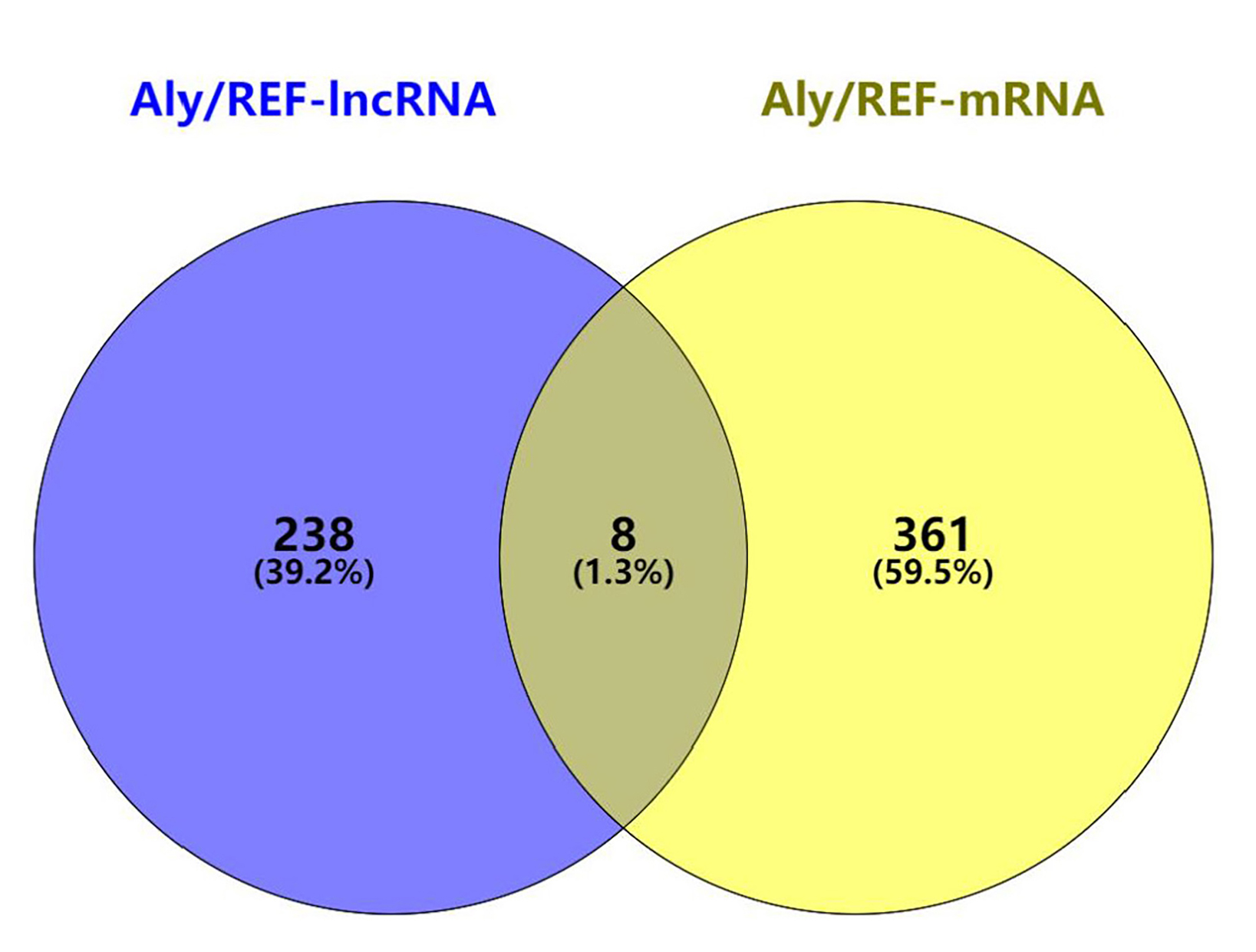
Firstly, the lncRNA-mRNA correlations were analyzed based on 369 differentially expressed mRNAs and 477 differentially expressed lncRNAs. For better investigating the associations among the coding genes, the NIANA approach [28] was used for regulatory network analysis, which exhibited the network objects for the 246 candidate lncRNAs.
Secondly, we screened the candidates in the lncRNA-mRNA
interaction network by the correlation
Similarly, 8 hub mRNA candidates (SLC3A2, CCNL1, WDR81, MLLT10, BCL2L1, FHL1, GON4L, MYO15B) also exerted vital parts in the regulatory network, as observed from Table 7. In addition, there were more genes and signaling pathways involved in the above constructed network, suggesting the complicated mechanisms by which Aly/REF-binding mRNAs and lncRNAs regulated the AAA pathogenic mechanism.
 Fig. 7.
Fig. 7.The Venn diagram of the candidates in the lncRNA-mRNA
interaction network.
lncRNA and mRNAs were selected with a threshold of correlation
| Transcript ID | Biotype | Strand | Gene ID | Locus | LncRNA Source | LncRNA length | Associated gene transID | Association gene name | |||
| ENST00000363981 | processed transcript | − | ENSG00000255717 | chr11:62619459-62623386 | Ensembl | 72 | ENST00000377892 | SLC3A2 | |||
| ENST00000364908 | misc RNA | + | ENSG00000201778 | chr3:156864296-156878549 | Ensembl | 93 | NM001308185 | CCNL1 | |||
| ENST00000571091 | lincRNA | − | ENSG00000186594 | chr17:1614804-1641893 | Ensembl | 525 | ENST00000446363 | WDR81 | |||
| uc001irb.3 | long noncoding | + | MLLT10 | chr10:21823093-22032559 | UCSC knowngene | 3611 | ENST00000307729 | MLLT10 | |||
| uc002wwk.3 | long noncoding | − | BCL2L1 | chr20:30252254-30311792 | UCSC knowngene | 2462 | ENST00000376062 | BCL2L1 | |||
| uc004ezm.2 | long noncoding | + | FHL1 | chrX:135228860-135293518 | UCSC knowngene | 2136 | ENST00000345434 | FHL1 | |||
| uc009wrg.1 | long noncoding | − | GON4L | chr1:155579566-155829191 | UCSC knowngene | 4500 | ENST00000271883 | GON4L | |||
| uc010dgi.1 | long noncoding | + | MYO15B | chr17:73584138-73704142 | UCSC knowngene | 3088 | ENST00000578462 | MYO15B | |||
| mRNA | |||||||||||
| Association gene name | Description | Deneid | Strand | Locus | RIP FPKM | Gene ID | Chromosome | Map location | |||
| SLC3A2 | solute carrier family 3 member 2 | ENSG00000168003 | + | chr11:62623517-62656355 | 14.1648 | 6520 | 11 | 11q13 | |||
| CCNL1 | cyclin L1 | ENSG00000163660 | − | chr3:156864296-156878549 | 27.0217 | 57018 | 3 | 3q25.31 | |||
| WDR81 | WD repeat domain 81 | ENSG00000167716 | + | chr17:1614804-1641893 | 46.204 | 124997 | 17 | 17p13.3 | |||
| MLLT10 | myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 | ENSG00000078403 | + | chr10:21823093-22032559 | 14.4303 | 8028 | 10 | 10p12 | |||
| BCL2L1 | BCL2 like 1 | ENSG00000171552 | − | chr20:30252254-30311792 | 26.9695 | 598 | 20 | 20q11.21 | |||
| FHL1 | four and a half LIM domains 1 | ENSG00000022267 | + | chrX:135228860-135293518 | 166.2 | 2273 | X | Xq26 | |||
| GON4L | gon-4-like (C. elegans) | ENSG00000116580 | − | chr1:155579566-155829191 | 42.58 | 54856 | 1 | 1q22 | |||
| MYO15B | myosin XVB | ENSG00000266714 | + | chr17:73584138-73704142 | 230.939 | 80022 | 17 | 17q25.1 | |||
| Note, RIP vs IgG. FPKM, fragments per kilobase of transcript per million mapped reads. | |||||||||||
AAA is one of the most severe vascular diseases in the vascular surgery [29, 30]. Because of the complex pathogenesis of AAA, efficient medical treatment for preventing the occurrence and rupture of AAA is lacking so far [31]. Among the researches on the AAA pathogenesis, epigenetic regulation occupies an increasing decisive position [32]. 5-methylcytosine modification on mRNA is a novel mRNA epigenetic modification, which have been found to play an essential role in other diseases [33, 34]. However, the evidence on the relationship between AAA and mRNA m5C modification is still lack. Therefore, this work aimed to examine the relationship of mRNA m5C expression with AAA. And for the first time, we observed that increased mRNA m5C modification occurred in AAA tissues, compared with the healthy controls.
Here, we observed up-regulating in the AAA tissue samples relative to matched
normal tissues (
Aly/REF is a specific mRNA m5C reader, which can bind to m5C sites in mRNA [33, 42], and contributes to the regulation of mRNA export [43]. Aly/REF plays a critical role on promoting the nuclear-cytoplasmic shuttling mRNA export in conjunction with NSUN2 [33]. It has been reported previously that either knock-down of NSUN2 or knock-down of Aly/REF affected the cytoplasmic to nuclear ratios of mRNAs [36]. In the current study, we found a higher level of Aly/REF in AAA group. Additionally, the expression of Aly/REF was strongly correlated with m5C status of mRNAs and the expression of NSUN2. Furthermore, according to the IHC analysis and IF analysis, NSUN2 and Aly/REF were co-expressed in the inflammatory cells in AAA tissues, which illustrated that NSUN2 and Aly/REF might mediate mRNA m5C modification in inflammatory cells. Yang et al. [33] have suggested a similar point about m5C formation in mRNAs is mainly catalyzed by the RNA methyltransferase NSUN2, and m5C is specifically recognized by the mRNA export adaptor Aly/REF as shown by in vitro and in vivo studies. NSUN2 modulates Aly/REF’s nuclear-cytoplasmic shuttling, RNA-binding affinity and associated mRNA export. The modification may be related with inflammatory infiltration in AAA. It has been reported that NSUN2 regulated AAA formation by promoting T cell recruitment. However, the research on the association of inflammation with m5C modification and Aly/REF is still lack. Phenotypic transformation of immune cells is an important mechanism of AAA progression. Further researches should focus on the potential role of m5C modification in the phenotypic transformation of immune cells.
Considering the higher level of Aly/REF in AAA and its specific binding to mRNA m5C sites as identified by other researches, Aly/REF may play a critical role in the mRNA m5C status in AAA. Therefore, we tried to perform RIP-Sequence of Aly/REF in AAA tissues to find out the downstream target lncRNA and mRNA of Aly/REF. According to our result, in AAA tissue samples, the downstream mRNAs were involved in several pathophysiological processes. As this study explored, Dnmt1 was candidate mRNAs of Aly/REF-interacting mRNA by RIP-seq identified in AAA tissue. Previous study demonstrated that high Aly/REF expression and low DNMT1 expression were both associated with poor head and neck squamous cell carcinoma prognosis [44]. Simultaneously, the target differential expression mRNA regulated by Aly/REF were exhibited by PPI network, four hub genes were obtained, including ALB, ATM, TRIP12, and HIF1A. As well as Aly/REF-HIF1A interaction, a bioprocesses of bladder cancer cells were demonstrated by a series of experiments in vitro, they found that hypoxia-inducible factor-1alpha (HIF-1A) indirectly up-regulated the expression of PKM2 by activating Aly/REF in addition to activating its transcription directly [45]. Future cellular and molecular studies are required to further validate our findings and to better understand the genes targeted for m5C modifications during AAA progression.
For example, Aly/REF-interacting lncRNAs were associated with various biological processes, including immune system process and macrophages infiltration, i.e., macrophage cytokine production, MHC class I protein complex, MHC class I protein binding, and MHC protein binding. Meanwhile, lncRNAs also participated in Phagosome pathway, PPAR signaling pathway and ECM-receptor interaction pathway, which have been identified to be modulated in the current study. Long non-coding RNAs (lncRNAs), which are the main non-coding RNAs, are transcripts longer than 200 nt. They are known to play a key role in chromatin remodeling, transcription, and post-transcriptional regulation [46]. At present, few studies exist on m5C related lncRNAs. Studies have performed quantitative mapping of the m5C sites in Arabidopsis thaliana on a transcriptome range, and found more than 1000 m5C sites in mRNA, long non-coding RNA [47]. NSUN2 may play a similar role in lncRNAs. Sun et al.[48] showed that NSUN2 deficiency significantly decreased the half-life of H19 RNA (lncRNA), which might be regulated by NSUN2-mediated m5C modification in hepatocellular carcinoma. Nevertheless, the mechanism of m5C methylation in lncRNAs promoting AAA progress is unclear, and deeper exploration would be helpful for understanding the pathogenesis of AAA.
Through bioinformation method, we also predicted eight hub mRNA candidates (SLC3A2, CCNL1, WDR81, MLLT10, BCL2L1, FHL1, GON4L, MYO15B) exerted vital parts in the lncRNAs-mRNAs regulation network. The sequence profile provided reliable basis for the mechanism of m5C methylation regulating AAA progression. However, further researches are needed to focus on the relationship of m5C target RNA and AAA, which may uncover a new cause of AAA occurrence.
We were first to observe m5A modification in human abdominal aortic aneurysm tissues. The results also reveal the important roles of m5C modulators, including NUSN2 and Aly/REF, in the pathogenesis of human AAA and provide a new view on m5C modification in AAA. Our findings suggest a potential mechanism of RNA methylation modification in clinical AAA. Understanding how these m5C RNA modifications occur, and the correlation between lncRNA changes in structure and function, may open up new therapeutic possibilities in AAA. Future molecular and cellular studies are required to further identify our findings and to better understand the genes targeted for m5C RNA methylation modifications during AAA progression.
Conceptualization—JZ and YSH; methodology—YCH, FXY and YHW; software—YSH and YCH; formal analysis—SYW, YSH and YCH; investigation—JZ and YSH; data curation— HZ and YCH; writing-original draft preparation—PPG, YCH and YSH; writing-review and editing—SJX and JZ; supervision—JZ and YSH; funding acquisition—YSH and JZ. All authors have read and agreed to the published version of the manuscript.
Human AAA samples collection conducted according to the Guidelines of the World Medical Association Declaration of Helsinki, and was approved by the Ethics Committees of First Hospital of China Medical University (ethical approval number: 2019-97-2).
We thank Cloud-Seq Biotech Ltd. Co. (Shanghai, China) for the RIP-transcriptome sequencing service and the subsequent bioinformatics analysis.
This work was supported by the Fundamental Research Funds for the Central Universities (grant number: DUT19RC(3)076), the National Natural Science Foundation of China (grant number: 81600370), and the China Postdoctoral Science Foundation (grant number: 2018M640270) for Yanshuo Han. This work was supported by the National Natural Science Foundation of China (grant: 81970402) for Jian Zhang.
The authors declare no conflict of interest.
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database (Accession Number: GSE 163615).
Supplementary material associated with this article can be found, in the online version, at https://www.fbscience.com/Landmark/articles/10.52586/5016.
AAA, abdominal aortic aneurysm; Ctrl, control group.