Frontiers in Bioscience-Landmark (FBL) is published by IMR Press from Volume 26 Issue 5 (2021). Previous articles were published by another publisher on a subscription basis, and they are hosted by IMR Press on imrpress.com as a courtesy and upon agreement with Frontiers in Bioscience.
1 Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, Univ Paris-Sud, Universite Paris-Saclay, F-91198, Gif-sur-Yvette cedex, France
Abstract
Mammalian NO-Synthases (NOSs) are the enzymatic sources of Nitric Oxide (NO°), a paradigmatic gasotransmitter involved in many (patho)-physiological processes. The increasing number of available genomes led to the identification of hundreds of new NOS proteins throughout the kingdoms of life, calling for a global investigation of this family of proteins. These new NOSs are commonly believed to share the same structure, functioning and role as mammalian NOSs. The scope of this article is to highlight the singularity of these NOSs and to describe their complex structural and functional diversity. NOS appears as a unique enzymatic machinery that exhibits a complex Structure – Activity – Function relationship. Its sophisticated redox mechanism and enzymatic regulation, coupled to the vast biological chemistry of reactive nitrogen species, leads to a specific cross-talk between NOS catalysis and its biological environment that implies a complex evolution of NOS function. This paper addresses the relationship between structure, function and evolution of NOS proteins using three NOS model families and advocates for an integrative and interdisciplinary approach that combines modelling studies, structural characterization, and in vitro/in vivo functional investigations.
Keywords
- NO-Synthases
- Nitric Oxide
- Phylogeny
- Mechanism
- Function
- Evolution
- Review
The biology of Nitric Oxide (NO°) started almost 250 years ago with the chemical synthesis of “Nitrous air” (NO°) by Joseph Priestley (1) and of its dephlogisticated or diminished product (now known as nitrous oxide, N2O). Although N2O biology was later on investigated by Humphry Davy (2), NO° itself did not gather much interest . 100 years later, another nitrogen oxide emerged in human physiology: Amyl nitrite was found to open the coronary arteries (Lauder Brunton, (3)) and derivatives such as nitroglycerin were rapidly used as medicine. It was even prescribed to Alfred Nobel who was suffering from intense chest pain – “Isn't it the irony of fate that I have been prescribed N/G 1, to be taken internally! They call it Trinitrin, so as not to scare the chemist and the public” (letter dated October 25, 1896 https://https://www.nobelprize.org/alfred_nobel/biographical/articles/ringertz/). Another century was necessary to unveil the major importance of NO° and other reactive nitrogen species in Biology.
Although its biosynthesis was already reported in bacteria as early as in the 50’s (4, 5), NO° has long been considered as a poisonous chemical. The concrete birth of NO° physiological history truly coincides with the unveiling of the nature of the Endothelium-Derived Relaxing Factor (EDRF (6, 7)). The discovery that such a major physiological mediator could be a highly reactive, hydrophobic, redox, diffusible gas somehow changed the way the community envisions signaling processes (8-11). This momentum has shaped the vision that we have of this molecule and changed its biological status: from a toxic, pollutant gas, it became a genuine biomolecule that diffuses and reacts with its expected targets, a unique mediator at the core of inter-cellular signaling. Because of this historical mold, NO° field mostly expands in mammals and reveals the involvement of NO° in an always larger number of physiological processes (12-15). Soon the enzymatic source of NO° was identified, and this new family of proteins was naturally named after their related production: NO-Synthases. Three isoforms were characterized: NOS1 and NOS3 are constitutively expressed NOSs named after the tissues they were isolated from: neuronal NOS (nNOS, (16)) and endothelial NOS (eNOS, (17)), respectively. NOS2 is inducibly expressed upon immune signaling and was therefore named inducible NOS (iNOS, (18)). These three types of NOSs were almost exclusively investigated from proteins issued from model mammals, respectively rattus norvegicus (19), mus musculus (20) and bos taurus (21). These three mammalian NOSs became the canonical NOSs (22-26) on which most of the enzymological, biochemical and structural investigations had been be achieved (with some data on human NOS isoforms, and splice variants (27-29)).
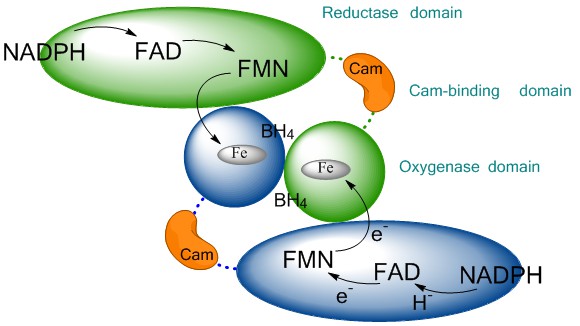
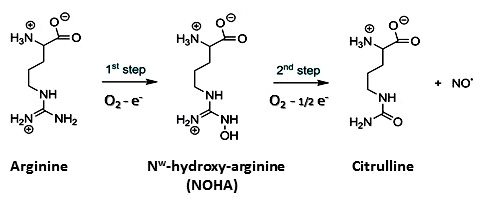
The standard knowledge acquired on NOSs structure, activity and function, that we will now briefly described, is entirely derived from investigations on these sole mammalian NOSs. As this article does not aim at providing a review on NOS structure, function or activity, we invite readers to look for the most recent reviews on that topics (30-36). Mammalian NOS isoforms share a high sequence homology (37, 38). All isoforms comprise an N-ter-oxygenase domain (Oxy), a Calmodulin-binding region (CaM) and a C-ter reductase domain (Figure 1), folded into a dynamic dimeric structure (39-41). NO° synthesis is achieved by the catalytic site, a heme-b cofactor buried within the oxygenase domain (42) that catalyzes two sequential oxidation reactions of L-arginine into NO° and citrulline (Figure 2). Electrons required for catalysis are conveyed by the reductase domain from the second substrate (NADPH), through two flavin cofactors (FMN and FAD), down to the heme catalytic site (34, 43, 44). Calmodulin binding, upon increase in Ca2+ concentration, allows a faster and coupled electron transfer (ET) from one monomer’s FMN to the other monomer’s oxygenase (Figure 1, (45-47)). Major structural differences between mammalian NOSs are both located in the N-ter and C-ter regions (24): i) whereas iNOSs remains cytosolic upon transcription and traduction, eNOS harbors palmitoylation and myristoylation sites in the N-ter region, that anchors it to the membrane of caveolae (48) and nNOS display as N-ter extension PDZ motifs that favors its binding to NMDAR complexes (49, 50); ii) whereas iNOS ET is mostly controlled by substrate binding (51), eNOS and nNOS reductase domains display different Auto-Inhibitory Elements (AIE) that finely tune and regulate ET and heme reduction (33, 52); iii) another major difference is the absence of sensitivity of CaM binding towards Ca2+ for iNOS (53, 54). These differences naturally fitted the distinction between highly regulated constitutive NOSs devoted to signal transduction – a process that requires fine catalytic tuning – and unleashed iNOS, associated to cytotoxic activity, for which a high and unregulated NO° production was expected. Thus, the cleavage between iNOS and constitutive NOSs functions has been mostly thought of in terms of differences in NO° flux, protein/protein interactions and post-translational modifications.
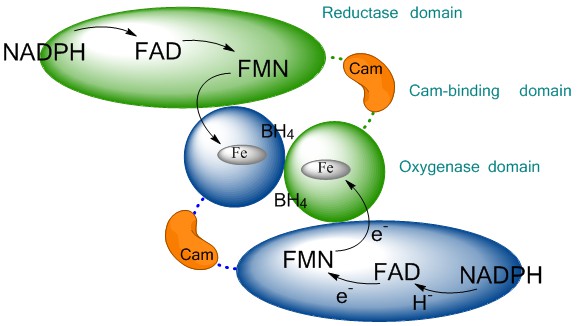 Figure 1
Figure 1Schematic representation of the NO-synthase quaternary structure. The reductase domain (ellipse) of one monomer (green) conveys the electron provided by NADPH to the oxygenase domain (circle) of the other monomer (blue) via successive electron transfers through FAD, FMN. The use of two sequential flavins allows the conversion of NADPH-hydride (H-) into regulated electron transfer up to the heme. This functional dimerization is insured by the binding of Calmodulin (CaM, orange) to the CaM-binding region and by the binding of BH4 cofactor at the oxygenase interface.
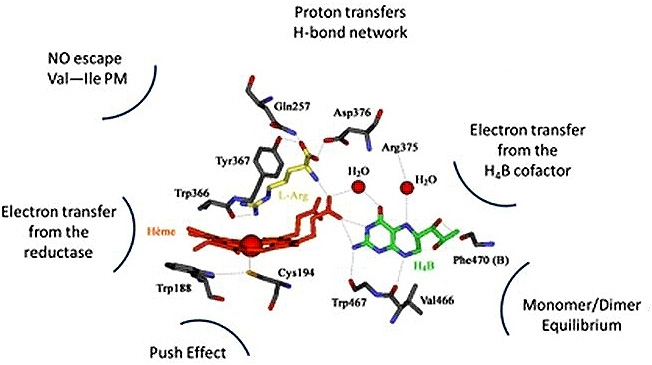
However, the study of NOSs molecular mechanism makes this dual partition more complex as it reveals sophisticated and specific catalytic regulation patterns. This is mostly due to the particularity of the subtle redox mechanism of NOS oxygenase: although NOS is fitted to produce NO°, NO° remains a strong inhibitor of NOS (Ki in the pM range, (55, 56)) that is engaged in fast geminate recombination (57). This NO rebinding modifies NOS catalytic production by generating two alternative catalytic cycles (31, 58, 59). In this regard, subtle changes in the values of key-kinetic steps will differentially modify the catalytic activity of NOS isoforms (55, 56). This is for example the case for the ratio between NO° off-rate versus heme reduction rates, the differences in ET rate between isoforms, the variations in FeIINO oxidation rates (55, 58, 60-63). These small variations, unpredictable from the 3D structures, induce differences in the NADPH/NO coupling and in the sensitivity to external factors (O2 concentration, Arg availability, redox status, (55, 58)). In the recent years, it has become obvious that NOSs are a complex machinery that might catalyze additional reactions (64-67) and produce many different reactive oxygen species such as superoxide, peroxynitrite…(60, 68-72).
Though, this knowledge has been overlooked. Data have been produced faster than they had been incorporated in an integrated picture of NOS/NO° activity and function. The impressive improvement of sequencing technologies has provided thousands of new genomes in which the presence of proteins homologous to NOS has been sought for. New NOSs emerged at first from genomes from bacteria, amoeba, mollusks, insects, and etc. X-Ray structures of the oxygenase domains were generated almost simultaneously (73, 74) or even before any functional characterization (75). A few enzymatic (73, 76-78) or mechanistic (79) assays confirmed the capacity of their catalytic site to produce NO°, leading to the conclusion that these many different NOSs were genuine NO-Synthases. This shaped the belief and the practice of the NOS/NO° community: any newly identified NOS that harbors a similar oxygenase domain is believed to behave like a mammalian NOS before any functional characterization. This assumption is mobilized in most of the investigations on NOSs. And even when these NOSs do not seem able to produce NO°, I’ve been replied, for example for bsNOS, that “they have to, it’s just that we don’t know how they do it”…
This article aims at clarifying and improving the way the NO° community deals with these many new NO-Synthases and addresses the question about “what NOS really stands for”. As we will try to show in this article, the NOSs do not constitute a homogenous protein family. This article provides a state-of-the-art picture of the presence of NOSs in the whole tree of life, highlighting their surprising structural diversity, their large of array of – often opposite -- functions and their inexplicable phylogenetic distribution. We believe that the current knowledge and concepts in the field are no longer adapted to investigate this new and vast scientific territory. In this regard, this article will briefly describe NOSs from three distinct model families: cyanobacteria, plants and basal metazoan and discuss the words, concepts, paradigms we may need to answer an apparently simple question: What is a NOS?
The historical circumstances of the unveiling of NO° physiological role – its implication in the regulation of mammalian vascular tone - have delineated the field in which NOS/NO° investigations were to be achieved and circumscribed the research on NO-Synthases mostly to the mammalian phylum. The first identified NO-Synthases were cloned from rats, mice, cows and humans and these NOSs have been used as the major investigation models so far (16-21). A handful of NOSs were cloned from other model organisms such as insects (Drosophila melanogaster, (80-83)), amoeba (Physarum polycephalum,(84, 85)) and mollusks (Limax valentianus, (86)), but the number of reports on these NOSs has remained negligible in contrast with the huge literature on mammalian NOS (mNOS). With the rapid evolution of the sequencing technologies, the presence of NOS-like proteins has been identified in many other genomes. Sequences homologous to NOS oxygenase domain were found at first in various bacteria, from the firmicutes and Deinococcus phyla: Bacillus subtilis, Staphylococcus aureus, Deinococcus radiodurans (2002, (73, 75, 76)) Bacillus Anthrax (2005, (87)). Later, similar sequence blasts led to the identification of NOSs in the genomes of bacteria from other phyla, such as Actinobacteria (Streptomyces turgidiscabies, (88)) or alpha-proteobacteria (Sorangium cellulosum, (89)). Although these bacterial NOSs (bacNOS) corresponded to a severely truncated form of the mammalian NOSs (90, 91) (see Figure 3), this new family of protein was spontaneously labelled bacterial NOS-like proteins and used as models for mammalian NOS study, which actually meant that they were believed to behave “like mammalian NOS”.
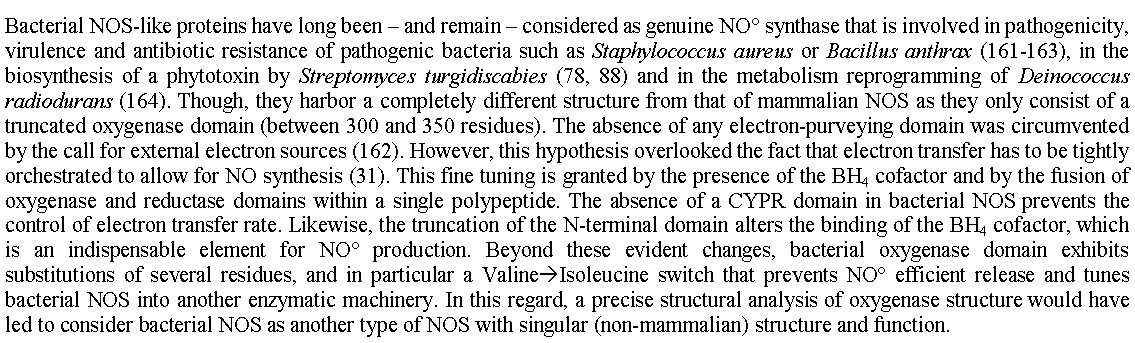 Figure 3
Figure 3Bacterial NOS-like proteins have long been – and remain – considered as genuine NO° synthase that is involved in pathogenicity, virulence and antibiotic resistance of pathogenic bacteria such as Staphylococcus aureus or Bacillus anthrax (161-163), in the biosynthesis of a phytotoxin by Streptomyces turgidiscabies (78, 88) and in the metabolism reprogramming of Deinococcus radiodurans (164). Though, they harbor a completely different structure from that of mammalian NOS as they only consist of a truncated oxygenase domain (between 300 and 350 residues). The absence of any electron-purveying domain was circumvented by the call for external electron sources (162). However, this hypothesis overlooked the fact that electron transfer has to be tightly orchestrated to allow for NO synthesis (31). This fine tuning is granted by the presence of the BH4 cofactor and by the fusion of oxygenase and reductase domains within a single polypeptide. The absence of a CYPR domain in bacterial NOS prevents the control of electron transfer rate. Likewise, the truncation of the N-terminal domain alters the binding of the BH4 cofactor, which is an indispensable element for NO° production. Beyond these evident changes, bacterial oxygenase domain exhibits substitutions of several residues, and in particular a Valine'Isoleucine switch that prevents NO° efficient release and tunes bacterial NOS into another enzymatic machinery. In this regard, a precise structural analysis of oxygenase structure would have led to consider bacterial NOS as another type of NOS with singular (non-mammalian) structure and function.
15 years later this reductive approach is no longer tenable: today, a simple blast of the available sequenced genomes led to the identification of hundreds of new NOS-like proteins in reptiles, turtles, amphibians, fishes, birds, insects, molluscs, arthropods and various primitive metazoans such as placozoan, amoeba, sponges, corals (see Tables). At least one thousand NOSs were found in the sole eubacteria kingdom (Table 1). However, the gene/protein banks annotation has often proven to be misleading as proteins were labelled as NOSs on the simple presence of small regulatory or non-essential structural motifs (if not linked to the presence of a small GTPase once associated to NO synthase in plants or snails, (92, 93)). Using two different templates we have achieved a blast against all available genomes and cleaned the resulting protein list (see legend of Table 1) in order to provide an update and accurate picture of this new family of protein (Tables 1-3). We will here describe the major phylogenetic groups where NOSs are to be found but also those from where NOS is absent.
| Firmicutes-Bacilli-Bacillaceae-Bacillus | |||
|---|---|---|---|
Bacillus subtilis Bacillus acidiceler Bacillus acidicola Bacillus aerophilus Bacillus akibai Bacillus alcalophilus Bacillus alkalitelluris Bacillus altitudinis Bacillus alveayuensis Bacillus aminovorans B. amyloliquefaciens Bacillus anthracis str. Bacillus aquimaris Bacillus aryabhattai Bacillus atrophaeus Bacillus aurantiacus Bacillus australimaris Bacillus axarquiensis Bacillus badius Bacillus beveridgei Bacillus bingmayongensis Bacillus bogoriensis Bacillus bombysepticus Bacillus butanolivorans Bacillus chagannorensis Bacillus cellulosilyticus Bacillus cellulasensis Bacillus cereus |
Bacillus cihuensis Bacillus clarkii Bacillus clausii Bacillus coahuilensis Bacillus cohnii Bacillus cytotoxicus Bacillus daliensis Bacillus eiseniae Bacillus encimensis Bacillus enclensis Bacillus endophyticus Bacillus farraginis Bacillus fastidiosus Bacillus firmus Bacillus flexus Bacillus gaemokensis Bacillus ginsengihumi Bacillus gobiensis Bacillus gottheilii B. glycinifermentans Bacillus halodurans Bacillus halotolerans Bacillus horikoshii Bacillus indicus Bacillus infantis Bacillus lonarensis Bacillus isronensis Bacillus koreensis |
Bacillus korlensis Bacillus krulwichiae Bacillus lehensis G1 Bacillus licheniformis Bacillus ligniniphilus Bacillus luciferensis] Bacillus macauensis Bacillus malacitensis Bacillus manliponensis Bacillus marisflavi Bacillus marmarensis B. massiliosenegalensis Bacillus massilioanorexius Bacillus massiliogorillae Bacillus megaterium Bacillus mojavensis Bacillus muralis Bacillus nealsonii Bacillus ndiopicus Bacillus obstructivus Bacillus okhensis Bacillus oleronius Bacillus mycoides Bacillus panaciterrae Bacillus paralicheniformis Bacillus patagoniensis Bacillus pseudofirmus Bacillus pseudalcaliphilus |
Bacillus pseudomycoides Bacillus pumilus B. psychrosaccharolyticus Bacillus rhizosphaerae Bacillus safensis Bacillus salsus Bacillus selenitireducens Bacillus shackletonii Bacillus simplex Bacillus siamensis Bacillus sonorensis Bacillus tequilensis Bacillus testis Bacillus thuringiensis Bacillus toyonensis Bacillus trypoxylicola Bacillus vallismotis Bacillus vietnamensis Bacillus velezensis Bacillus vireti Bacillus weihaiensis B. weihenstephanensis Bacillus wiedmannii Bacillus wakoensis Bacillus xiamenensis Bacillus zhangzhouensis Brevibacterium frigoritolerans |
| Firmicutes-Bacilli-Bacillaceae-Lysinibacillus | |||
Lysinibacillus massiliensis Lysinibacillus sphaericus Lysinibacillus acetophenoni Lysinibacillus xylanilyticus |
Lysinibacillus fusiformis ZC1 Lysinibacillus contaminans Lysinibacillus manganicus Lysinibacillus sinduriensis |
Lysinibacillus xyleni Lysinibacillus boronitolerans Lysinibacillus varians Lysinibacillus saudimassiliensis |
|
| Firmicutes-Bacilli-Bacillaceae-Anoxybacillus | |||
Anoxybacillus flavithermus Anoxybacillus thermarum Anoxybacillus amylolyticus |
Anoxybacillus aydernesis Anoxybacillus suryakudensis Anoxybacillus pushchinoensis |
Anoxybacillus tepidamans | |
| Firmicutes-Bacilli-Bacillaceae-Pontibacillus | |||
Pontibacillus marinus Pontibacillus chungwhensis |
Pontibacillus yanchengensis Y32 Pontibacillus litoralis |
Pontibacillus halophilus | |
| Firmicutes-Bacilli-Bacillaceae-Geobacillus | |||
Geobacillus Stearothermophilus Geobacillus thermoleovorans Geobacillus kaustophilus |
Geobacillus vulcani Geobacillus jurassicus Geobacillus thermocatenulatus |
Geobacillus subterraneus Geobacillus zalihae Geobacillus uzenensis | |
| Firmicutes-Bacilli-Bacillaceae-Oceanobacillus | |||
Oceanobacillus picturae Oceanobacillus oncorhynchi Oceanobacillus sojae |
Oceanobacillus iheyensis Oceanobacillus manasiensis Oceanobacillus massiliensis |
Oceanobacillus jeddahense Oceanobacillus timonensis | |
| Firmicutes-Bacilli-Bacillaceae-Halobacillus | |||
Halobacillus aidingensis Halobacillus alkaliphilus Halobacillus dabanensis |
Halobacillus halophilus Halobacillus karajensis Halobacillus kuroshimensis |
Halobacillus mangrovi Halobacillus massiliensis Halobacillus salinus | |
| Firmicutes-Bacilli-Bacillaceae | |||
Anaerobacillus arseniciselenatis Anaerobacillus macyae Salsuginibacillus kocurii Salipaludibacillus aurantiacus Sediminibacillus halophilus Caldalkalibacillus thermarum Aquibacillus sp. Edaphobacillus lindanitolerans Salimicrobium halophilum Salimicrobium jeotgali Salimicrobium album |
Terribacillus saccharophilus Terribacillus halophilus Terribacillus aidingensis Marinococcus halotolerans Marinococcus luteus Marinococcus halophilus Fictibacillus phosphorivorans Fictibacillus macauensis Fictibacillus arsenicus Paucisalibacillus globulus Thalassobacillus cyri |
Domibacillus indicus Domibacillus aminovorans Domibacillus iocasae Domibacillus enclensis Virgibacillus proomii Virgibacillus halodenitrificans Virgibacillus pantothenticus Virgibacillus chiguensis Virgibacillus dokdonensis Psychrobacillus psychrotolerans Psychrobacillus psychrodurans | |
| Firmicutes-Bacilli-Paenibacillae | |||
Brevibacillus brevis Brevibacillus parabrevis Brevibacillus formosus Brevibacillus borstelensis |
Brevibacillus choshinensis Brevibacillus borstelensis Brevibacillus panacihumiSaccharibacillus sacchari Brevibacillus reuszeri |
Saccharibacillus sacchari Saccharibacillus kuerlensis Cohnella thermotolerans Cohnella laeviribosi | |
| Firmicutes-Bacilli-Paenibacillae-Paenibacillus | |||
Paenibacillus alginolyticus Paenibacillus algorifonticola Paenibacillus alvei Paenibacillus amylolyticus Paenibacillus antarcticus Paenibacillus assamensis Paenibacillaceae bacterium GAS479 Paenibacillus beijingensis Paenibacillus borealis Paenibacillus bovis Paenibacillus camerounensis Paenibacillus campinasensis Paenibacillus catalpae Paenibacillus chitinolyticus Paenibacillus chondroitinus Paenibacillus curdlanolyticus Paenibacillus daejeonensis Paenibacillus dauci Paenibacillus dendritiformis Paenibacillus ehimensis |
Paenibacillus elgii Paenibacillus etheri Paenibacillus ferrarius Paenibacillus fonticola Paenibacillus glacialis Paenibacillus glucanolyticus Paenibacillus harenae Paenibacillus gorillae Paenibacillus ihbetae Paenibacillus jamilae Paenibacillus kribbensis Paenibacillus lactis Paenibacillus lautus Paenibacillus larvae subsp. larvae Paenibacillus macquariensis Paenibacillus massiliensis Paenibacillus naphthalenovorans Paenibacillus odorifer Paenibacillus pabuli Paenibacillus panacisoli Paenibacillus pectinilyticus |
Paenibacillus peoriae Paenibacillus pini Paenibacillus pinihumi Paenibacillus polymyxa Paenibacillus popilliae Paenibacillus rhizosphaerae Paenibacillus senegalensis Paenibacillus selenitireducens Paenibacillus swuensis Paenibacillus taiwanensis Paenibacillus terrae Paenibacillus terrigena Paenibacillus thiaminolyticus Paenibacillus tianmuensis Paenibacillus taichungensis Paenibacillus typhae Paenibacillus tyrfis Paenibacillus uliginis Paenibacillus vortex Paenibacillus xylanexedens Paenibacillus yonginensis | |
| Firmicutes-Bacilli-Alicyclobacillae | |||
Alicyclobacillus acidocaldarius Alicyclobacillus acidiphilus Alicyclobacillus sendaiensis Alicyclobacillus mali |
Alicyclobacillus contaminans Alicyclobacillus pomorum Alicyclobacillus vulcanalis Alicyclobacillus tengchongensis |
Alicyclobacillus hesperidum Alicyclobacillus acidoterrestris | |
| Firmicutes-Bacilli-Planococcaceae | |||
Sporosarcina globispora Sporosarcina newyorkensis Sporosarcina psychrophila Sporosarcina ureae Paenisporosarcina sp. Planomicrobium okeanokoites Planomicrobium glaciei Viridibacillus arvi Viridibacillus arenosi |
Jeotgalibacillus soli Jeotgalibacillus alimentarius Jeotgalibacillus campisalis Jeotgalibacillus malaysiensis Jeotgalicoccus psychrophilus Planococcaceae bacterium VT-49 Solibacillus silvestris StLB046 Solibacillus kalamii |
Bhargavaea cecembensis Bhargavaea ginseng Bhargavaea beijingensis Rummeliibacillus stabekisii Caryophanon tenue Caryophanon latum Kurthia huakuii Kurthia gibsonii Kurthia massiliensis | |
| Firmicutes-Bacilli-Planococcaceae-Planococcus | |||
Planococcus halocryophilus Planococcus faecalis Planococcus maritimus |
Planococcus antarcticus Planococcus donghaensis Planococcus plakortidis |
Planococcus massiliensis Planococcus rifietoensis | |
| Firmicutes-Bacilli-Thermoactinomycetaceae | |||
Shimazuella kribbensis Thermoactinomyces vulgaris |
Marininema halotolerans Marininema mesophilum |
Risungbinella massiliensis | |
| Firmicutes-Bacilli-Listeriaceae | |||
Listeria grayi Listeria riparia Listeria newyorkensis |
Listeria cornellensis Listeria grandensis Listeria rocourtiae Listeria booriae |
Listeria weihenstephanensis Listeriaceae bacterium Brochothrix campestris Brochothrix thermosphacta | |
| Firmicutes-Bacilli-Staphylococcaceae | |||
Staphylococcus arlettae Staphylococcus agnetis Staphylococcus argenteus Staphylococcus aureus Staphylococcus caprae Staphylococcus capitis Staphylococcus carnosus Staphylococcus cohnii Staphylococcus chromogenes Staphylococcus delphini Staphylococcus edaphicus Staphylococcus equorum Staphylococcus epidermidis Staphylococcus fleurettii Staphylococcus gallinarum |
Staphylococcus haemolyticus Staphylococcus hominis Staphylococcus hyicus Staphylococcus intermedius Staphylococcus lugdunensis Staphylococcus lutrae Staphylococcus microti Staphylococcus muscae Staphylococcus nepalensis Staphylococcus pasteuri Staphylococcus piscifermentans Staphylococcus pseudintermedius Staphylococcus massiliensis Staphylococcus saprophyticus Staphylococcus schleiferi |
Staphylococcus schweitzeri Staphylococcus sciuri Staphylococcus simiae Staphylococcus simulans Staphylococcus stepanovicii Staphylococcus warneri Staphylococcus pettenkoferi Staphylococcus xylosus Staphylococcus vitulinus Staphylococcus succinus Macrococcus caseolyticus Macrococcus canis Macrococcus sp Nosocomiicoccus massiliensis Nosocomiicoccus ampullae | |
| Firmicutes-Bacilli-Bacillales-unclassified | |||
Acidibacillus ferrooxidans Geomicrobium sp. JCM 19038 Exiguobacterium undae Exiguobacterium enclense Exiguobacterium alkaliphilum |
Exiguobacterium indicum Exiguobacterium acetylicum Exiguobacterium aurantiacum Exiguobacterium sibiricum 255-15 Exiguobacterium antarcticum B7 |
Exiguobacterium oxidotolerans Exiguobacterium chiriqhucha Exiguobacterium mexicanum Exiguobacterium marinum Exiguobacterium sp. | |
| Firmicutes-Bacilli-Lactobacillales-Streptococcaceae | |||
Streptococcus pneumoniae | |||
| Actinobacteria-Actinomycetales-Pseudonocardiaceae-Pseudonocardia | |||
Pseudonocardia dioxanivorans Pseudonocardia ammonioxydans Pseudonocardia acaciae |
Pseudonocardia autotrophica Pseudonocardia spinosispora |
Pseudonocardia oroxyli Pseudonocardia thermophile Pseudonocardia sp. | |
| Actinobacteria-Actinomycetales-Pseudonocardiaceae-Amycolatopsis | |||
Amycolatopsis mediterranei U32 Amycolatopsis azurea DSM 43854 Amycolatopsis orientalis Amycolatopsis japonica Amycolatopsis alba Amycolatopsis decaplanina Amycolatopsis keratiniphila |
Amycolatopsis vancoresmycina Amycolatopsis lurida NRRL 2430 Amycolatopsis lexingtonensis Amycolatopsis rifamycinica Amycolatopsis balhimycina Amycolatopsis tolypomycina Amycolatopsis xylanica |
Amycolatopsis pretoriensis Amycolatopsis regifaucium Amycolatopsis lurida Amycolatopsis coloradensis Amycolatopsis australiensis Amycolatopsis saalfeldensis Amycolatopsis sp. | |
| Actinobacteria-Actinomycetales-Pseudonocardiaceae | |||
Saccharopolyspora erythraea NRRL Saccharopolyspora sp(i)a NRRL Saccharopolyspora spinosa Saccharopolyspora hirsuta Saccharopolyspora flava Saccharopolyspora antimicrobica Saccharopolyspora shandongensis Saccharomonospora xinjiangensis Saccharothrix espanaensis Saccharothrix syringae Saccharothrix sp. Saccharothrix sp. Saccharothrix sp. Kibdelosporangium phytohabitans Kibdelosporangium aridum Kibdelosporangium sp. Actinomycetospora chiangmaiensis |
Actinoalloteichus cyanogriseus Actinoalloteichus spitiensis Actinoalloteichus sp. Actinoalloteichus hymeniacidonis Actinokineospora inagensis Actinokineospora bangkokensis Actinokineospora terrae Actinokineospora enzanensis A. spheciospongiae Actinosynnema mirum Actinosynnema pretiosum Actinophytocola xanthii Alloactinosynnema album Alloactinosynnema iranicum Alloactinosynnema sp. Actinosynnema sp. Actinophytocola xinjiangensis |
Lechevalieria aerocolonigenes Lechevalieria xinjiangensi Lechevalieria fradiae Allokutzneria albata Allokutzneria sp. NRRL B-24872 Kutzneria albida Kutzneria sp. 744 Catenulispora acidiphila Crossiella equi Lentzea albida Lentzea albidocapillata Lentzea kentuckyensis Lentzea flaviverrucosa Lentzea guizhouensis Lentzea jiangxiensis Lentzea violacea Lentzea waywayandensis | |
| Actinobacteria-Actinomycetales-Micromonosporaceae-Micronospora | |||
Micromonospora carbonacea Micromonospora globosa Micromonospora matsumotoense Micromonospora echinofusca Micromonospora mirobrigensis |
Micromonospora peucetia Micromonospora rosaria Micromonospora rifamycinica Micromonospora siamensis Micromonospora yangpuensis |
Micromonospora coxensis Micromonospora nigra M. purpureochromogenes Micromonospora globosa Micromonospora pattaloongensis | |
| Actinobacteria-Actinomycetales-Micromonosporaceae | |||
Actinoplanes missouriensis Actinoplanes utahensis Actinoplanes globisporus Actinoplanes subtropicus Actinoplanes friuliensis Actinoplanes philippinensis Actinoplanes derwentensis Actinoplanes awajinensis |
Salinispora arenicola Salinispora pacifica Salinispora tropica Actinoplanes atraurantiacus Actinoplanes rectilineatus Actinoplanes sp. Catenuloplanes japonicus Dactylosporangium aurantiacum |
Hamadaea tsunoensis Catelliglobosispora koreensis Couchioplanes caeruleus Hamadaea tsunoensis Asanoa ishikariensis Asanoa hainanensis Verrucosispora sp. | |
| Actinobacteria-Actinomycetales-Streptomycetaceae | |||
Streptacidiphilus jeojiense Streptacidiphilus albus Streptacidiphilus jiangxiensis Streptacidiphilus carbonis |
Streptacidiphilus melanogenes Streptacidiphilus neutrinimicus Streptacidiphilus anmyonensis Streptomycetaceae bacterium |
Streptoalloteichus hindustanus Kitasatospora mediocidica Kitasatospora azatica | |
| Actinobacteria-Actinomycetales-Streptomycetaceae-Streptomyces | |||
Streptomyces scabiei 87.22 Streptomyces alboverticillatus Streptomyces abyssalis Streptomyces acidiscabies 84-104 Streptomyces alboviridis Streptomyces alni Streptomyces aureofaciens Streptomyces avermitilis MA-4680 Streptomyces avicenniae Streptomyces bikiniensis Streptomyces diastatochromogenes Streptomyces erythrochromogenes Streptomyces exfoliatus Streptomyces flavochromogenes Streptomyces fulvoviolaceus Streptomyces glaucescens Streptomyces glauciniger Streptomyces griseus |
Streptomyces qinglanensis Streptomyces ipomoeae Streptomyces katrae Streptomyces lavendulae Streptomyces mirabilis Streptomyces globisporus Streptomyces griseoluteus Streptomyces griseochromogenes Streptomyces harbinensis Streptomyces indicus Streptomyces nanshensis Streptomyces olivochromogenes Streptomyces pathocidini Streptomyces puniciscabiei Streptomyces purpeofuscus Streptomyces roseus Streptomyces roseoverticillatus |
Streptomyces rubidus Streptomyces rubellomurinus Streptomyces turgidiscabies Streptomyces uncialis Streptomyces virginiae Streptomyces viridochromogenes Streptomyces vietnamensis Streptomyces scabrisporus Streptomyces specialis Streptomyces viridifaciens Streptomyces venezuelae Streptomyces wedmorensis Streptomyces xanthophaeus Streptomyces yanglinensis Streptomyces yokosukanensis Streptomyces zhaozhouensis | |
| Actinobacteria-Actinomycetales-Nocardiaceae-Nocardia | |||
Nocardia amamiensis Nocardia asiatica Nocardia altamirensis Nocardia asteroides NBRC Nocardia abscessus Nocardia arthritidis Nocardia anaemiae Nocardia acidivorans Nocardia arizonensis Nocardia brasiliensis Nocardia amikacinitolerans Nocardia brevicatena Nocardia araoensis |
Nocardia beijingensis Nocardia exalbida Nocardia gamkensis Nocardia lijiangensis Nocardia harenae Nocardia crassostreae Nocardia alba Nocardia niwae Nocardia otitidiscaviarum Nocardia pseudovaccinii Nocardia pseudobrasiliensis Nocardia puris Nocardia mexicana |
Nocardia salmonicida Nocardia soli Nocardia thailandica Nocardia transvalensis Nocardia takedensis Nocardia tenerifensis Nocardia terpenica Nocardia vulneris Nocardia vinacea Nocardia xishanensis Nocardia yamanashiensis | |
| Actinobacteria-Actinomycetales-Nocardiaceae-Rhodococcus | |||
Rhodococcus opacus B4/ M213 Rhodococcus imtechensis RKJ300 Rhodococcus jostii RHA1 Rhodococcus erythropolis Rhodococcus enclensis Rhodococcus wratislaviensis |
Rhodococcus rhodnii Rhodococcus fasciansRhodococcus rhodochrous Rhodococcus qingshengii Rhodococcus yunnanensis Rhodococcus koreensis |
Rhodococcus kyotonensis Rhodococcus marinonascens Rhodococcus maanshanensis Rhodococcus tukisamuensis | |
| Actinobacteria-Actinomycetales-Streptosporangiaceae | |||
Streptosporangium roseum DSM Streptosporangium amethystogenes Streptosporangium canum Streptosporangium amethystogenes Streptosporangium sp. Herbidospora cretacea Herbidospora daliensis Herbidospora sakaeratensis |
Nonomuraea coxensis Nonomuraea candida Nonomuraea solani Nonomuraea coxensis Nonomuraea wenchangensis Nonomuraea jiangxiensis Nonomuraea maritima Herbidospora mongoliensis |
Microbispora rosea Herbidospora yilanensis Microtetraspora glauca Planobispora rosea Planomonospora sphaerica Sinosporangium album | |
| Other Actinobacteria | |||
Actinobacteria-Corynebacteriales Skermania piniformis Mycobacterium abscessus. Corynebacterium sp. OG2 Actinobacteria-Micrococcales Brachybacterium. faecium Actinobacteria-Propionobacteriales Kribbella catacumbae Kribbella sp. ALI-6-A Actinobacteria-Frankiales Cryptosporangium arvum Sporichthya polymorpha Actinobacteria-Nakamurellales Nakamurella multipartita DSM |
Actinobacteria-Streptosporangiales Actinomadura flavalba Actinomadura hibisca Actinomadura formosensis Streptosporangium subroseum Actinobacteria-Jiangellales Jiangella gansuensis Jiangella alkaliphila Jiangella muralis Jiangella alba Unclassified Actinobacteria Actinobacteria bacterium OV450 | ||
| Cyanobacteria | |||
|
Aphanocapsa montana Scytonema hofmanni Aphanizomenon flos-aquae Dolichospermum circinale Calothrix sp PCC 7103 Microchaete sp PCC |
Aliterella atlantica CENA595 Neosynechococcus sphagnicola Nostoc sp Anabaena sp Chlorogloeopsis fritschii Mastigocoleus testarum |
Microcoleus sp Microcoleus vaginatus FGP-2 Crinalium epipsamum PCC9333 Planktothrix sp. Planktothrix paucivesiculata PCC9631 Synechococcus sp | |
| Bacteroidetes | |||
Marivirga sericea B Phaeodactylibacter xiamenensis Algoriphagus sp. M8-2 Bacteroidetes bacterium MED-G21 |
Spirosoma montaniterrae Spirosoma radiotolerans Spirosoma fluviale |
Spirosoma linguale Spirosoma rigui Spirosoma sp. | |
| Deinococcus | |||
Deinococcus maricopensis Deinococcus geothermalis Deinococcus peraridilitoris Deinococcus pimensis Deinococcus sp. Deinococcus actinosclerus |
Deinococcus aquatilis Deinococcus gobiensis Deinococcus deserti VCD115 Deinococcus proteolyticus MRP Deinococcus misasensis |
Deinococcus radiodurans Deinococcus murrayi Deinococcus pimensis Deinococcus grandis Deinococcus reticulitermitis Deinococcus wulumuqiensis | |
| Proteobacteria | |||
Proteobacteria (alpha)-Rhodobacterales Silicibacter sp. TrichCH4B Jannaschia seosinensis Henriciella sp. Pseudooceanicola sp. Pseudooceanicola marinus Pseudooceanicola antarticus Proteobacteria (alpha)-Rhodospirillales Rhodospirillaceae bacterium TMED140 Caenispirillum bisanense Proteobacteria (alpha)-Sneathiellales Sneathiella sp. |
Proteobacteria (gamma)-Enterobacterales Klebsiella pneumoniae Proteobacteria (beta) -Burkholderiales Hydrogenophaga sp. A37 Proteobacteria (alpha)-Sphingomonadales Erythrobacter sp. Sphingomonas elodea Sphingomonas pituitosa Sphingomonas sp. Leaf Novosphingobium subterraneum : Alphaproteobacteria bacterium TMED87 PA4 Proteobacteria (delta)-Myxococcales Sorangium cellulosum | ||
| Verrucomicrobia | |||
Verrucomicrobia bacterium SCGC AAA168-F10 Verrucomicrobia subdivision 6 bacterium BACL9 Verrucomicrobia bacterium SCGC AAA168F10 Verrucomicrobiaceae bacterium TMED137 Verrucomicrobiaceae bacterium TMED76 Verrucomicrobiaceae bacterium TMED86 |
Opitutae bacterium TMED149 Roseibacillus sp Rubritalea squalenifaciens DSM 18772 | ||
| Other Eubacteria | |||
Chloroflexi Ktedonobacter racemifer Coraliomargarita sp. TMED73 |
Balneolaeota Balneola vulgaris Balneolaceae bacterium TMED105 | ||
| Archaea-Euryarchaeota-Halobacteria-Natrialbales | |||
Natronobacterium gregoryi SP2 Natronobacterium texcoconense Natrialbaceae archaeon |
Halobiforma nitratireducens JCM 10879 Halostagnicola kamekurae Halostagnicola sp. | ||
| Archaea-Euryarchaeota-Halobacteria-Halobacteriales | |||
Natronomonas pharaonis Halorientalis sp. IM1011 |
Halovenus aranensis Halalkalicoccus paucihalophilus | ||
| Census has been achieved using the online NCBI Blastp suite ©. Two distinct Query sequences have been used (bsNOS from Bacillus subtilis subsp. spizizenii TU-B-10, and iNOSoxy ref) using BLOSUM 62 as matrix and low Gap Costs (Existence 6, Extension 2). Validity of retrieved sequences was verified via the presence of the NOS-specific heme binding WRNxxxC motif. The list was updated on the 23rd of November 2017. Used nomenclature is derived from the NCBI version of Lifemap © | |||
| Viridiplantae | ||
|---|---|---|
Klebsormidium nitens Klebsormidium flaccidum Chaetosphaeridium globosum Cosmarium subtumidum Planophila terrestris Chlamydomonas cribrum Phacotus lenticularis |
Bathycoccus prasinos Gonium pectorale Bolbocoleon piliferum Volvox aureus Pleurastrum insigne Golenkinia longispicula Scherffelia dubia |
Helicodictyon planctonicum Leptosira obovata Pandorina morum Nephroselmis pyriformis Pteromonas sp Ostreococcus tauri Ostreococcus lucimarinus |
| Stramenopiles-Bacillariophyta-Coscinodiscophyceae-Thalassiosirales | ||
| Thalassiosira oceanica | ||
| Stramenopiles-Oomycetes-Saprolegniales-Saprolegniaceae | ||
| Thraustotheca clavata | ||
| Opisthokonta-Fungi-Dikarya-Ascomycota | ||
Aspergillus arachidicola Aspergillus bombycis Aspergillus carbonarius ITEM Aspergillus kawachii Aspergillus luchuensis Aspergillus nomius Aspergillus oryzae Aspergillus niger Aspergillus flavus Aspergillus parasiticus SU-1 Aspergillus welwitschiae Dothistroma septosporum Elaphomyces granulatus Endocarpon pusillum Z07020 |
Colletotrichum chlorophyti Colletotrichum fioriniae PJ7 Colletotrichum graminicola M1.001 Colletotrichum higginsianum Colletotrichum gloeosporioides Colletotrichum incanum] Colletotrichum orbiculare MAFF 240422 Colletotrichum orchidophilum Colletotrichum salicis Colletotrichum simmondsii Colletotrichum sublineola Colletotrichum tofieldiae Coniosporium apollinis CBS 100218 Coniochaeta ligniaria NRRL 30616 |
Glomerella graminicola M1.001 Lepidopterella palustris CBS 45 Macrophomina phaseolina Neofusicoccum parvum UCRNP2 Oidiodendron maius Zn Ophiocordyceps sinensis CO18 Ophiocordyceps unilateralis Phialocephala scopiformis Phialocephala subalpina Pseudomassariella vexata Umbilicaria pustulata Verruconis gallopava Zymoseptoria tritici IPO323 Zymoseptoria brevis |
| Opisthokonta-Fungi-Dikarya-Basidiomycota | ||
| Rhizoctonia solani | ||
| Opisthokonta-Fungi-Chytridiomycota | ||
| Gonapodya prolifera JEL478 | ||
| Opisthokonta-Opisthokonta incertae sedis-Ichthyosporea-Ichthyophonida | ||
| Sphaeroforma arctica JP610 | ||
| Amoebozoa | ||
| Physarum polycephalum | ||
| Heterolobosea | ||
| Naegleria gruberi strain NEG-M | ||
| Haptophyceae-Prymnesiales-Chrysochromulinaceae | ||
| Chrysochromulina sp. CCMP291 | ||
| The same protocol as in Table 1 was used. Additional species in viridiplantae come from Jeandroz et al. (94). | ||
| Placozoa | ||
|---|---|---|
| Trichoplax adhaerens | ||
| Porifera | ||
| Amphimedon queenslandica | ||
| Cnidaria-Anthozoa | ||
Nematostella vectensis Acropora digitifera |
Discosoma striata Orbicella faveolata |
Stylophora pistillata Exaiptasia pallida |
| Cnidaria-Hydrozoa | ||
| Hydra magnipapillata | Hydra vulgaris | |
| Lophotrochozoa-Brachyopoda | ||
| Lingula anatina | ||
| Lophotrochozoa-Bryozoa | ||
| Bugula neritina | ||
| Lophotrochozoa-Annelida | ||
| Capitella teleta | ||
| Lophotrochozoa-Mollusca-Bivalvia | ||
Crassostrea gigas Crassostrea virginica |
Azumapecten farreri Mizuhopecten yessoensis |
Mytilus galloprovincialis |
| Lophotrochozoa-Mollusca | ||
| Sepia officinalis | Octopus bimaculoides | |
| Lophotrochozoa-Mollusca-Gastropoda | ||
Aplysia californica Ambigolimax valentianus Biomphalaria glabrata Haliotis asinina |
Lottia gigantea Lymnaea stagnalis Limulus polyphemus |
Planorbella trivolvis Stramonita canicula Stramonita haemastoma |
| Ecdysozoa-Panarthropoda-Tardigrada | ||
| Hypsibius dujardini | Ramazzottius varieornatus | |
| Ecdysozoa-Arthropoda-Chelicerata-Arachnida | ||
Ixodes scapularis Stegodyphus mimosarum |
Parasteatoda tepidariorum Tetranychus urticae |
Sarcoptes scabiei Euroglyphus maynei |
| Ecdysozoa-Arthropoda-Crustacea-Maxillopoda | ||
| Tigriopus japonicus | ||
| Ecdysozoa-Arthropoda-Crustacea-Branchiopoda | ||
| Daphnia pulex | Daphnia magna | |
| Ecdysozoa-Arthropoda-Crustacea-Eumalacostraca-Amphipoda | ||
| Hyalella azteca | ||
| Ecdysozoa-Arthropoda-Crustacea-Eumalacostraca-Decapoda | ||
Panulirus argus Carcinus maenas Gecarcinus lateralis |
Marsupenaeus japonicus Fenneropenaeus chinensis Penaeus monodon |
Litopenaeus vannamei Scylla paramamosain Portunus trituberculatus |
| Ecdysozoa-Arthropoda-Hexapoda-Collembola | ||
| Orchesella cincta | Folsomia candida | |
| Ecdysozoa-Arthropoda-Hexapoda-Insecta-Diptera | ||
Anopheles darlingi Anopheles dirus Anopheles gambiae Anopheles stephensi Anopheles sinensis Aedes aegypti Aedes albopictus Bactrocera dorsalis Bactrocera cucurbitae Bactrocera latifrons Bactrocera oleae Ceratitis capitata Clunio marinus Drosophila yakuba Drosophila erecta |
Drosophila mojavensis Drosophila grimshawi Drosophila simulans Drosophila willistoni Drosophila ananassae Drosophila melanogaster Drosophila persimilis Drosophila pseudoobscura Drosophila eugracilis Drosophila ficusphila Drosophila miranda Drosophila virilis Drosophila obscura Drosophila sechellia |
Drosophila biarmipes Drosophila takahashii Drosophila elegans Drosophila serrata Drosophila suzukii Drosophila kikkawai Drosophila bipectinata Drosophila arizonae Drosophila rhopaloa Drosophila busckii Lucilia cuprina Musca domestica Rhagoletis zephyria Stomoxys calcitrans Zeugodacus cucurbitae |
| Ecdysozoa-Arthropoda-Hexapoda-Insecta-Hemiptera | ||
Acyrthosiphon pisum Cimex lectularius Diuraphis noxia |
Bemisia tabaci Halyomorpha halys Diaphorina citri |
Rhodnius prolixus Nilaparvata lugens Myzus persicae |
| Ecdysozoa-Arthropoda-exapoda-Insecta-Coleoptera | ||
|
Aethina tumida Dendroctonus ponderosae Aquatica lateralis Oryctes borbonicus |
Luciola cruciate Luciola lateralis Agrilus planipennis Anoplophora glabripennis |
Nicrophorus vespilloides Tribolium castaneum Lucidina biplagiata |
| Ecdysozoa-Arthropoda-Hexapoda-Insecta-Lepidopterea | ||
|
Amyelois transitella Danaus plexippus Mythimna separate Plutella xylostella |
Manduca sexta Bombyx mori Helicoverpa armigera Operophtera brumata Pieris rapae |
Papilio Xuthus Papilio polytes Papilio machaon Papilio polytes Spodoptera exigua |
| Ecdysozoa-Arthropoda-Hexapoda-Insecta-Hymenoptera | ||
|
Acromyrmex echinatior Apis cerana (cerana) Apis mellifera Apis florea Apis dorsata Athalia rosae Atta colombica Atta cephalotes Bombus terrestris Bombus impatiens Camponotus floridanus Cephus cinctus Cerapachys biroi Ceratina calcarata Ceratosolen solmsi marchali |
Copidosoma floridanum Cyphomyrmex costatus Eufriesea Mexicana Fopius arisanus Diachasma alloeum Dinoponera quadriceps Dufourea novaeangliae Harpegnathos saltator Habropoda laboriosa Lasius niger Linepithema humile Megachile rotundata Melipona quadrifasciata Microplitis demolitor Monomorium pharaonis |
Nasonia vitripennis Neodiprion lecontei Orussus abietinus Polistes dominula Polistes canadensis Philanthus triangulum Solenopsis invicta Pogonomyrmex barbatus Pseudomyrmex gracilis Trachymyrmex cornetzi Trachymyrmex septentrionalis Trichogramma pretiosum Vollenhovia emeryi Wasmannia auropunctata |
| Ecdysozoa-Arthropoda-Hexapoda-Insecta-Orthoptera | ||
| Gryllus bimaculatus | Acheta domesticus | |
| Ecdysozoa-Arthropoda-Hexapoda-Insecta-Phthiraptera | ||
| Pediculus humanus corporis | ||
| Ecdysozoa-Arthropoda- Hexapoda-Insecta-Blattodea | ||
| Zootermopsis nevadensis | ||
| Echinodermata | ||
| Acanthaster planci Strongylocentrotus purpuratus Apostichopus japonicus | ||
| Cephalochordata | ||
| Branchiostoma floridae | Branchiostoma belcheri | |
| Tunicata | ||
| Ciona intestinalis | ||
| Craniata-Actinopterygii-Chondrichthyes | ||
| Callorhinchus milii | Scyliorhinus canicula | Rhincodon typus |
| Craniata-Actinopterygii-Clupeocephala | ||
Astyanax mexicanus Acanthochromis polyacanthus Austrofundulus limnaeus Boleophthalmus pectinirostris Clarias sp. SM-2014 Clarias batrachus Carassius auratus Carassius carassius Clupea harengus Ctenopharyngodon idella Cyprinodon variegatus Cyprinus carpio Cynoglossus semilaevis Danio rerio Esox lucius Fundulus heteroclitus Hippocampus kuda Hippocampus comes Haplochromis burtoni |
Ictalurus punctatus Kryptolebias marmoratus Labrus bergylta Larimichthys crocea Lates calcarifer Megalobrama amblycephala Maylandia zebra Micropogonias undulatus Monopterus albus Neolamprologus brichardi Nothobranchius furzeri Notothenia coriiceps Oreochromis niloticus Oncorhynchus mykiss Oncorhynchus kisutch Oryzias latipes Paralichthys olivaceus Platichthys flesus |
Pundamilia nyererei Poecilia formosa Poecilia latipinna Poecilia mexicana Poecilia reticulata Pygocentrus nattereri Salmo salar Sciaenops ocellatus Seriola dumerili Scophthalmus maximus Stegastes partitus Sinocyclocheilus rhinocerous Sinocyclocheilus grahami Sinocyclocheilus anshuiensis Tetraodon nigroviridis Takifugu poecilonotus Takifugu rubripes Xiphophorus maculatus |
| Craniata-Actinopterygii-Semionotiformes | ||
Lepisosteus oculatus | ||
| Craniata-Actinopterygii-Osteoglossiformes | ||
Scleropages formosus Latimeria chalumnae | ||
| Craniata-Sarcopterygii-Tetrapoda-Sauria-Aves | ||
Acanthisitta chloris Amazona aestiva] Anas platyrhynchos Anser sp. Anser anser Anser cygnoides domesticus Antrostomus carolinensis Apaloderma vittatum Aptenodytes forsteri Apteryx australis mantelli Aquila chrysaetos canadensis Balearica regulorum gibbericeps Buceros rhinoceros silvestris Calidris pugnax Callipepla squamata Calypte anna Caprimulgus carolinensis Cariama cristata Cathartes aura Chaetura pelagica Charadrius vociferus Chlamydotis macqueenii Colinus virginianus Colius striatus |
Columba livia Corvus brachyrhynchos Corvus cornix cornix Colius striatus Coturnix japonica Cuculus canorus Egretta garzetta Eurypyga helias, Falco peregrinus Falco cherrug Ficedula albicollis Fulmarus glacialis Gallus gallus Gavia stellata Geospiza fortis Haliaeetus leucocephalus [Haliaeetus albicilla Leptosomus discolor Lepidothrix coronate Lonchura striata domestica Manacus vitellinus Meleagris gallopavo Melopsittacus undulatus Merops nubicus |
Mesitornis unicolor Nestor notabilis Nipponia nippon Numida meleagris Opisthocomus hoazin Parus major Patagioenas fasciata monilis Pelecanus crispus Phaethon lepturus Phalacrocorax carbo Phoenicopterus ruber ruber Picoides pubescens Podiceps cristatus Pseudopodoces humilis Pterocles gutturalis Pygoscelis adeliae Serinus canaria Sturnus vulgaris Struthio camelus australis Taeniopygia guttata Tauraco erythrolophus Tinamus guttatus Tyto alba Zonotrichia albicollis |
| Craniata-Sarcopterygii-Tetrapoda-Sauria-Crocodylia | ||
Alligator mississippiensis Alligator sinensis |
Crocodylus porosus Gavialis gangeticus | |
| Craniata-Sarcopterygii-Tetrapoda-Sauria-Lepidosaure | ||
Anolis carolinensis Aspidoscelis uniparens Python bivittatus Phrynocephalus erythrurus |
Ophiophagus hannah Protobothrops mucrosquamatus Gekko japonicus |
Pogona vitticeps Phrynocephalus przewalskii Phrynocephalus erythrurus Thamnophis sirtalis |
| Craniata-Sarcopterygii-Tetrapoda-Sauria-Testudines | ||
| Chrysemys picta bellii | Pelodiscus sinensis | Chelonia mydas |
| Craniata-Sarcopterygii-Tetrapoda-Amphibia | ||
Xenopus (Silurana) tropicalis Xenopus laevis |
Rhinella marina Nanorana parkeri | |
| Craniata-Sarcopterygii-Tetrapoda-Metatheria | ||
| Monodelphis domestica | Phascolarctos cinereus | Sarcophilus harrisii |
| Craniata-Sarcopterygii-Tetrapoda-Prototheria | ||
| Ornithorhynchus anatinus | ||
| Craniata-Sarcopterygii-Tetrapoda-Eutheria-Xenarthra | ||
| Dasypus novemcinctus | ||
| Craniata-Sarcopterygii-Tetrapoda-Eutheria-Glires-Rodentia | ||
Castor canadensis Cavia porcellus Chinchilla lanigera Cricetulus griseus Dipodomys ordii Eospalax fontanierii baileyi Fukomys damarensis Heterocephalus glaber |
Ictidomys tridecemlineatus Jaculus jaculus Marmota marmota marmota Mesocricetus auratus Meriones unguiculatus Microtus ochrogaster Mus musculus Mus pahari |
Mus caroli Nannospalax galili Octodon degus Neotoma lepida Peromyscus maniculatus bairdii Rattus rattus Rattus norvegicus |
| Craniata-Sarcopterygii-Tetrapoda-Eutheria-Glires-Lagomorphes | ||
Ochotona princeps Oryctolagus cuniculus |
Ochotona curzoniae Ochotona collaris | |
| Craniata-Sarcopterygii-Tetrapoda-Eutheria-Ruminants | ||
Bison bison bison Bos taurus Bos bovis Bos grunniens mutus Bos indicus |
Bos mutus Bubalus bubalis Cervus elaphus hippelaphus Ophiophagus hannah] Capra hircus |
Odocoileus virginianus texanus Ovis aries Ovis aries musimon Pantholops hodgsonii |
| Craniata-Sarcopterygii-Tetrapoda-Eutheria-Insectivores | ||
| Condylura cristata | Sorex araneus | Erinaceus europaeus |
| Craniata-Sarcopterygii-Tetrapoda-Eutheria-Carnivores | ||
Acinonyx jubatus Ailuropoda melanoleuca Canis lupus familiaris Panthera pardus Neomonachus schauinslandi |
Felis catus Mustela putorius furo Panthera tigris altaica Odobenus rosmarus divergens |
Ursus maritimus Acinonyx jubatus Leptonychotes weddellii Enhydra lutris kenyoni |
| Craniata-Sarcopterygii-Tetrapoda-Eutheria-Cetartiodactyla | ||
Physeter catodon Delphinapterus leucas Camelus dromedaries Vicugna pacos |
Balaenoptera acutorostrata scammoni Camelus ferus Camelus bactrianus |
Sus scrofa Lipotes vexillifer Orcinus orca Tursiops truncatus |
Craniata-Sarcopterygii-Tetrapoda-Eutheria-Perissodactyles | ||
Equus caballus Equus przewalskii Cheval |
Ceratotherium simum simum Equus asinus | |
| Craniata-Sarcopterygii-Tetrapoda-Eutheria-Chiropteres | ||
Pteropus alecto Hipposideros armiger Miniopterus natalensis |
Rousettus aegyptiacus Pteropus vampyrus Myotis davidii |
Eptesicus fuscus Myotis lucifugus Rhinolophus sinicus |
| Craniata-Sarcopterygii-Tetrapoda-Eutheria-Dermopteres | ||
| Galeopterus variegatus | Chrysochloris asiatica | |
| Craniata-Sarcopterygii-Tetrapoda-Eutheria-Pholidota | ||
Loxodonta Africana Echinops telfairi |
Elephantulus edwardii Orycteropus afer afer |
Tupaia chinensis Manis javanica |
| Craniata-Sarcopterygii-Tetrapoda-Eutheria-Primates | ||
Aotus nancymaae Callithrix jacchus Carlito syrichta Cebus capucinus imitator Cercocebus atys Chlorocebus sabaeus Colobus angolensis palliatus Gorilla gorilla gorilla |
Homo sapiens Macaca mulatta Macaca fascicularis Macaca nemestrina] Mandrillus leucophaeus Microcebus murinus Nomascus leucogenys Otolemur garnettii |
Pan troglodytes Pongo abelii Pan paniscus Papio Anubis Propithecus coquereli Rhinopithecus roxellana Rhinopithecus bieti Saimiri boliviensis |
| Same protocol as in Table 1. Species might include various isoforms. | ||
In Prokaryotes. In the Eubacteria Domain, NOSs were found in many phyla (Table 1): mostly Firmicutes (>580 sequences), Actinobacteria (>320) but also in Cyanobacteria (36), Deinococcus (20), Proteobacteria (18), Bacteroidetes (11), Verrumicrobia (10), Chloroflexi (2). Due to a bias in the choice of the organisms to be sequenced, this list cannot reflect a true distribution of NOSs in Eubacteria. However, it shows how NOSs are spread throughout the bacterial kingdom. NOSs are mostly found in the Terrabacteria group. They are rarely found in Proteobacteria and are absent from the genomes of other bacteria (Acidobacteria, Aquificae, Chlorobi, Chlamydiae, Fusobacteria, Spirochaetes, Thermotogae….). Though, in the phyla where NOSs are highly present, such as in Gram+ bacteria, NOSs are only present in certain classes and absent from some major ones. This is the case for Firmicutes: NOSs are present in Bacilli but not in Clostridia. In the Bacilli class, NOSs are found in all species of the Bacillales order but are completely absent from the Lactobacillales order (except for the unique Streptococcus pneumoniae). This patchiness is well illustrated for Cyanobacteria: around 36 NOS sequences (in species and subspecies) were found (over at least 300 sequenced genomes) but the rationale of their distribution does seem obvious. NOSs are found throughout the cyanobacterial phyla (in Nostocales, Oscillatoriales or Chroococales) in species that correspond to different physiologies (subsections 1-5) or ecologies (fresh-water, marine, terrestrial…). Which is highlighted in Firmicutes and Cyanobacteria holds for the whole Eubacteria kingdom. The presence of NOSs in only 13 distinct (alpha-, beta-, gamma- or delta-) proteobacteria requires a combination of various distinct explanations: loss from a common eubacterial ancestor in this phyla and discrete horizontal genome transfer (HGT) in very specific niches. As Hydrobacteria and Terrabacteria might have diverged around three billion years ago, implying a common ancestor long before the Great Oxidation Event (GOE), a plural, ramified and complex evolution scenario needs to be found. The same heterogeneity is observed in the Archae Domain. Only 10 sequences of NOS-related proteins (over several hundreds of available genomes) were found in species corresponding to only two orders (Halobacteriales and Natrialbales) from the unique Halobacteria class. Though, only a few archae species from this very class exhibit a NOS-like sequence.
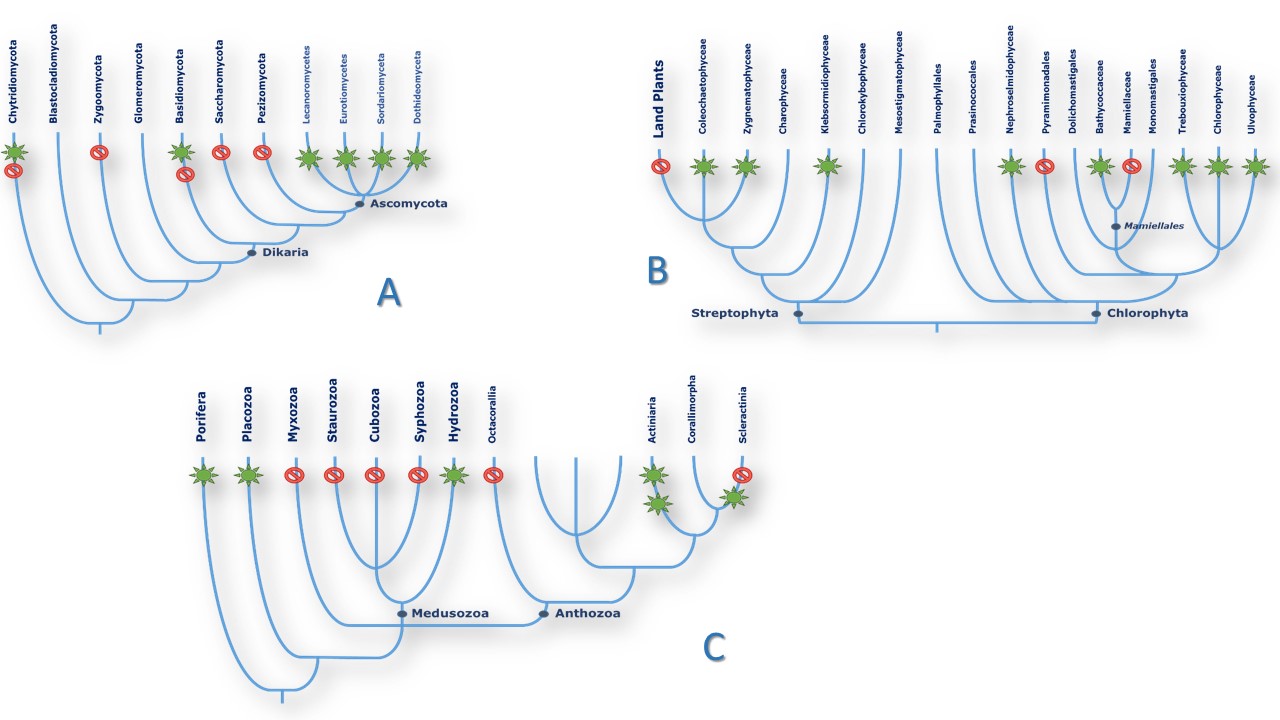
In Eukaryotes: Fungi and Plants. The picture remains much contrasted in the Eukaryote domain (Table 2). NOSs seem to be absent from the Bikontes group, (Excavata, Rhizaria, Alveolata) although it can be found in two Stramenopiles species and in at least two dozen of algae (94, 95). On the other hand, NOSs seem more present in Unikontes, although only one NOS-related sequence has been found in Amoebae and none in Choenaflagellata. Fungal NOSs were only found in the Ascomycota phylum (38 species) with the intriguing presence of NOS sequence in one Basidiomycota and one Chytridiomycota fungi, and no related sequences in Glomeromycota nor in Zygomycota. NOS sequences were clustered in only 4 classes and 13 different orders of fungi (among dozens) belonging to the same Pezizomycotina subphylum. However, the absence of NOSs in Saccharomycotina yeasts and in most of the sequenced species of these classes (Penicillium, Microsporum, Blastomyces and etc.) highlight the same patchy distribution of NOS in the fungal tree of life (Figure 4A). The pattern in the Green lineage is more puzzling: whereas land plants ubiquitously uses NO° for a wide range of purposes as different as immunity, stress response, growth or mycorrhizal symbiosis regulation ((96-100), no NOS sequence is to be found in land plants, which suggest that they might have lost their NO-Synthase in the course of evolution. NOS-related proteins today can only be found in a discrete number of green algae from both Chlorophytes and Streptophytes phyla (94, 95). Though, NOSs are not ubiquitous in green algae since only 23 NOSs were identified among the several dozens of algal genomes that have been sequenced so far. For example, NOS seems omnipresent in the Bathycoccaceae family but is absent from the Mamiellaceae family that yet belongs to the same Mamiellales order. Likewise, many NOSs sequences have been found in the Chlamydomonadales order although NOS is clearly absent from the model alga Chlamydomonas reinhardtii (Figure 4B). Once again, the distribution of NOSs found in algae and fungi remains apparently inexplicable.
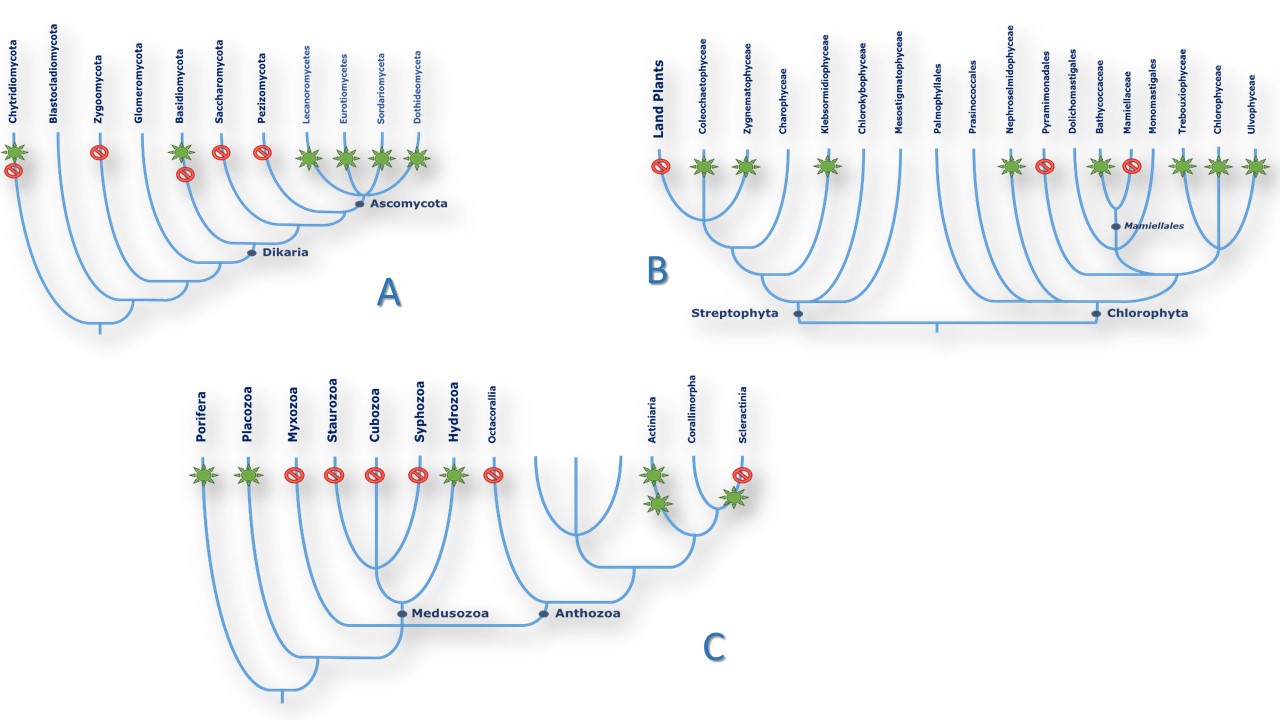 Figure4
Figure4Schematic patterns of NOS distribution in three representative clades. Species phylogeny was drawn from NCBI and (165). Green stars that NOS-like proteins have been found in (almost) all species of the branch. Red sign means that no homologous sequences were found in the branch. A. Fungi. B. Algae. C. Basal metazoans.
A few articles have reviewed in the past years the presence of NOSs in metazoan and in particular in invertebrate and marine organisms (101-103). However, the number of sequenced genomes has exponentially increased since then, giving rise to a wealth of new data that we will summarize and analyze here (Table 3). NOSs are found in Porifera and in all radiates phyla such as Cnidaria and Placozoa. In Cnidaria, NOSs are mostly found in Anthozoan. NOSs are absent from the sequenced genomes of Myxozoan. NOSs have not yet been identified in jellyfishes or seawasps, but they are not completely absent from the Medusozoa family as NOSs have been found in Hydra vulgaris (104). Besides, NOSs are not ubiquitous in Anthozoan and seem absent from Octocorallia species. Once again, the distribution of NOS in the Cnidaria Phylum does not seem homogeneous (Figure 4C). This patchy distribution is still observed in Bilateria and in particular in Protostomes. NOSs are absent from major worm’s phyla (Platyhelminthes, Acanthocephala, Rotifera, Nemertea, Nematodes, Gastrotriches…), with only one NOS in the genome of one annelid worm (Capitella teleta). In Ecdysozoan, NOSs are found in arthropods and tardigrades. In Lophotrochozoan, NOSs are primarily found in mollusks in, with singlet presence in Bryozoa and Brachyopods (Figure 4B). NOS presence becomes ubiquitous only at the level of Chordata: the genome of all Chordata species seems to harbor a NOS-related sequence so far.
This long “topological” list of NOSs (that remains to be frequently updated) conveys three major conclusions: i) this new, large and diverse family of proteins calls for a thorough characterization; ii) the considerable number of NOS proteins throughout the tree of life makes impossible the systematic structural and functional characterization of each of these NOSs; iii) mammalian NOSs do not represent the most important class of species that harbor a NOS in their genome and as such mNOS can no longer be considered as the archetypal NOS. It is tempting to achieve a phylogenetic analysis of this large population of proteins in order to see if some rational evolutive story can emerge from their patchy distribution, as it has been done many times in the past (102). We achieved such an analysis by using a careful sampling of NOSs that would be representative of the heterogeneity of NOS distribution: we selected the sequence of 93 different NOSs from various prokaryotes and eukaryotes. The multiple-alignment of these sequences was used to generate a phylogenetic tree (Figure 5, see legend). The major analyses of this tree are reported in Figure 6. It seems that this tree can be decomposed in distinct parts that each display different patterns. In vertebrates, NOSs phylogeny seems to unfold along the type of the isoforms (eNOS, nNOS, iNOS). This is not the case for the other metazoans and in particular for invertebrate’s NOSs that seem to follow a species-based phylogeny. In Eukaryotes (Plants, Fungi, Stramenopiles…), the phylogeny seems more related to ecological/physiological factors: the NOS from the photosynthetic Stramenopiles is on the same branch as algae’s NOSs, whereas the NOS from the oomycetes Thraustotheca clavata (a Stramenopiles species too) is located within the branch of Fungi NOSs. The same multiple rationales account for NOS phylogeny in prokaryotes that balances between a structure-based phylogeny (Glob-NOSs branch includes NOSs from Cyanobacteria and Bacteroidetes, whereas cyanobacterial EF-NOSs cluster with Fe/S-NOSs from Proteobacteria; see below for structural explanations), and species-based phylogeny (in the case of Archae or Actinobacteria for instance). Besides, the distribution of NOS is not homogeneous within most of these clades, with a discrete presence in some phyla, and major absences in other ones (see above). A rationale that would try to address the evolution of the NOS family as a whole would have to imply many additional scenarii and many singular events (HGT, loss, gain…) to account for such a heterogeneous distribution and complex phylogeny.
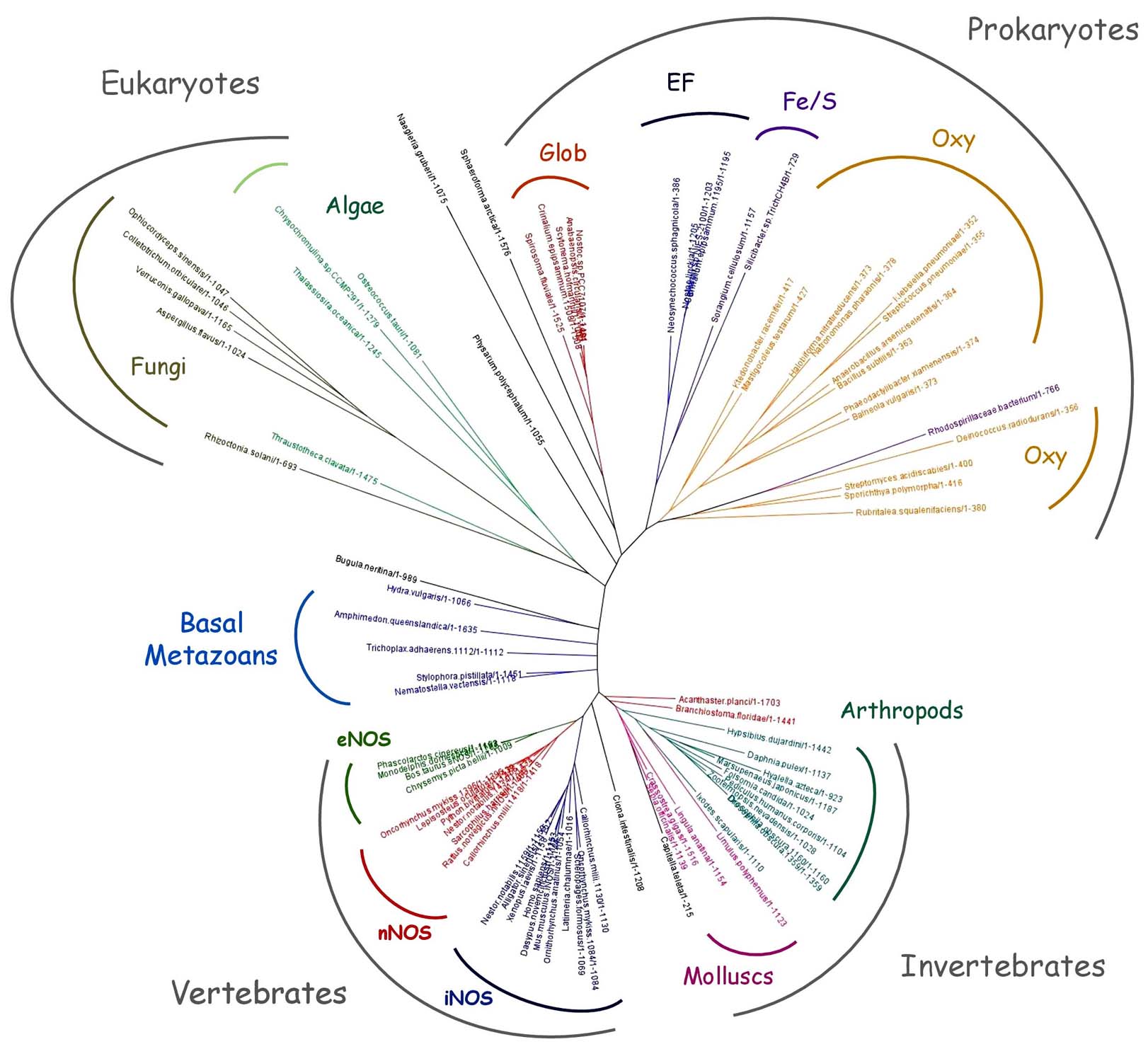 Figure 5
Figure 5Tentative phylogenetic tree of NOS protein family. Phylogenetic tree of a selection of 93 NOSs representative of NOS phyletic diversity, including NOSs from Cyanobacteria (Scytonema hofmanni, Crinalium epipsammum, Anabaenopsis circularis, Neosynechococcus sphagnicola, Nostoc linckia, Nostoc sp. PCC 7107, Mastigocoleus testarum, Calothrix sp. NIES-2100), Firmicutes (Anaerobacillus arseniciselenatis, Bacillus subtilis, streptococcus pneumoniae), Actinobacteria (Streptomyces acidiscabies, Sporichthya polymorpha), Proteobacteria (Silicibacter sp. TrichCH4B, Sorangium cellulosum, Rhodospirillaceae bacterium, Klebsiella pneumoniae), Bacteroidetes (Spirosoma fluviale, Phaeodactylibacter xiamenensi), Archae (Halobiforma nitratireducens, Natronomonas pharaonis DSM 2160) and other Prokaryotes (Deinococcus radiodurans, Ktedonobacter racemifer, Balneola vulgaris, Rubritalea squalenifaciens). Sequences include NOSs from various Eukaryotes such as Chlorophytes (Ostreococcus tauri), Haptophytes (Chrysochromulina sp.), Stramenopiles (Thalassiosira oceanica, Thraustotheca clavata), Amoeba (Physarum polycephalum), Heterobolosea (Naegleria gruberi), Fungi (Ophiocordyceps sinensis CO18, Rhizoctonia solani AG-1 IB, Verruconis gallopava, Colletotrichum orbiculare MAFF 240422, Aspergillus flavus NRRL3357), Ichtyosporea (Sphaeroforma arctica). We used also NOSs from various animals such as Porifera (Amphimedon queenslandica), Placozoa (Trichoplax B), Cnidaria (Stylophora pistillata, Nematostella vectensis, Hydra vulgaris), Bryozoa (Bugula neritina), Brachyopoda (Lingula anatina), Annelid (Capitella teleta), Mollusc (Limulus Polyphemus, Sepia officinalis, Crassostrea gigas), Panarthropod (Hypsibius dujardini), Arachnid (Ixodes scapularis), Collembolla (Folsomia candida), Crustacea (Marsupenaeus japonicas, Hyalella Azteca, Daphnia pulex), Insect (Drosophila obscura, Pediculus humanus corporis, Zootermopsis nevadensis), Echinoderm (Acanthaster planci), Tunicate (Ciona intestinalis), Cephalochordate (Branchiostoma floridae) and from various vertebrates such as Fish (Oncorhynchus mykiss, Callorhinchus milii, Lepisosteus oculatus, Scleropages formosus, Latimeria chalumnae), Bird (Nestor notabilis), Crocodile (Alligator sinensis), Turtle (Chrysemys picta bellii), Reptile (Python bivittatus), Amphibian (Xenopus laevis) and various Mammals (Dasypus novemcinctus, Monodelphis domestica, Ornithorhynchus anatinus, Phascolarctos cinereus, Sarcophilus harrisii) including the three canonical NOSs from Mus musculus (iNOS), Bos taurus (eNOS) and Rattus norvegicus (nNOS). The types of chosen isoforms (when several did exist) was made randomly to increase the heterogeneity of the sampling. In several cases, multiple isoforms were used for a single species. Labels include the species names and the sequence length in order to ease the identification of the NOS type. Phylogenetic branches were colored based on the type of NOSs (eNOS, nNOS and iNOS, but also Oxy-NOS, Glob-NOS, EF-NOS and FeS-NOS) or on the nature of the clade (arthropods, fungi…). Sequence alignment of the full-length proteins has been achieved using Jalview 2.7. © as multiple alignment editor (166) and PROBCONS © with two rounds of pre-training, 300 passes of iterative refinement and 3 passes of consistency transformation (see supplementary file. Phylogenetic tree was generated using Seaview 4.5. © graphical interface using PhyML algorithm (with Blosum62 model and 100 replicates bootstrapping).
We believe that NOS phylogeny and distribution cannot be explained by considering all these NOSs as the same protein, and that this phylogeny is unable reflect the evolutive history of this family. We think that the only way to draw some sense out of this picture is to consider several different groups of NOSs, corresponding to strictly different (structural and functional) types of proteins, and following distinct evolutive tracks.
The main difficulty when addressing NOS evolution is linked to the “mammal bias” that prevails in the NO° field and that imposes a unique mammalian-centred vision of NOSs. Grossly, all new NOSs are believed to behave like one of the three mammalian isoforms (see Figure 3). As seen through this phylogenetic tree, this vision only holds for vertebrates NOSs and has no heuristic value outside this phylum. In fact, because of its very nature, an oxygenase that uses a very sophisticated and sensitive redox mechanism to produce a radical, gaseous, and thus extremely reactive molecule (NO°) with a large array of biological reactivity, NOS appears as a versatile enzyme: no fixed biochemical activity can be assigned to it, NOS biochemical activity can give rise to various biological effects, depending on the cellular or biological environment, these effects can lead to distinct and often opposed biological outcomes. This versatility has major implication on the way we should investigate NOS structure, function and evolution.
There is some confusion in the way we address NOS function. It often encompasses three distinct phenomena: NOS chemical activity, the biochemical effects of its catalytic production and their ensuing biological function. NOS function has mostly been understood as “NO°” function, whereas there is no univocal relationship between NOS and NO°. Indeed, NOSs have the ability to achieve different chemistries, to produce various reactive species that in turn exert distinct biological effects, which may be employed for various purposes. The “problem” of plant NO-Synthases illustrates this confusion very well (105, 106). As NO° is a ubiquitous and important physiological mediator in plants, the presence of a plant NOS was beyond any doubt. This led to the publication of two articles in Cell and Science, confirming the common idea that plants were meant to have a NOSs. These articles were soon retracted, and the sequencing of numerous land plants genomes confirmed the absence of a “genuine” mammalian-like NO-synthase (94). On the opposite many algae and photosynthetic organisms (stramenopiles, haptophytes, bacteria) harbor a NOS. So why land plants would have lost NOS as they still need and use NO°, what could be the function of plant NOSs if not related to land plant NO° physiology? This question seems paradoxical if one considers a univocal relationship between NOS and NO°. But there is no paradox if one considers that NO° can be produced by many alternative sources and that NOS might have other activities than producing NO°. One must therefore clearly distinguish between NOS and NO°; understanding NOS function requires to analyze the nature of NOS catalytic production, the biochemical effects of this RNOS, and their biological impact.
This phylogenetic tree is supposed to account for the evolution of the structure of a designated entity (NOS) in relation with its function in a particular environment. It relies on a certain stability of the “activity” that enacts the selection and evolution process. However, NOS enzymatic activity is not stable in time: NOS today, i.e. mammalian NOSs as we know them, are oxydo-reductase that uses a gas (oxygen) to produce another gas (nitric oxide), both major redox reagents. As NOSs probably emerge long before the Great Oxidation Event (GOE), in an anoxic and highly reductive environment, it is natural to think that its initial biochemistry was unrelated to the oxidation of Arginine and that its activity was not NO° production (107). This is reminiscent of the “exaptation” concept proposed almost forty years ago by S.J. Gould and R. Lewontin (108) and defined by Gould and Vrba as “ such characters evolved for other usages – or for no function at all – later coopted for their current role” (109). Thus, the original NOS structure might have served another (or no) purpose than the current ones described in mammals. As a consequence, as NOS environment has deeply evolved in the last three billion years, NOS chemistry has probably changed several times. Likewise, as NOS chemistry is strongly related to its physico-chemical environment (O2 concentration, redox status, Nitrogen cycle…), NOS activity is also a function of its biological milieu. As NOS physico-chemical environment strongly varies between, for instance, a halophilic Archae (such as Halobiforma nitratireducens), an anaerobe Bacilli (such as Anoxybacillus pushchinensis), an insect NOS or a macrophage NOS, it is likely that their catalytic activity will vary likewise (58, 61). In this regard, one could wonder what could be the function of NOS in anoxic organisms/tissues, or even if some of its biological activity could still be related to specific anoxic conditions. NOS catalytic activity is not fixed as it varies with the physico-chemical conditions of its milieu, that itself varies with the geochemical history and with the ecological niche of the organism.
As NOS environment has experienced many physico-chemical changes in the last three billions of year, many different chemistries, effects and functions have probably emerged in the course evolution. This is illustrated by the various catalytic activities of mammalian and bacterial NOSs: Arg oxidation (signaling), NO° dioxygenase (110), RN0S isomerisation (detoxification (66)), heavy production of RNOS ((67, 111-113)), nitrite reduction (hypoxic signaling, (64, 65, 114)) and etc. The balance is determined both by the milieu and many sophisticated molecular regulations and each NOS might be apt to achieve different activities simultaneously (Figure 7). It is therefore extremely difficult to determine “ex nihilo” which function each of these NOSs is actually exerting. Besides, the evolution of conditions with time also holds at the organismal level and the same “NOS” (even as an individual protein) might experience different catalytic activity. As a consequence of its long history, each NOS is able to exert distinct biochemical activities that could superimpose and lead to distinct biological effects.
 Figure 7
Figure 7Illustration of the variability of NOS catalytic activity as a function of its environment and of the nature of its biochemistry.
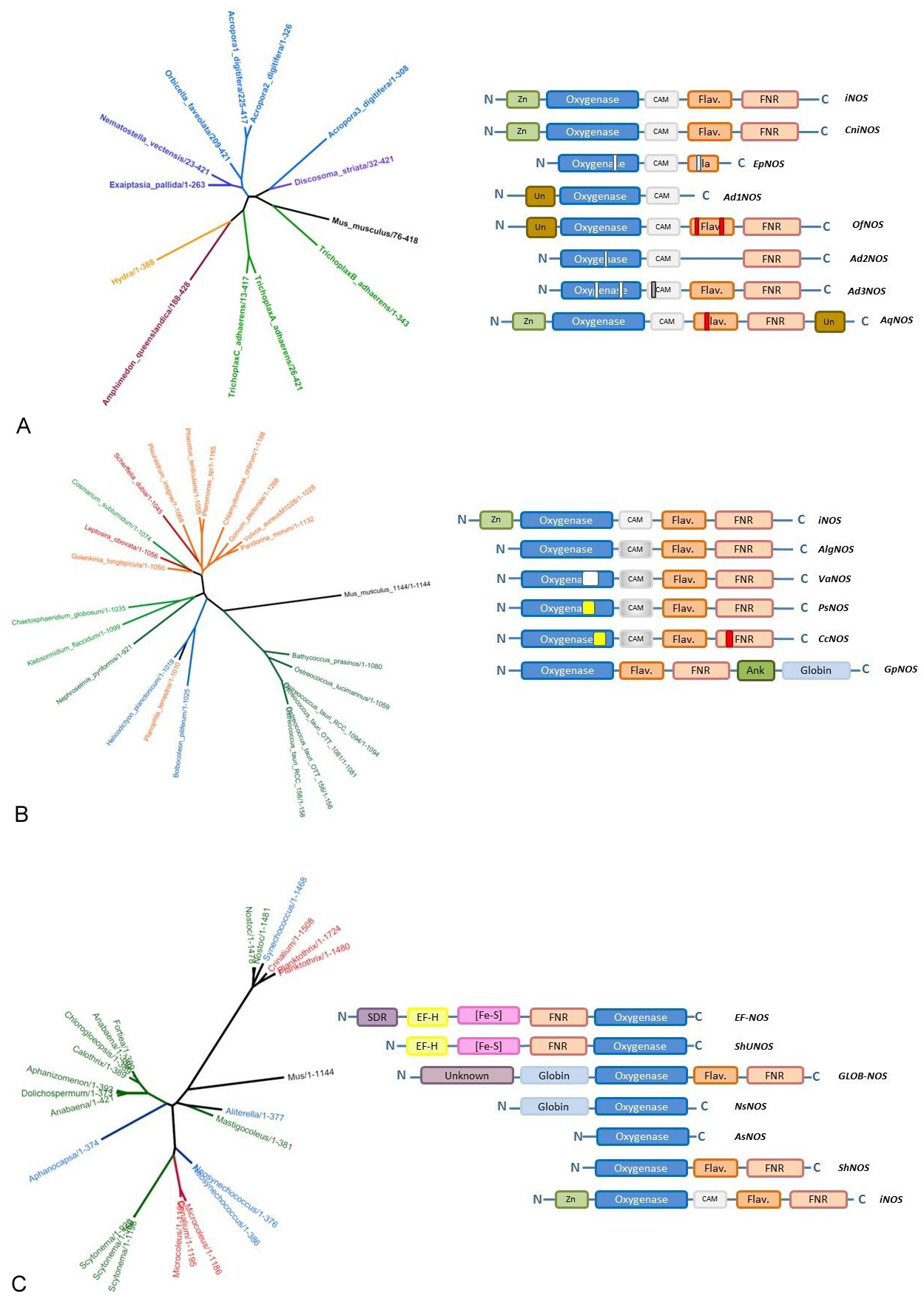
This analysis becomes even more complex when this evolution concerns not only one single NOSs but several different types of NOS per organism. Indeed, in mammals, three different NOSs have been found, with different structures and various – if not opposite – functions (115, 116). This standard picture is found throughout the vertebrate clade but does not hold beyond it. As we highlighted it, only one NOS-related sequence is to be found in the genome of most organisms such as insects, bacteria, fungi, plants… However, many different patterns (of NOS distribution) are found throughout the tree of life. For example, mollusks harbor at least two types of NOSs with different structures (NOSX1 and NOSX2 for example with 1387 and 1163 residues for Aplysia californica). Whereas cnidaria species seem to host only one NOS, Acropora digitifera harbor three highly different NOSs (Table 3, Figure 8A). This is the same for the placozoan Trichoplax adherens that displays three similar NOSs. This heterogeneous picture is not limited to metazoans but extends to plants and bacteria. For example, two different NOSs (one full-length and one truncated form) are found in the genome of Ostreococcus tauri (Figure 8B). Two extremely different NOSs are also found in the cyanobacterial Crinalium epipsamum (Figure 8C), whereas all the other cyanobacterial genomes harbor only one NOS-related sequence. This co-existence of different number and types of NOSs in many different species suggests the co-evolution of parallel regimes of activity and functions.
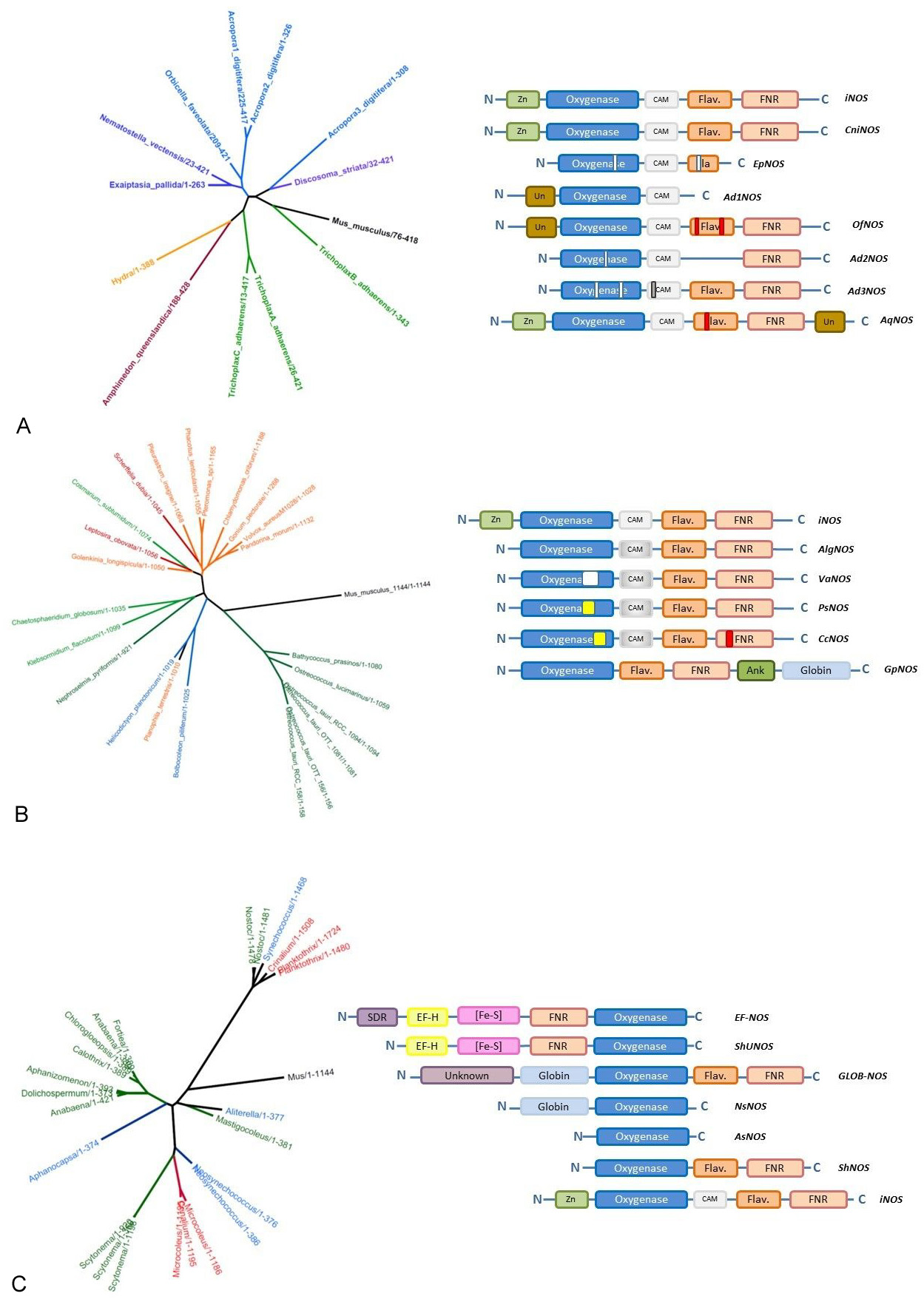 Figure 8
Figure 8Examples of the evolutionary and structural diversity of NOS family for three phylogenetic groups. Phylogenetic trees have been generated using the same procedure as in Figure 5; modules used to depict the structures of various NOSs are the same as in Figure 9. White rectangles represent additional gaps observed in the protein sequence whereas colored rectangles correspond to large inserts. Panel A: Basal metazoan NOSs. Panel B. Plant NOSs. Panel C. Cyanobacterial NOSs. Schemes and Figures
As we try to describe the function and evolution of this family of proteins, we should wonder which protein we are actually dealing with and address the heterogeneity of this family that goes beyond the mammalian NOSs. Indeed, NOS family consists of many different proteins with sequence lengths between 230 and 1950 residues that are composed of various modules and share only one conserved domain, the oxygenase domain (see below for structural details). As this domain only represents 15 to 20 % of the functional holoenzyme, it might not be sufficient to delineate a standard “NO-Synthase” protein. Besides, the strong homology of these catalytic domain does not imply that the chemistry is similar, and that the activity remains identical. In any case, the NOS family is not a structurally homogenous family. The disparities in NOSs structure indicate likely variations in their activity and function.
In this context, it seems difficult to analyse the evolution of one “standard” NOS, when its number and structure vary unpredictably between phyla and within phyla, when its structure is not linked to a common and single activity, producing NO°, and when this activity remains variable in time and space. The numerous and different molecular structures, the variety of their chemistry (due to structural but also environmental changes), the multiplicity of RNOS effects and thus of NOS potential function impedes any straightforward phylogenetic analysis.
The complex relationship between the structure, the activity and the function of proteins within the NOS family calls for an adapted phylogenetic approach. As the genomic sequence of any NOS does not correspond to a standard function, one must classify and analyze more precisely this family of proteins. This should be based on a better knowledge of the structure that could help characterizing their probable biochemical activity and might provide a more suitable vision of their biological function. For that matter one should take into account the great diversity of NOSs structure and try to relate it to specific patterns of activity. We’d like to present how this approach could be achieved on three representative phyla: Cyanobacteria, Algae and basal Metazoans.
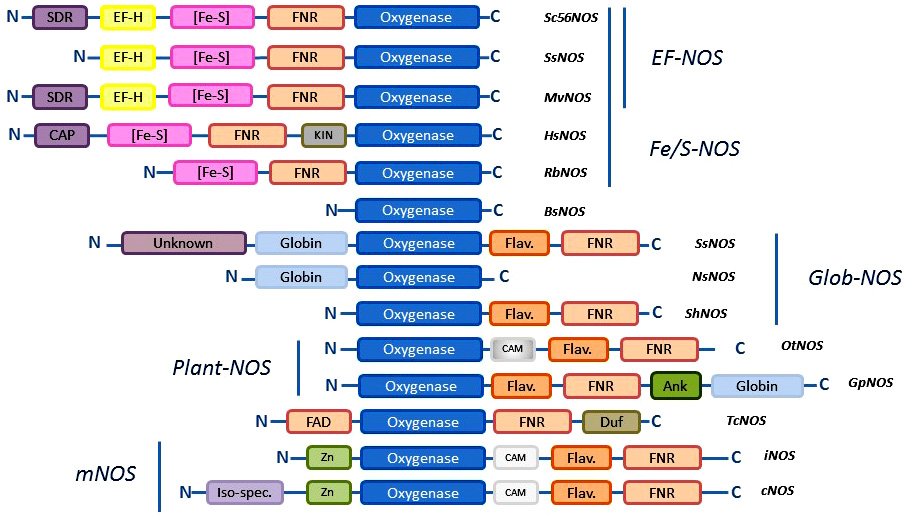
The diversity of NOS proteins resides not only in the number of NOSs that are present in any organisms but also in the types of NOSs that are identified. Until now, a short number of structural motifs involved in the control of the structure and mechanism of NOSs have been used to characterize the sequences of new NOS-like proteins: the catalytic site that lies within the oxygenase domain; the Calmodulin-binding domain; the auto-inhibitory elements (AIEs) that regulate electron transfer and control NO° production; the N-terminal extension that determine subcellular localization (24). Based on these patterns, four types of NOSs have been used as “canonical” NOSs: i) neuronal NOS that is a full-length NOS with an N-terminal PDZ domain that allows nNOS to anchor partner proteins; ii) other constitutive NOSs (such as eNOS) that mostly differ from nNOS by their N-terminal extension (palmytoylation/myrystoilation sites for eNOS); iii) inducible NOSs that are deprived of N-terminal extension and lacks most of the auto-inhibitory elements suggesting an unleashed NO° production; iv) bacterial NOSs that consists only of the truncated oxygenase domain (Figure 9). These categories have been commonly used to achieve a first and gross classification of NOS-like proteins. Based on the first sequence analyses made on mollusks, insects and other animals, many articles have considered that metazoan NOSs would globally correspond to mammalian NOSs and could be classified along these categories (102) (101, 103). As a consequence, it was proposed that metazoan NOS might have a common ancestor, presumably a neuronal NOS (101). Though, as noted by Andreakis and colleagues, “the presence of domains de?ning the three isoforms—PDZ domain, inhibitory loop, myristoylation, and palmitoylation motifs — was differently observed though not always ascertainable” (101). It should also be noted that the classification based on genomic data is instable as some of the available genomes remain partial and the provided sequences are often truncated. Besides, the potential existence of multiple transcription initiation sites and of alternative splicing variants, as suggested for Limax, Physarum, and Drosophila for example (85, 86, 117, 118), calls for cautiousness when analyzing the genomic data of these NOS-like proteins. Indeed, although our census of NOS seems to confirm the predominance of standard “full-length” NOSs in metazoans (in tetrapods the three mammalian isoforms are found ubiquitously, nNOS and iNOS are found in fishes, and the cephalochordate present several neuronal NOS-like proteins), a simple classification based on mammalian NOSs categories, does not provide a pertinent framework to apprehend NOS family.
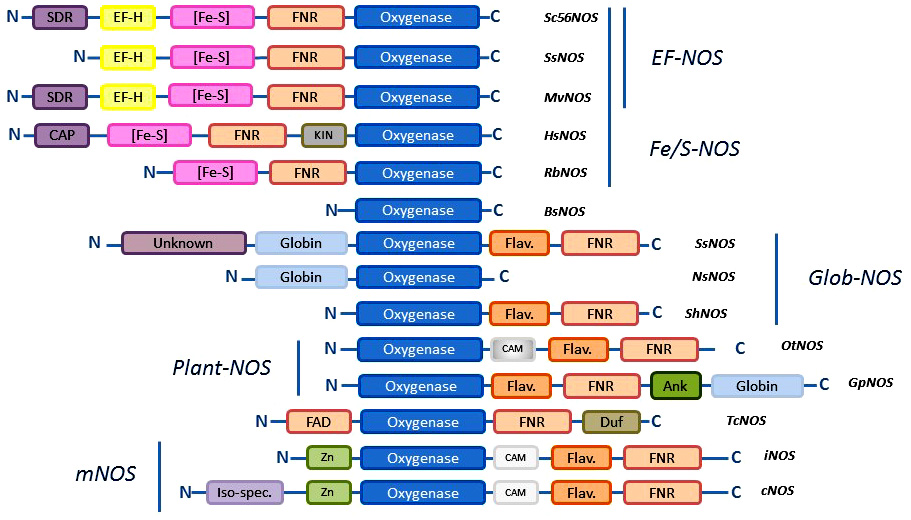 Figure 9
Figure 9Examples of different structural families of NOS proteins. Protein sequences of various NOSs are schematized as the arrangements of various structural, catalytic or regulatory modules. All NOSs share a common oxygenase domain (blue). FMN domain is represented by a dark orange Flavodoxin module (Flav.) whereas the FAD/NADPH domain is represented by a light orange FNR box. Calmodulin binding region is depicted by a light grey rectangle (CaM), dark grey when the region is putative. Zn/S motif is associated to a light green box. Additional domains (see main text) comprise a globin domain (light blue box) a PEF motif (EF-H, yellow) ) a Short-chain Dehydrogenases/Reductases motif (SDR, violet), a bacterioferritin-associated ferredoxin domain ((Fe-S), pink), a kinesin domain (KIN, grey), a CAP-ED domain (CAP, violet), an ankyrin-repeat domain (Ank, green) and some domains of unknown function (Unk, DUF…). Examples of NOSs (SsNOS, HsNOS…) are associated to each type of structure.
New types of NOSs, different from mammalian NOSs are increasingly found. The first unconventional NOS was found in a Gram- bacterium, Sorangium cellulosum (Sc56NOS, (89)). This Sc56NOS illustrates three major discrepancies between the structure of mammalian NOS and that of new NOS-related proteins. Although these NOSs exhibit a strong homology of their oxygenase domain with that of mNOS, they are characterized by : i) a distinct electron transfer chain: in that case the flavodoxin domain is substituted by another electron purveyor, presumably a bacterioferritin-associated ferredoxin domain (Bfd – (2Fe-2S), Figure 9); ii) the absence of major regulating domains such as the CaM-binding domain the zinc tetrathiolate motif and the N-ter BH4 (tetrahydrobiopterin) binding region; iii) the presence of unexpected enzymatic or regulatory units: a Short-chain Dehydrogenases/Reductases motif (SDR) and a PEF motif (Penta-EF-hand family), that suggest new and additional biochemical activity; iv) a modified sequence topology, with a complete inversion of the place of the whole electron transfer chain that is here located on the opposite (N-Ter) side of the oxygenase, suggesting a completely different fold and remodeled electron transfer process. In this regard, it becomes unreasonable to think that this enzyme could behave as a genuine NO-Synthase.
This atypical picture can now be observed for many other new NOSs. Indeed, the analysis of the primary sequence of many prokaryotic NOSs reveal the existence of a new structural family of NOSs that are all devoid of the standard reductase domain and present an Iron-Sulfur cluster as part of the electron transfer chain. These NOSs, schematized in Fig.3 (Fe/S-NOS), seem evolutionary related (Figure 5). Some present alternative reductase motifs but lack the SDR motif, such as NOSs from the proteobacteria Rhodospirillaceae bacterium TMED140 or Silicibacter sp. TrichCH4B. Some NOSs do not harbor a PEF-regulatory motif or, like the NOS from Hyphomonas sp. TMED17, have alternative domains such as a kinesin-associated domain or a CAP-ED domain (for Effector Domain of the CAP family of transcription factors). Many Fe/S-NOSs are found in cyanobacteria such as Microcoleus sp PCC7113, Microcoleus vaginatus FGP-2, Crinalium epipsamum (Oscillatoriales) or Scytonema hofmanni UTEX B 1581 and various Nostoc and Calothrix species (Nostocales). These NOSs are highly homologous and are evolutionary related (Figure 5). They exhibit the same Fe/S motifs in the N-ter region, and the presence of an EF-Hand motif that suggests some Ca2+ sensitivity (family EF-NOS, Figure 9).
Symmetrically, another family of atypical NOSs can be found in cyanobacteria. We and others have identified another family of NOS that present the same Oxygenase-Reductase combination, without the CaM-binding or the zinc tetrathiolate motifs but supplemented by a globin-domain and an undefined N-ter extension (119). These NOSs are mostly found in cyanobacteria with distinct morphological properties (subsection I, III, IV) such as Nostoc sp PCC 7107 (NsNOS), Synechococcus sp PCC7335 (SpNOS) and Scytonema hofmanni (ShNOS) (Figure 9). They are also present in some Bacteroidetes species of the Spirosoma genus.
But structurally different NOSs can even be found in phyla more closely related to mammals. This is the case for plant NOSs that are devoid of the N-terminal Zn/S and BH4-binding region and do not harbor a mNOS-like CaM-binding domain (119, 120). We also report here the presence of a singular NOS in the Stramenopiles Thraustotheca clavate with a split reductase domain and an additional C-ter region comprising a domain of unknown function (Figure 9). Truncated forms of standard NOSs are also often observed, such as the ones found in the coral Acropora digitifera, or in the algae Ostreococcus tauri. Inversely, many NOSs present additional sequences (Figure 8). For example, NOSs from Echinodermata (Acanthaster planci or Strongylocentrotus purpuratus) present large N-ter insertions (around 350 residues between the oxygenase and the PDZ domains), whereas NOSs from the algae Gonium pectorale display a large (over 400 residues) C-ter extension that includes an ANK motif (ankyrin repeats) and a globin domain (Figure 8B).
This rapid overview of various structural types of NOSs does not aim at producing an exhaustive list of all potential types of NOSs to be found. We just want to stress the great diversity of NOS structures that is not limited to the three-canonical mammalian NOSs. All these new regulatory or catalytic motifs, along with their specific and different combinations, will ineluctably modify the nature of the chemical activity of these NOSs.
Phylogenetic analyses of NOS most often aimed at producing one common and global evolutive story for this family of protein. This implies that these NOSs belong to a single type of protein, at least in the considered clade. If the NOSs present within the tetrapod superclass are relatively homogeneous, this is not the case in many other phyla where the number and nature of NOSs is constantly heterogeneous. For example, the nNOS-like NOSs are not the only NOSs in the mollusks phylum: whereas bivalves exhibit only nNOS-like proteins, cephalopods have another type of constitutive NOS (without PDZ domain). Besides gastropods harbor 4 different types of NOS: nNOS-like proteins, two additional types of constitutive NOSs devoid of PDZ domain plus one iNOS-like protein (not shown). The same pattern can be observed in cnidarians (Figure 8A). Whereas only a small part of cnidarians harbors a NOSs in their genomes, many different types of NOSs can be identified, such as iNOS-like (Discosoma striata), an nNOS-like (Orbicella faveolata), both (Stylophora pistillata) or various truncated forms (Acropora digitifera). In these conditions, it seems extremely difficult to elaborate an evolutive story common for all NOSs when there are so many different types of NOSs, apparently randomly distributed. For instance, as iNOS-like proteins are found in basal metazoans such as cnidaria and placozoa and are the sole NOSs found in other eukaryotes such as plant, amoeba and fungi, it seems unlikely that the common ancestor of metazoan could be a neuronal NOS-like protein. In fact, the heterogeneity of NOSs types and distribution within the tree of life prevents any simple and straightforward explanation of NOS family evolution.
The wealth of genomic data will ineluctably increase the number of NOS types and the heterogeneity of their distribution. The limited mammalian categories already proved insufficient to characterize and class these new NOSs in a stable and heuristic manner that could provide with a “simple” explanation for NOS evolution. There is a need of a better classification of NOSs, based on a more precise knowledge on the structure and function of NOS. Until now, NOS classification has been based on structural motifs related to subcellular localization (PDZ, myristoylation sites) and electron transfer regulation (CaM domain, autoinhibitory loop) but none were related to NOS catalytic activity. This absence of concerns about the fine structure of the oxygenase domain is due to the conviction that any oxygenase domain is structurally identical and will uniformly product NO° for any new NO-Synthase (Figure 6). In this regard, the knowledge acquired on mNOS structure and function, and particularly on their oxygenase domain, should be thoroughly used to generate a more precise description of the various distinct types of NOSs. It is not our goal here to describe the subtle variations of the oxygenase domains of the many hundreds of NO-Synthases. However, we will describe this kind of analysis for three representative examples: NOSs from basal metazoans, plants and cyanobacteria.
 Figure 6
Figure 6This phylogenetic tree does not aim at classifying the huge diversity of NOSs (that encompass several hundreds of proteins) but at illustrating the difficulty in achieving this kind of analysis with a family of proteins that exhibit a strong diversity of molecular structures, catalytic functions in various ecological and physiological conditions. In this NOS phylogenetic tree, NOSs are whether distributed along their specific structure or along the species phylogeny. In vertebrates, NOSs are split in three branches that correspond to the three mammalian isoforms (eNOS, nNOS and iNOS). Surprisingly, this distribution does not hold for the other chordates such as cephalochordates or echinoderms. Even if the invertebrates and the basal metazoan exhibit various isoforms they do not partition between nNOS-like or iNOS-like but follow their species phylogeny. This suggests a completely distinct evolution of NOS family outside the vertebrates’ lineage. Sometimes, NOSs gather along morphological/physiological similarities. This is the case for the NOS from the oomycetes Thraustotheca clavata that cluster with the Fungi NOSs. Likewise, NOSs in the “Algae” branch belong to photosynthetic organisms from completely distinct phyla: plants (Ostreococcus tauri), haptophytes (Chrysochromulina sp.) and stramenopiles (Thalassiosira oceanica). In prokaryotes, NOSs first gather along their type of structures: Oxy, Glob-NOS, EF-NOS, Fe/S-NOS (see Figure 3), independently on the phylogenetic distribution of the species. For example, the “Glob-NOS” branch includes NOSs from various classes of Cyanobacteria and Bacteroidetes, whereas the (EF-NOS-Fe/S-NOS) branch correspond to Cyanobacteria and Proteobacteria. Apart for protists, this phylogenetic tree grossly overlaps with a species-based Tree of Life.
We have identified NOS-related sequences in six cnidarians, in one porifera and in the placozoan Trichoplax adhaerens (Table 3 and Figure 8A). Different sequences were obtained for Trichoplax (isoforms TaNOSA, TaNOSB and TaNOSC) and Acropora digitifera (AdNOS1, AdNOS2, AdNOS3). Although the retrieved sequences might be artefactually truncated, their alignment shows that they correspond to three different proteins (not shown). It shows various N-ter-truncated regions AdNOS2, AdNOS3 TaNOSA and TaNOSB, but a significant extension for AdNOS1. We also observed several gaps in the sequence of AdNOSs and Exaiptasia pallida NOS (EpNOS). We focused on the part of the oxygenase domain overlapping with iNOS domain that we use as numbering reference. Percentage of sequence identity of the oxygenase domain of these NOSs with iNOSoxy range between 50% (AdNOS and HvNOS from Hydra vulgaris) and 60% (EpNOS, TaNOSB and NvNOS and OfNOS, NOSs from Nematostella vectensis and Orbicella faveolata TAB). AdNOS1 and AdNOS2, along with TaNOSA and TaNOSC, seem closely related (PI >80 %). A rapid phylogenetic analysis (Figure 8A, see legend) confirms the homology information. It stresses the peculiar behavior of TaNOSB (distant from the other TaNOS isoforms but closely related to iNOS) and of AdNOS3 (distant from the other AdNOSs and the other Scleractinia NOSs and close to NOSs from Corallimorpharia). It also highlights the existence of four NOSs groups corresponding to species phylogeny (Anthozoans, Hydrozoans, Sponges and Placozoans) with no obvious link with their structural specificities (PDZ domain, truncations, N-ter extension…). We further refine this gross analysis by looking at major residues of the catalytic sites, involved in substrate binding, heme environment, BH4 binding, dimer interface and substrate channel (not shown). This comparative analysis shows that all Arg-binding and BH4-binding residues are conserved for these NOSs. Likewise, the hydrophobic pocket that surrounds the heme is conserved. No specific feature of the oxygenase domain can discriminate between the NOSs of these 4 families. The same phylogenetic analysis was obtained for full-length proteins, suggesting that no specific oxygenase-related feature can account for these distinct families and the particularity of AdNOS3 and TaNOSB. Though, whereas all NOSs exhibit the mNOS-like valine that allows NO release, NOS from hydra vulgaris shows a substitution into an Isoleucine, similarly to what has been reported for bacNOS (121), which suggest that HvNOS is not fit to release NO (61, 122).
NOSs are absent from land plants but were identified in the genomes of 20 different algae, from the Streptophyta and Chlorophyta phyla, distributed in 12 orders from 9 distinct classes (Table 2, (94, 95)). Apart from the large N-Terminal extension of Gonium pectorale NOS (see above), plant NOS exhibit a homologous global structure (Figure 8B). We report only a few number of gaps (100 residues in Volvox aureus NOS sequence) but several insertions in structurally and functionally relevant regions, such as a 120-residues insert in Pteromonas sp. (PsNOS) and a 53-residue insert for Chlamydomonas cribrum (CcNOS). Focusing on the oxygenase domain, we report a weak homology with murine inducible NOS (percentage of sequence identity (PI) between 32% for CcNOS and 38% for Prasinophytes NOSs) with weak intra-class (around 50% PI) or intra-phyla (below 50% PI) homology. Despite the global sequence homology, the phylogenetic analysis of these NOSs do not overlap with a species phylogeny (Figure 8B). Although NOS within major classes (Mammiellales, Chlamydomonales) form distinct branches, some associations remain unexpected such as the grouping of NOSs from Streptophyta, Chlorophyceae and Trebouxiophyceae species, or of NOSs from the Chlorophyceae and Ulvophyceae species (Figure 8B). The analysis of the oxygenase domain sequence shows the absence of the Zn/S cluster and of the N-terminal BH4 binding region (not shown, (95, 120)). Arg-binding residues are well conserved except for Bolbocoleon piliferum NOS (BpNOS) that shows large variations in the 340-350 and 360-370 region (iNOS-derived numbering). Likewise, the hydrophobic core that surrounds heme binding is extremely well conserved for plant NOSs, except again for BpNOS. Unlike any other NOSs, this NOS also exhibits the bacterial NOS-like Isoleucine, suggesting an incapacity to efficiently release NO (121, 122). Surprisingly NOS from Cosmarium subtidinum (CsNOS) does not exhibit a Valine, nor a Leucine but a Phenylalanine, which questions the actual functioning of CsNOS. Apart from the highly conserved Arginine (iNOS 375) that h-bonds with BH4 moiety, only few residues involved in BH4 binding are conserved. This is again the case for BpNOS and CsNOS that are deprived of the essential W457 and F470 residues (W457 is engaged in p-stacking interaction with BH4 ring and controls its redox properties (123), and both W457 and F470 are engaged in H-bonds with BH4 moiety that controls its precise positioning at the edge of the heme (124)). NOSs from plants seem to form a distinct family from mammalian NOS with no Zn/S cluster, no CaM domain, a weak global homology and an altered BH4-binding capacity. However, some of these NOSs exhibit singular structural feature (CcNOS and PsNOS display major insertions, BpNOS and CsNOS show dramatic changes in core catalytic residues) that will definitely affect their actual enzymatic functioning. Besides, the phylogenetic tree of plant NOS displays unexpected partitioning that does not coincide with algae phylogeny. Alternative explanations are thus required to address the evolution and functioning of this peculiar NOS family.
We have identified 35 different NOSs in various cyanobacterial species and subspecies (Table 1). Many different types of NOSs are observed, sometimes in the same species. If most of the NOSs correspond to the “oxygenase” isoform (Figure 9 and 5C), several Glob-NOS and EF-NOSs have been also identified. Additionally, truncated forms of these EF-NOS (without the SDR domain) or of the Glob-NOS (without the reductase or the globin domains) can also been noted. A rapid phylogenetic analysis suggests three different families: Glob-NOS, EF-NOS (and related) and Oxy-NOSs (Figure 8C). Identity percentage of the shared oxygenase domain with iNOSs range between 40 and 48 %, which suggests that cyanobacterial NOSs are more closely related to mammalian NOSs than plant NOSs. All Glob-NOSs (and EF-NOS alike) are extremely similar, despite the fact that the species belong to very different branches of the cyanobacteria tree and to distinct morphological subsections. This is confirmed by the analysis of crucial catalytic site residues (not shown). Whereas most of the Arg-binding residues are conserved, the mammalian NOS-like Valine is only observed in EF-NOSs whereas all the Glob-NOS and most of the Oxy-NOSs instead exhibit an Isoleucine that prevents NO° release. Likewise, Glob-NOSs exhibit a slightly less well-conserved heme hydrophobic pocket but a more conserved BH4-binding capacity. The C-ter region of all these NOSs is clearly remodeled with a more open heme edge (alike what was observed for plant NOS). Along with the truncation of the N-ter region, this surely affects BH4 binding. This comparative analysis of major structural motifs of the oxygenase domains of cyanobacterial NOSs is in line with the phylogenetic analysis, which suggests a diverging evolution of the three major families of cyanobacterial NOSs within the phyla. The distribution of each type of NOSs in all kinds of phylogenetic/morphological groups of cyanobacteria could imply that these three very different NOSs were already present in the cyanobacterial common ancestor.
It is obviously impossible to achieve an exhaustive analysis of every structural variations that occur for all NOSs. However, using three different model clades (from cyanobacteria, plants, and cnidaria) we highlighted the facts that oxygenase sequences were experiencing many different sorts of truncations, gaps or inserts, but also many important single-residue modifications that surely would affect substrate and cofactor binding, and in turn NO° synthesis and release. This kind of analysis is just an example that should be extended to the many other residues/motifs that are crucial in the chemistry of NO synthesis.
This article aimed at showing the complexity that lies behind NO-Synthases. We wanted to stress the importance of the relationship between the molecular structure of any single NOS, its function, its environment and the evolution of the whole NOS family.
Genomics has modified the way we address the question of function and evolution of proteins. It has allowed the identification of hundreds of proteins that were automatically annotated “NO-Synthases” based on the presence in their sequence of the oxygenase domain. This oxygenase domain, that is specific to NOSs, is highly homologous between all these proteins, from bacteria to humans, which led to the assembly of this family of proteins. Based on the great knowledge gathered over the last thirty years on NO° and NO-Synthases in mammals, all these proteins are today genuinely (but improperly) believed to catalyze the production of NO and to participate to signaling functions.
The problem resides here in the belief that a strong (but uncomplete) sequence homology – and most of the time a simple database annotation – can characterize the nature of such proteins and lead to the assumption that all these proteins carry similar structure and functions. Though, a name does not make a function, not even a structure. The name “NO-Synthase» hides a great diversity of objects. The NOS family gather very different kinds of proteins that, behind a common catalytic site, are characterized by a great structural heterogeneity. Variations include the absence of crucial regulatory modules, overhaul of the electron transfer chain – including new reductase domains, modified 3D architecture… – new regulatory motifs and even new catalytic domains. In this regard, it is reasonable to suggest that these NOSs are not genuine NO-Synthases and might carry a completely different activity and function.
The high structural homology of NOS catalytic site is believed to preserve a common enzymatic activity between all these NOSs, namely producing NO°. This is where the special nature of NO-Synthases comes into line. Catalysis of NO synthesis is not just a question of structure that would naturally produce NO from its substrates L-arginine, NADPH and dioxygen. This redox chemistry requires an extraordinarily sophisticated mechanism that involves a fine regulation of the proximal and distal heme environment along with a complex sequence of proton and electron transfers (Figure 10, (30, 125, 126)).
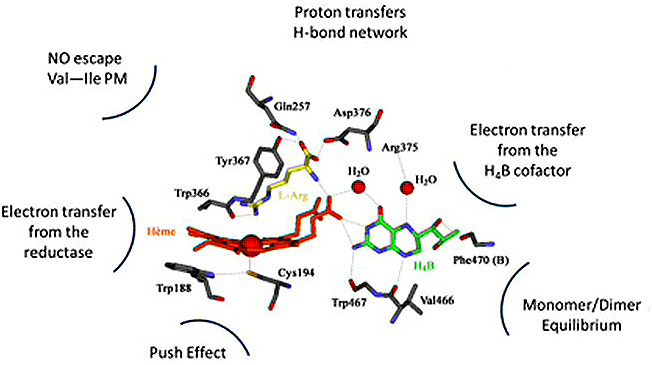 Figure 10
Figure 10Schematic representation of NOS catalytic site and of the structural parameters that control NOS mechanism. 3D structures derived from iNOS 1NOD PDB structure.
At first, it is to be noted that NOSs catalyze two sequential and different sets of reactions (Figure 2). The hydroxylation of L-arginine involved a P450-like mechanism (38, 127), whereas NOHA oxidation is achieved through a NOS-specific mechanism (26, 32). The molecular mechanisms of these two reaction sequences are clearly different and require different processes of electron and proton transfers. The necessary balance between these two reaction steps is allowed by the rigid structure of the catalytic site that ensures an adequate substrate positioning (distinct between Arg and NOHA) within a complex distal H-bond network, which in turn controls the switch between a P450- (Step 1) and a NOS-specific (Step 2) mechanism (30, 42, 63, 128-130). Subtle structural changes might modify this the redox chemistry and divert NOS from NO° production (131). Besides, NO geminate recombination on NOS heme leads to the co-existence of two distinct catalytic cycles (56, 58): an NO°-producing cycle, that corresponds to an efficient NO° release, and an RNOS-releasing cycle linked to the reduction of heme-bound NO and its conversion into other RNOS (such as peroxynitrite (PN) or nitrate). We and others have shown that small variations in some central catalytic parameters can modify the balance between futile and efficient cycles, and deeply modify NOS biochemical activity. Here-again, small structural variations might modify the extent of geminate recombination (132), the rates of ET (133, 134) or NO° release (121, 122) and lead to different catalytic production (61, 110).
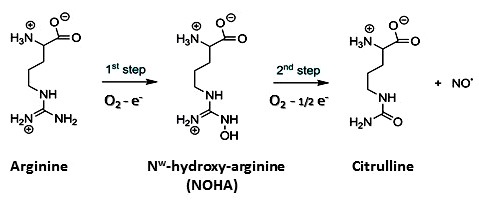 Figure 2
Figure 2The two consecutive catalytic steps of NO-synthase. L-Arginine hydroxylation requires one molecule of oxygen and one equivalent NADPH leading to the formation of Nω-hydroxyl-arginine (NOHA). Mechanism is supposed to be similar to that of Cytochromes P450. NOHA oxidation follows a NOS-specific mechanism that requires a second molecule of oxygen and a single external electron (0.5. NADPH).
The impact of negligible structural changes on NOS activity can be illustrated by two other minor modifications. We showed that subtle changes in H-bonding that can be observed between bacNOS and mNOS, modify the balance between an effective O2 activation and a futile autoxidation (135, 136). Small variations in the H-bond between the proximal thiolate and the vicinal tryptophan could slow down NOS activity or turn it into a superoxide anion synthase (137). An even more subtle variation in the structure can radically modify NOS activity. A Val'Ile substitution, that would go unnoticed in phylogenetic analyses, switch one protein from a NO° synthesis device, to a RNOS (nitrate but perhaps peroxynitrite) synthase. Indeed, this mutation slows down NO° off rate by two orders of magnitude, preventing NO° efficient release and favoring its dioxygenation (66, 121). It has been shown that this simple mutation in iNOS prevents NO° synthesis and switch it to nitrate synthesis (61, 110). All the NOSs that carry this V/I substitution thus appear unfit to produce NO°, not mentioning the ones that exhibit another type of residues (Ala, Phe). The proton transfers have been poorly investigated and the exact number, sequence and source of protons remains undetermined. It is known however that an impaired proton transfer during Step1 would prevent the build-up of an Oxoferryl complex (necessary for Arg hydroxylation) and lead to the production of H2O2. Thus, small changes in the distal H-bond network, or in the substrate binding modes could convert NO-Synthases into H2O2 producing enzyme (138).
The electron transfer processes have been thoroughly investigated and are today much more documented (34, 36, 44). NOSs are characterized by two complementary electron donors: i) the reductase domain is the original source of the electrons required to reduce the heme and the BH4 radical. Despite strong homology, the reductase domains of mammalian NOSs harbor different properties, leading to major differences in the electron transfer rates (133). We and others showed that NO° production is the result of a fine tuning between ET and other catalytic parameters and that changes in electron transfer rate modify NOS catalysis outcome (58, 63). Obviously, the new reductase motifs that we described here-above, or even external reductase proteins, will show a completely different tuning of ET rate (if any), leading to alternative catalysis.
Another major marker of NOS mechanism is the use of a redox cofactor, BH4. Indeed, NO° synthesis by NOS requires a fast ET (to prevent FeIIO2 autoxidation, (36)), but a transient and reversible one, in order to preserve the capacity to release NO° (a fast ET would lead to NO-/PN/NO3- release (36, 130)). This peculiar ET is provided by BH4 that activates FeIIO2 intermediate but re-oxidizes Fe-NO complex to allow NO° release. In this regard NOSs that do not harbor the same BH4 association should be unable to produce NO°. Though, many bacteria that harbor a NOS lack the BH4 biosynthesis machinery. It has been proposed that BH4 could be replaced by another redox cofactor, such as tetrahydrofolate ((139), FH4). Though, data on FeIIO2 activation by FH4 are lacking, such as the actual role of FH4 in NO° production. On the opposite, we observe here a strong variation of BH4-binding elements. Apart from the Arg375, the N-ter binding region and the C-ter elements involved in BH4 binding site are generally not conserved in non-metazoan NOSs, suggesting that BH4 or even other redox cofactor might not intervene in NOS catalysis.
As shown here, NOS molecular mechanism is extremely complex and versatile. Although all NOS proteins exhibit similar structure for the oxygenase domain, they do not automatically carry on the same chemistry, and their function might be extremely variable.
NO-Synthases are very peculiar oxygenase proteins. Beyond being a redox protein, NOSs have the particularity to catalyze the conversion of one gas (O2) into another gas (NO°) through an extremely sophisticated redox mechanism, which confers to NOS a strong dependency to its physico-chemical environment. For instance, NOS activity will greatly depend on the redox status of its close cellular environment: oxidative conditions (inflammation, immune responses…) will lead to the oxidation of NOS pterin cofactor that will turn NOS into a superoxide synthase (140-142). NOS activity also depends on O2 cellular concentrations: we showed that NOS apparent KmO2 was greatly different between mammalian NOSs (from 2 to 400 µM), suggesting different sensitivities to O2 concentration changes and different biochemistry in hypoxic, and even anoxic conditions (55). As an illustration, it has been proposed that eNOS, in hypoxic conditions, will maintain its role in vasodilation by producing NO° by other means: in these conditions, NOS could convert significant concentrations of nitrite into NO° by achieving another kind of chemistry, Nitrite reduction (114). Symmetrically, in oxidative stress conditions, NOS activity could be switched to other enzymatic activity such as catalase in the presence of high concentrations of H2O2 (72), PN isomerase (66) or NO° dioxygenase when high NO° concentrations lead to direct major NO° binding to NOS heme (60, 61, 110).
Beyond NOS multiple faces, the adaptive evolution of NO° function has been already described (143). As NO° is a diffusible and extremely reactive gas, its biochemical activity will mostly depend on its biological environment: i) NO first emerged in a reductive environment and was most likely mobilized in antioxidant defense (107, 143). This is still the case today in mammals, where NO° is a major contributor to the redox homeostasis, prevents oxidative stress in the cardiovascular system (115); ii) in relation with its anti-oxidant use and its gas properties, NO° eventually became the central element of a sensing/signaling machinery; at distinct organismal/cellular level, NO° plays the role of a signaling molecule mobilized in intercellular signaling or symbiosis regulation (98, 99, 144-148); iii) in a more oxidative context, NO potentiates the reactivity and toxicity of many RNOS (such as superoxide anion), leading to alternative uses in immunity. It clearly appears that NO° is involved in a large range of physiological processes that extends beyond its native function in mammals. However, this variation of NO° biochemistry does not always take place in a selected/evolutive frame but is often at the core of various pathological dysfunctions. In mammals for example, oxidative conditions will lead to the uncoupling of NOSs, switching its biological activity from signalling to inflammation (149). It is therefore reasonable to assume that the variability of the ecological and physiological environment of NO° might have led to a broadening of its biological function.
This statement must extend to the large panel of reactive nitrogen and oxygen species (RNOS), deriving from NO° bio-synthesis, such as peroxynitrite (150-152) or nitrosothiols (153, 154). Indeed, each RNOS displays a specific and distinct chemical biology (155, 156), that was first evoked to explain the multiplicity of “NO°” biological role (150). However the «good» RNOS, such as NO° and nitrosothiols, have been increasingly associated to pathological activity (157-159), whereas the “ugly” ones such as nitrogen dioxide or peroxynitrite are tentatively associated to signalling processes (160). Biological effects of RNOS are sequentially depending on their reaction with other biomolecules (and conversion into different RNOS), on their interaction with their physiological targets, on the cellular and metabolic status of the organisms, and more globally on the physico-chemical properties of their milieu.
To summarize, NOS is able to catalyze the production of multiple RNOS. The balance between these distinct catalytic reactions is tuned by the properties of the milieu. At a second level, outcome of NOS activity is made more complex by the various and variable reactivity of each of these RNOSs. At last, the pattern of RNOS production and reaction is itself controlled by the physiological status and the physico-chemical environment of NOS. This incredibly complex biology of NOS is often screened by the confusion that persists in the apprehension of NOS, NO° and RNOS. In most of the reports, there is a confusion in the respective nature of these agents: NOS is believed to produce NO°, “NO°” name is actually recovering many different nitrogen oxides. This is illustrated by the extensive and maladroit use of DAF as a NO° probe, and of cPTIO as an NO° inhibitor that overlooks the actual current chemistry of cPTIO with NO° and of DAF with other RNOS. If we assume that “NOS = NO° = RNOS”, there is no way to address the diversity of NOS and NO° biological effects and their interaction with its physiological and chemical environment. NOS function is not all included in its genomic sequence nor in its 3D structure but in the complex interaction between its mechanism, the RNOS chemistry and the physico-chemical environment. If we want to address NOS function and its evolution, we must take into account this complex diversity of NOS structure, activity and effect, and systematically investigate the relationship between NOS structure, its enzymatic activity, the biological effect of its catalytic production and the impact of its environment at each level.
After this series of questionings, it appears difficult to assign a definitive function to any new NOS. Is there a unique, selected function or a multiplicity of biochemical activity? When mammalian NOSs are to be considered, one would unambiguously respond: one function, signaling versus immunity. However, iNOS might be involved in the signaling and promotion of the immune response. eNOS and nNOS might be both exerting local antioxidant activity. In pathological conditions, constitutive NOSs will eventually be involved in deleterious biochemical processes. The frontiers between distinct functions, and beyond that between pathological and physiological effects, do not seem so obvious when it comes to NOS and NO°, even in mammals. This could have been foreseen as NO° emerged as an atypical signaling molecule, a new type of mediator: “a gas was indeed a surprise for an endogenous role, and a labile and toxic gas even more so. As the first surprise of such an unlikely agent” as it was highlighted by D.E. Koshland in its editorial of the 1992 Science Molecule of the Year (9). Unlike the conventional signaling mediators, NO° was a toxic, labile and extremely reactive molecule that thirty years after its discovery seems to exert its signaling activity in an uncontrolled and even random manner. Its role in signaling processes along with its strong and toxic reactivity could have led to reconsider the definition of “Signaling” and even to question the frontiers between a physiological activity and a noxious role, between transducing a signal and exerting deleterious and irreversible modifications. If we consider the large biological chemistry of RNOS and the versatility of NOS enzymology, these questions become even more puzzling. The multiple facets of NOS structure and activity could be addressed by considering its structure, and in particular the oxygenase domain, as a template that can catalyze various enzymatic reactions and that can be used for different purposes. In this context, the function seems undeniably related to both the enzymatic possibilities of NOS, to its physico-chemical environment and to the biological conditions. These three levels should be tackled simultaneously in order to assign a function in the frame of a biological purpose.
Although environment adaptation might have driven the evolution of NOS structure, the versatility of its enzymology, the complexity of RNOS biological chemistry and overall its impact on its own cellular/biological environment might make the rationale(s) of such an evolution difficult to unveil. The loose connection between Structure and Function makes the phylogenetic approach much more arduous. The superimposition of many functional stories, that cross, overlap and co-act impedes a simple straightforward analysis of NOS phylogeny. Which specific NOS activity is to be focused on when a new activity (and function) emerges and co-exists with an already efficient one? What is the selected structure and for which evolutive gain? Considering the important diversity of NOS structures and the multiplicity of their potential activity, NOS phylogenetic analysis will have to disentangle many parallel evolutive stories and address the co-evolution of distinct structure/activity patterns. We believe that this complex structure-function-evolution relationship is not unique to NOSs. What we describe here for NOSs should apply to many other proteins, and in particular to metalloproteins that are more susceptible to carry multiple biochemical capacities. This is illustrated for example by Globins that display a large variety of biochemical activities (O2 storage and transport, signaling, NO detoxification…) that are related both to fine tuning of their active sites and to their physiological and ecological environment.
In this context, a specific methodological approach seems needed to tackle the complex relationship between NOS structure, catalytic functioning, environmental niche and biological function. The large diversity of NOS family precludes any systematic exploration and necessitates the use of model families that display significant structural diversity. By combining modelling studies, structural characterization, and in vitro/in vivo functional investigations, an integrative and interdisciplinary approach might give some insight into the structural and ecological parameters that determine NOS activity, and provide with some predictions about the function of any different NOS protein. This task will also have to take into account the ever-changing environment of NOS, through time but also milieu. The role of NOS milieu here is major for it not only serves as a selection riddle, but for it conditions NOS actual activity. NOS environment will determine its activity, its function and the way NOS structure will evolve. This is complementary to the concept of exaptation as here the environment might contribute to the fashioning of “aptation”, participate to the design of the function and co-determine its evolution.
In any case, we believe it is now timely to ask what NOS stands for. In order to answer this question, we must avoid reducing the categories by which we apprehend NOS to the sole mammalian concepts and try to write complex evolutive stories that are not restricted to a small number of NOS archetypes but that take into account the vast multiplicity of NOS structure and function, and their singular interaction with their environment.
Abbreviations: Nitric Oxide (NO°); NO-Synthases (NOSs); Nitrous oxide, N2O; Endothelium-Derived Relaxing Factor (EDRF); neuronal NOS (nNOS); endothelial NOS (eNOS); inducible NOS (iNOS); Calmodulin-binding region (CaM); Electron transfer (ET); Auto-Inhibitory Elements (AIE); peroxynitrite (PN); bacterial NOSs (bacNOS); mammalian NOS (mNOS); horizontal genome transfer (HGT); Great Oxidation Event (GOE); Reactive Nitrogen and oxygen Species (RNOS); Reactive Oxygen Species (ROS); L-arginine (Arg); Tetrahydrobiopterin (BH4); tetrahydrofolate (FH4); All NOSs from various species are labelled XyNOS where X and y are the initial of the species.