1 Federal Williams Research Center of Forage Production & Agroecology, Nauchniy Gorodok, 141055 Lobnya, Moscow Region, Russia
Abstract
Red clover (Trifolium pratense L.) is an important forage crop throughout the world due to its high forage quality, nitrogen fixation capacity and beneficial effects on the soil fertility. But aluminum (Al) toxicity limits significantly red clover production in acid soils, which represent more than one third of the world's agricultural lands. Natural variation for Al3+ ions resistance has been identified in many crop species so development of tolerant accessions and varieties is a promising approach for red clover breeding. In this context the objectives of this article were to select in vitro and evaluate using different DNA markers the tolerant to toxic aluminum breeding samples of red clover.
Seeds of the experimental hybrid population were germinated under various aluminum concentrations, including control without aluminum. Epicotyls of seedlings without roots and with roots not less than 4–5 mm were subcultivated on agar's medium of Gamborg B5 with 2 mg/L of 6-benzylaminopurine and 100 mg/L of Al3+ and then planted in the cassettes with soil. Seedlings and adult plants F2, tolerant to 50 and 100 mg/L of Al3+ were selected, grown in vegetative pots and used further for molecular analyses. Genetic variability between tolerant and susceptible red clover genotypes was evaluated based on DNA markers: sequence-related amplified polymorphism (SRAP), retrotransposon microsatellite amplified polymorphism (REMAP) and inter-primer binding site polymorphism (iPBS).
Aluminum-tolerant red clover samples were obtained by in vitro selection on the medium with toxic aluminum ions. F2 seedlings in the variants with 50 mg/L and 100 mg/L of Al3+ were characterized by longer length and roots size compared with F1 seedlings and variety-standard at the same aluminum concentration. Subsequent molecular analysis showed that REMAP and iPBS were efficient markers to detect distinguishes among red clover accessions. The average level of polymorphism was identified as 45.8 using REMAP and 68.2% with iPBS; the average values of polymorphism information content (PIC) were 0.764 and 0.746 accordingly, higher compared to SRAP (0.741).
Combination of the biotechnology methods and the current DNA-technologies based on REMAP and iPBS markers is effective approach to improve precision and reliability of selection and assessing of red clover genotypes with tolerance to toxic aluminum ions (Al3+). Breeding samples identified in this study, can be used as a promising initial material for development the new varieties with stable inheritability of the aimed trait.
Keywords
- red clover
- in vitro culture
- retrotransposons
- REMAP
- iPBS
- SRAP
- aluminum tolerance
Red clover is a traditional forage crop throughout the world. Its wide distribution is due to its high forage quality, the ability of symbiotic nitrogen fixation, and the effect on improving soil fertility. However, at cultivation on acidic soils red clover yield decreases under the influence of toxic aluminum (Al3+) ions. In Russia, the problem of increased aluminum concentrations is especially actual due to the large number of clay and loamy soils characterized by excessive acidity. The area of acidic soils with pH of less than 5.5 is about 65 mln ha; in some regions of the country, their share reaches 70% [1]. Toxic aluminum in soils of this kind is usually found in the form of insoluble compounds, but it can also be in an exchange-absorbed state [2]. At high concentrations, it is rapidly absorbed by the roots and localized in cell membranes. As a result, the growth of the root system is inhibited, the number of root hairs decreases, and the ability to absorb water and minerals from the soil declines [3]. Toxic aluminum ions also slow down the growth of the aboveground part of plants and photosynthesis processes, which leads to a decrease in productivity and crop quality. At a concentration of mobile aluminum from 2 to 5 mg/100 g of soil, inhibition of growth and deformation of plant organs are observed, and at 10 mg/100 g of soil the plants are partially die [4]. There is evidence that aluminum has the ability to bind to nucleic acids. In this cases DNA topology may change, its rigidity increases, and the replication process becomes more difficult. As a result, DNA and protein synthesis is disrupted [5]. At the present moment there are a number of well studied major gene families involved in aluminum tolerance in higher plants [6]. Some of them such as multidrug and toxic compound extrusion (MATE), Al-activated malate transporter family (ALMT) are responsible for exudation of citrate or malate from the root to chelate and detoxify Al. The third major family is ATP-binding cassette (ABC) transporters acts in intracellular redistribution of Al and it sequestration into vacuole [7].
Agrochemical methods that improve the properties of soils with high acidity and high concentrations of aluminum (liming, chelating, and addition of organic and mineral fertilizers) require large financial investments, labor and time costs. In this regard, the issues of creating acid-tolerant breeding samples and varieties, adapted to the toxic effects of aluminum ions are relevant.
To date, in the State Register of Breeding Achievements of the Russian Federation, there is a single variety of red clover “Topaz” selected by Federal Williams Research Center of Forage Production & Agroecology with increased resistance to acidic soils. However, intensive breeding work is underway in this direction. Samples tolerant to toxic Al3+ ions are obtained on selective media in vitro, which are studied in laboratory, vegetation, and field experiments to develop varieties with high resistance, productivity and longevity [8, 9]. In the light of recent advances in molecular biology, breeders are looking for the possibility of selecting of aluminum-tolerant forms using different types of DNA markers that can identify genetic modifications of plants. Markers for these purposes should have necessary characteristics—accessibility for identification of phenotypic expressions of allelic variants, uniformity of distribution in the genome, reproducibility of the analysis results, and the possibility of its automation. Polymerase chain reaction (PCR) is a simple and fast way to detect variability between stable and susceptible genotypes in a population or between the original forms and samples selected on medium with a high content of aluminum in different forms. Currently, single nucleotide polymorphism (SNP), restriction fragment length polymorphism (RFLP), simple sequence repeat (SSR) and gene-specific molecular markers have been used to identify aluminum-resistant plants [10]. Previously, in crops such as barley, wheat, sorghum, wheat, rye, corn and tomato, it was shown that the MATE, ALMT transporters contain transposable elements in their sequence [11]. However, information on the mechanisms of aluminum resistance in red clover is lacking, as well as data on informative genetic markers.
The aim of the present study was to evaluate based on retrotransposon microsatellite amplified polymorphism (REMAP), inter-primer binding site polymorphism (iPBS) and sequence-related amplified polymorphism (SRAP) analyses the genetic peculiarities of red clover breeding samples, obtained in vitro on selective medium with toxic aluminum (Al3+).
The work was carried out in the laboratories of agricultural biotechnology and molecular and genetic studies of forage crops of the Russian State Center of Forage Production and Agroecology.
The object of research was the genotypes of an experimental hybrid population of red clover, developed in the department of the gene pool of the Federal Williams Research Center of Forage Production and Agroecology and selected in vitro for resistance to the action of Al3+. As a control, the genotypes of the Russian variety VIK 7 were included in the analysis, which, as a rule, is used as a standard in selection tests of this crop (Table 1).
| Genotype | Al3+ concentration, mg/L | Sample |
| №1 (Standard) | 0 | RC1 (0) |
| 50 | RC1 (50) | |
| 100 | Seedlings did not survive | |
| Experimental hybrid population | ||
| №2 | 0 | RC2 (0) |
| 50 | RC2 (50) | |
| 100 | RC2 (100) | |
| №3 | 0 | RC3 (0) |
| 50 | RC3 (50) | |
| 100 | RC3 (100) | |
Al, aluminum; RC, Red clover.
The seeds of the experimental samples were germinated for 30 days on filter paper in Petri dishes with 50 mL of Gamborg B5 medium (G0210, Duchefa Biochemie B.V., Haarlem, The Netherlands) and 2.0 mg/L of 6-benzylaminopurine (B0904, Duchefa Biochemie B.V., Haarlem, The Netherlands) without Al3+ and with its addition in different concentrations (50 and 100 mg/L). The criteria for evaluation and selection on tolerance to increased acidity were differences in the length of seedlings and the peculiarities of their root formation under selective conditions [12]. Epicotyls of seedlings without roots and with roots not less than 4–5 mm were subcultivated on agarose medium of Gamborg B5 with 2 mg/L of 6-benzylaminopurine and 100 mg/L of Al3+ and then on the same medium composition, with the exception of Al3+ [13]. The remaining seedlings of each sample F1 were planted in cassettes with soil of pH 4.6 and grown until plants with 5–7 leaflets were formed, which were transplanted into soil with normal acidity and F2 seeds were obtained from them. In vegetation and field experiments, for at least 3 years of vegetation, the features of development and the ability to survive after 3–4 mowing of plants F1 and F2 with different tolerance to soil acidity were studied [14]. Least significant difference at 5% significance level (LSD05) was calculated according to B.A. Dospekhov [15].
The in vitro and in vivo sampling scheme is shown in Fig. 1.
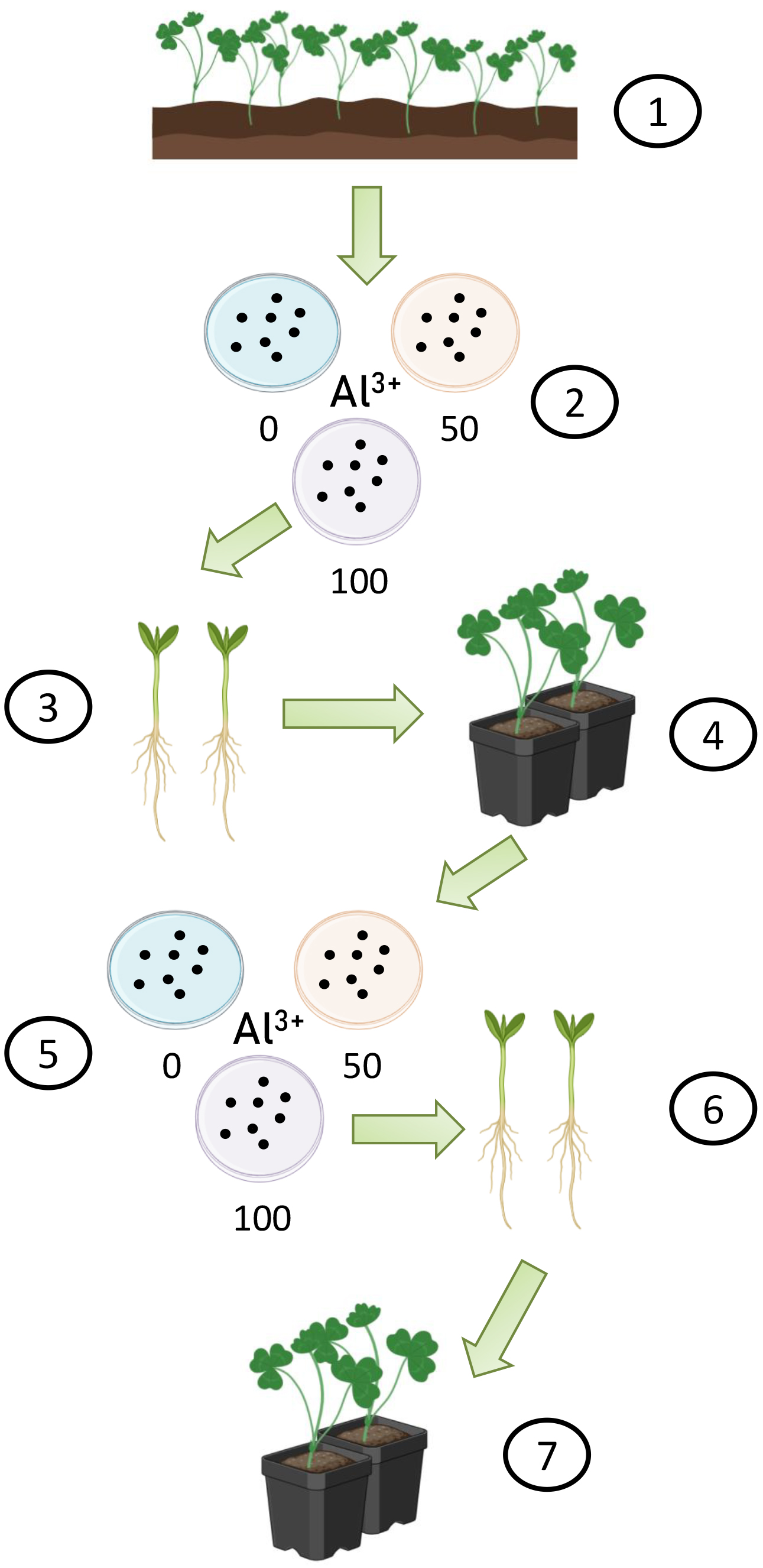 Fig. 1.
Fig. 1. A scheme for obtaining acid-tolerant forms of red clover. (1) Selection of genotypes from an experimental hybrid population in the field; (2) Obtaining F1 seeds and germination with different concentrations of aluminum and on Al-free media; (3) Obtaining F1 seedlings, tolerant to 100 mg/L of Al3+; (4) Red clover samples F1, planted in cassettes with soil; (5) Obtaining F2 seeds and germination on media with Al3+ in comparison with the control; (6) Obtaining F2 seedlings, tolerant to 100 mg/L of Al3+; (7) Experimental red clover plants, cultivated in the vegetative pots. Figure created with Biorender (https://www.biorender.com).
The REMAP method (retrotransposon-microsatellite amplification polymorphism) involves the use of a combination of long terminal repeats (LTRs) of retrotransposons, which are the most common class of mobile genetic elements in plants, and SSR loci as markers for detecting mutations in various genotypes [16, 17]. This type of markers is of undoubted interest due to their multiplicity and wide distribution across chromosomes, including coding regions and heterochromatin [18, 19]. In this study we used one primer for LTR of gypsy-like env retrotransposon R173 and a second primer for microsatellite MS17 (Fig. 2A, Ref. [20, 21, 22]).
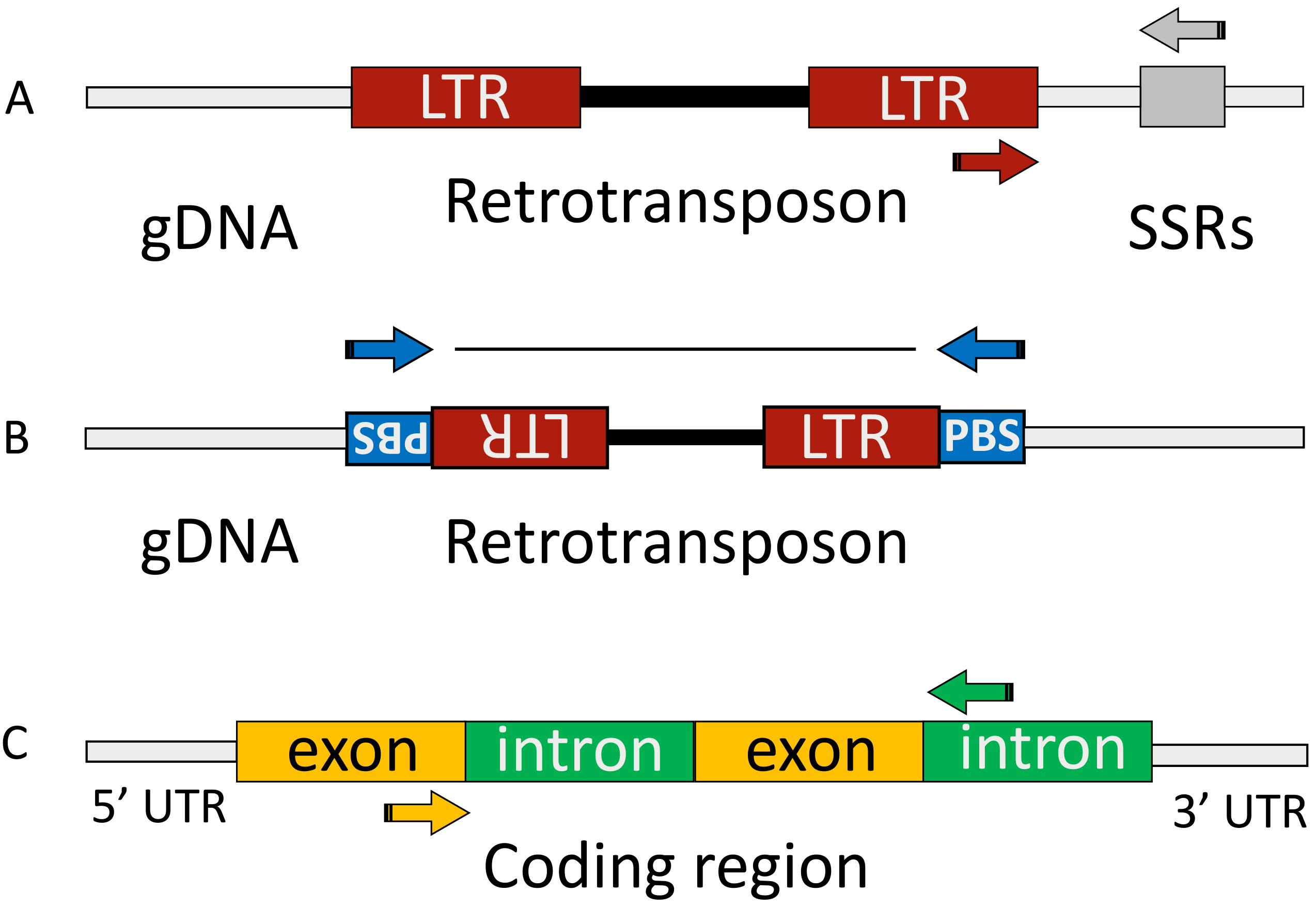 Fig. 2.
Fig. 2. Outline of REMAP, iPBS and SRAP. This shows the genomic features with positions of the priming sites for retrotransposon-based methods and SRAP marker system. (A) In the REMAP method, amplification is performed using primers complementary to both the LTR sequence and a microsatellite region (SSRs). (B) The iPBS approach involves two retrotransposons that need to be arranged in opposing orientations and must be in close proximity with each other. The diagram showcases two essential structural components of retrotransposons: the LTR (long terminal repeat) and the PBS (primer binding site). (C) In the SRAP method amplification is carried out between primers matching on exons and introns within one gene or between nearby genes. The figure was modified from [20, 21, 22]. REMAP, retrotransposon microsatellite amplified polymorphism; iPBS, inter-primer binding site polymorphism; SRAP, sequence-related amplified polymorphism; LTR, long terminal repeat.
iPBS (inter-primer binding site polymorphism) is a relatively new effective method for detecting LTR sequences [20, 23]. LTRs of retrotransposons have insertional activity and are often a source of mutagenesis, which causes an increase in the size of some genome regions or changes in gene expression [24]. Here we used single primer to amplify sequence between two primer binding sites (PBS) of inverted LTR-retrotransposons (Fig. 2B).
SRAP markers (sequence-related amplified polymorphism) determine the variability of amplified DNA fragments in intron-exon regions of the genome [25]. This group of highly polymorphic markers is widely used to assess genetic diversity, identify genes responsible for the target traits expressions, and for comparative genomic analysis [26, 27, 28]. For our research we used combination of GC-rich forward and AT-rich reverse primer, previously elaborated for amplification of open reading frames (Fig. 2C).
For REMAP and SRAP analysis, DNA was isolated from a total sample of epicotyls of 30 seedlings per accession (bulked sample), using a modified SDS method [29, 30]. At application iPBS technology total DNA was extracted of F2 plants in pots with soil of normal acidity in green house. The DNA concentration was measured using a NanoNabi spectrophotometer 3.455 (microDigital Co., Ltd, Seongnam, South Korea) and the quality of the isolated DNA was checked by electrophoresis in 0.8% agarose gel.
The synthesis of primers and reagents for PCR was carried out in the commercial company “Eurogen” (Evrogen LLC, Moscow, Russia). All primers are listed in Supplementary Table 1. The quantitative composition of PCR mixture was optimized in the laboratory and standardized for all types of markers. Amplifications were performed in a 20 µL reaction mixture containing: 5U Taq DNA polymerase—0.4 µL (Evrogen LLC, Moscow, Russia), 50
The principal coordinate analysis (PCoA) was performed using the GenAlEx 6.5 program (https://biology-assets.anu.edu.au/GenAlEx/Welcome.html) [33]. The polymorphism information content (PIC) was calculated using the formula:
where i—the i-th allele of the j-th marker, n—the number of alleles of the j-th marker, P—the frequency of alleles. The frequency of occurrence of each allele was determined (for a specific combination of primers), and then the sum of the squares of the frequencies of occurrence of all alleles was found and the resulting number was subtracted from one [34].
When studying the adaptability of plants to unfavorable environmental conditions, including increased soil acidity, the main attention is paid to the root zone, since they ensure the reliability of the processes of phytocenosis and rational consumption of nutrients by plants [35]. The most sensitive to the toxic effect of aluminum part of seedlings is the tip of the root. In this regard, at the first stage of our research we determined the differences in the length and features of root formation of seedlings, obtained on media with Al3+ (50 mg/L) and Al3+ (100 mg/L), as well as seedlings on aluminum-free media (0 mg/L) as control. On a medium without aluminum, a significant difference in the length of seedlings was noted only between standard variety RC1 and control sample RC3 in both generations. It was found that the length of seedlings in samples F1 RC2 and F2 RC2 with 50 mg/L Al3+ exceeded the value of this indicator in the control variant of the RC1 (50) standard variety, both in the F1 and F2 generations, respectively (Fig. 3). The seedlings F2 of the RC2 sample in the variant with 50 mg/L of Al3+ were also characterized by a longer length compared with F1 at the same aluminum concentration. The F2 RC2 seedlings, obtained on a medium with the addition of 100 mg/L of Al3+ exceeded the length of the initial F1 RC2 (100), whereas the seedlings of the variety-standard genotypes F1 RC1 and F2 RC1 did not survive at such a high concentration of toxic aluminum ions. This proved the effectiveness of our selective system using 100 mg/L of Al3+ in culture medium and was consisted with the data of literary sources about dependence the intensity of seedlings development in an acidic environment of the individual genotype genetic tolerance and concentration of the selective factor [12, 36, 37].
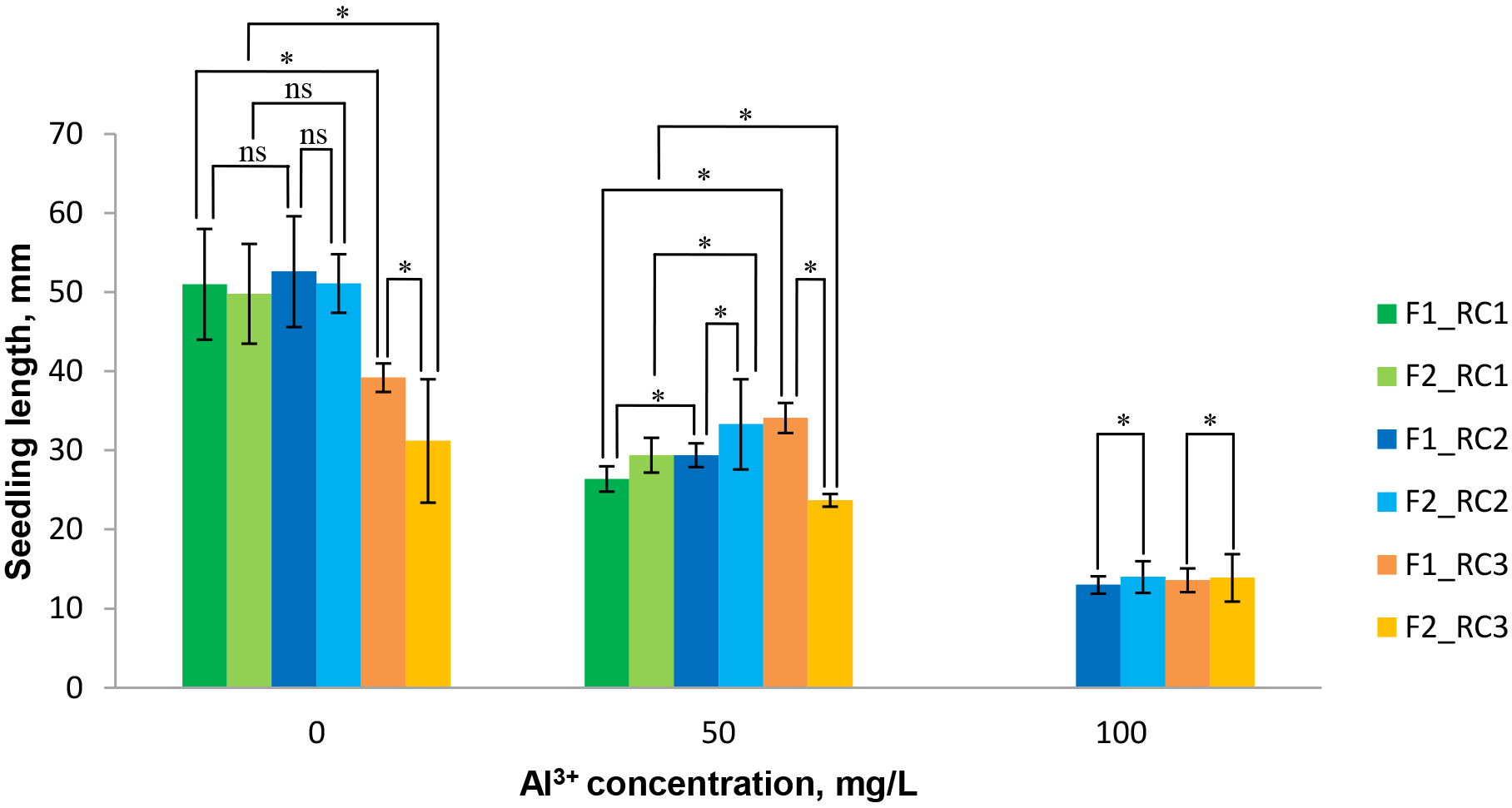 Fig. 3.
Fig. 3. Diagrams illustrating differences in the length of seedlings of red clover samples selected in vitro on medium with Al3+, compared with the control (Al3+-free medium). Least significant difference (LSD05) was determined according to Student-Newman-Keuls test at *p
When studying the peculiarities of root formation in experimental samples, it was determined that the length of the roots of seedlings F2 RC2 (100) and F2 RC3 (100) when selected on a medium with a concentration of 100 mg/L of Al3+ exceeded this parameter in seedlings of the initial F1 population (Fig. 4).
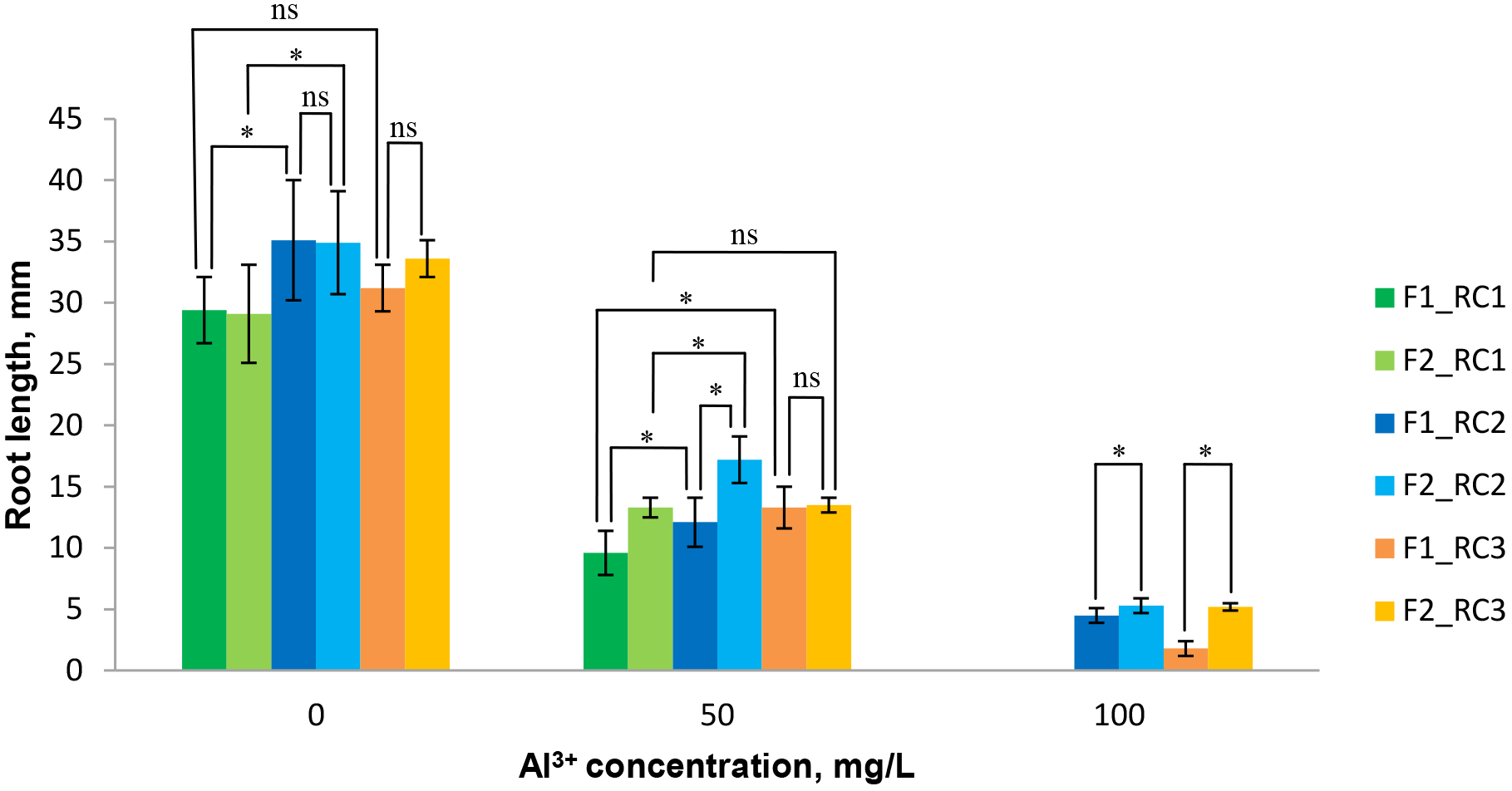 Fig. 4.
Fig. 4. Diagrams illustrating differences in the length of seedlings roots for red clover samples, selected on media with Al3+, compared with the control (Al3+-free medium). Least significant difference (LSD05) was determined according to Student-Newman-Keuls test at *p
Samples obtained on medium with different concentrations of selective factor (in vitro) and evaluated by the length of seedlings and the nature of root formation were analyzed on the basis of PCR technology in order to determine the most effective types of markers for selecting forms with a target trait based on differences in the nature of DNA spectra. Examples of electrophoregrams with different groups of markers are shown in Fig. 5.
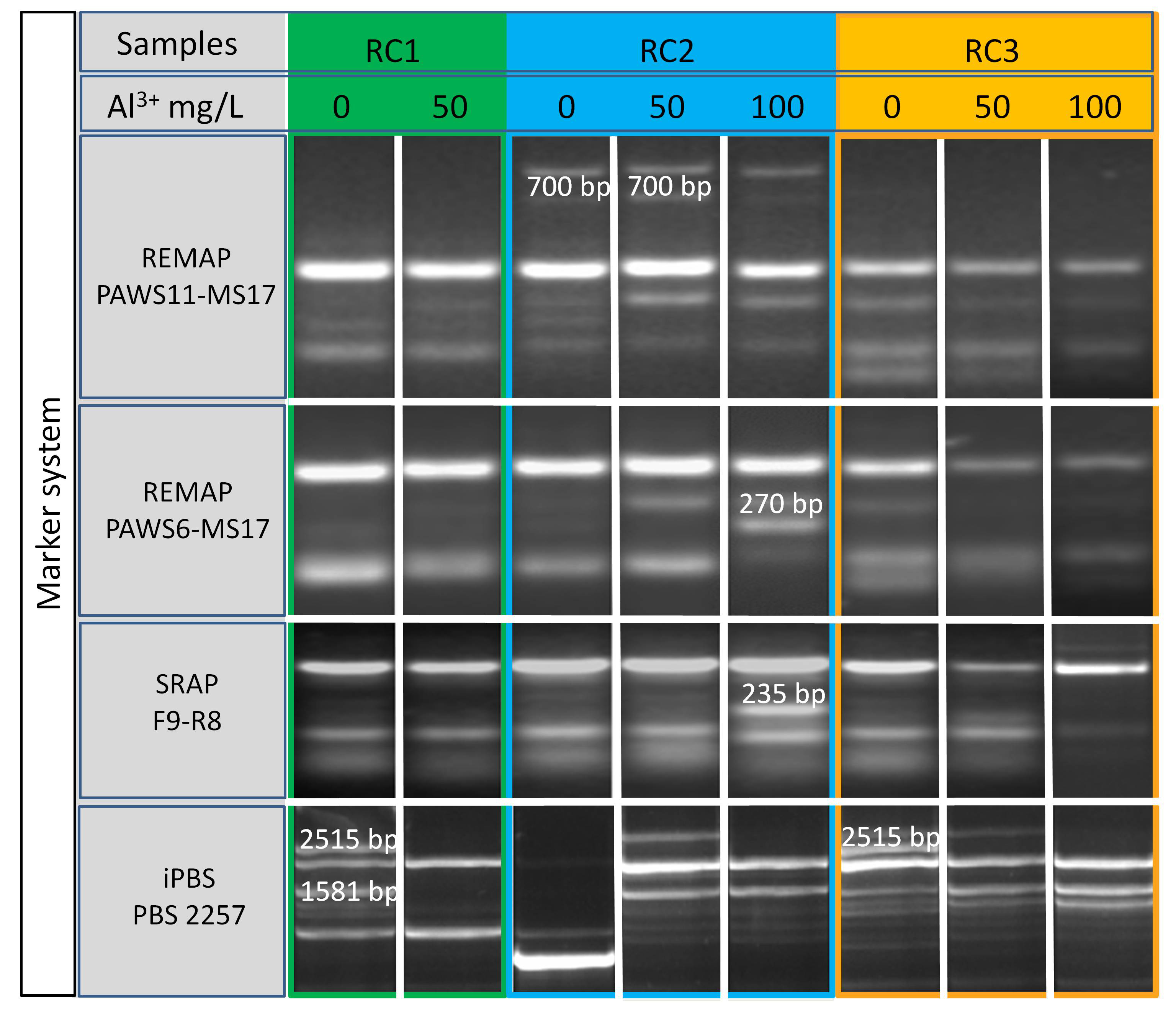 Fig. 5.
Fig. 5. Electrophoregrams of polymerase chain reaction (PCR) analysis on selection the red clover samples, tolerant to increased concentrations of Al3+ based on REMAP, SRAP, and iPBS markers.
The choice of REMAP and iPBS markers based on retrotransposons (mobile genetic elements) for our research is due to the fact that the features of their location and activation in the plant genome depend not only on the genotype, but also on external conditions, as a response to various biotic and abiotic stresses, which include the introduction into culture in vitro [38, 39, 40]. For REMAP analysis in combination with different PawS markers we used the SSR locus MS17 with a motif of 5′(CT)10G [31]. DNA fingerprinting of bulk samples of seedlings showed that the RC2 (100) genotype was distinguished with PawS6+MS17 primer pair by the presence of a 270 bp amplicon in the DNA profile, which was absent in the RC2 (0) control variant and with the addition of 50 mg/L of Al3+ (see Fig. 5). With PawS11+MS17 markers, the stable form of RC2 (100) differed from the samples without aluminum RC2 (0) and with a lower concentration of aluminum ions RC2 (50) by the absence of a 700 bp DNA fragment. The PawS5, PawS16, and PawS17 markers, when used in combination with the MS17 marker, revealed a high degree of homology in the obtained amplification products, therefore, they are unsuitable for evaluation on the basis of resistance to high acidity.
The use of the SRAP marker system is justified by their high informativeness, cost-effectiveness, and versatility in analyzing the different types of forage crops, as evidenced by the results of previous studies [41, 42, 43]. Comparative analysis of DNA profiles of red clover samples revealed that with a combination of SRAP markers F9-R8, the RC2 genotype obtained on the medium with 100 mg/L of Al3+, an additional amplicon with a size of 235 bp appeared, which was not present in the initial form or on the medium with the addition of 50 mg/L of Al3+ (see Fig. 5). Amplification was successful with other tested combinations of SRAP markers, however, no significant differences were found in the intron-exon regions of the genome in seedlings, grown under normal and selective conditions.
Among the 10 tested single primers for iPBS markers [23], 6 turned out to be informative: specific amplicons were identified with each of them, distinguishing aluminum-resistant forms from control options. In particular, when using the PBS 2257 primer, RC1 (0)—genotype of standard variety, differed from RC1 (50) by the presence of two additional amplicons of 1581 and 2515 bp in size. With the same marker, the RC2 (0) sample did not have common amplicons in the DNA spectrum with the forms RC2 (50) and RC2 (100). The RC3 sample at aluminum concentrations of 50 mg/L and 100 mg/L was characterized by the absence of an amplicon with a size of 2515 bp in comparison with the initial form of RC3 (0) (see Fig. 5).
Thus, experimental data of the analysis using markers based on retrotransposons of the LTR group (REMAP and iPBS) confirmed their increased transcriptional activity (reactivation) caused by exogenous stresses, in our case, subcultivation and regeneration in vitro. According to some researchers, genome fluctuations due to changes in the localization of retrotransposons can occur within one generation of plants, but sometimes they are reversible [20, 44, 45]. This points on the importance of the next research for screening the forms obtained by biotechnology methods in subsequent generations of offspring (F3 and beyond) with the involvement of an expanded set of informative DNA markers.
Based on the data of binary matrices, formed according to the results of analysis with markers of three types, the polymorphism of DNA spectra for different forms of red clover was calculated (Table 2, Ref. [23, 25, 31, 46]).
| Marker | Percentage polymorphism, % | PIC | References* |
| REMAP markers | |||
| PawS11-MS17 | 44.8 | 0.790 | [31, 46] |
| PawS16-MS17 | 38.5 | 0.757 | |
| PawS17-MS17 | 40.7 | 0.763 | |
| PawS5-MS17 | 42.9 | 0.786 | |
| PawS6-MS17 | 61.9 | 0.726 | |
| Average | 45.8 | 0.764 | – |
| SRAP markers | |||
| F9-R7 | 60.9 | 0.855 | [25] |
| F9-R8 | 5.8 | 0.554 | |
| F9-R9 | 44.2 | 0.880 | |
| F13-R8 | 50.0 | 0.648 | |
| F13-Em2 | 57.9 | 0.770 | |
| Average | 43.8 | 0.741 | – |
| iPBS markers | |||
| PBS 2257 | 100.0 | 0.750 | [23] |
| PBS 2253 | 66.6 | 0.774 | |
| PBS 2248 | 57.9 | 0.704 | |
| PBS 2232 | 65.2 | 0.794 | |
| PBS 2300 | 69.2 | 0.831 | |
| PBS 2239 | 50.0 | 0.625 | |
| Average | 68.2 | 0.746 | – |
Note: *—literature sources with information on primer sequences and amplification conditions. PIC, polymorphism information content; MS, microsatellite marker.
The data of Table 2 indicate a greater discriminatory potential of genetic markers based on retrotransposons (REMAP and iPBS) for the assessment and selection of acid-tolerant red clover samples: the average percentage of polymorphic loci in DNA profiles was 45.8 and 68.2 versus 43.8 in SRAP analysis. Higher average PIC values (0.764 and 0.741) also confirmed the advantage of using primers to REMAP and iPBS markers for the purposes of our study.
For visual differentiation of stable forms and control ones (which were not selected on an aluminum containing medium), an analysis was performed using PCoA method, which makes possible graphically represent the genetic relationships with minimal deviations. The values of the first two coordinates of the diagram were 37% and 21%, which together explains 58% of the total molecular variability (Fig. 6).
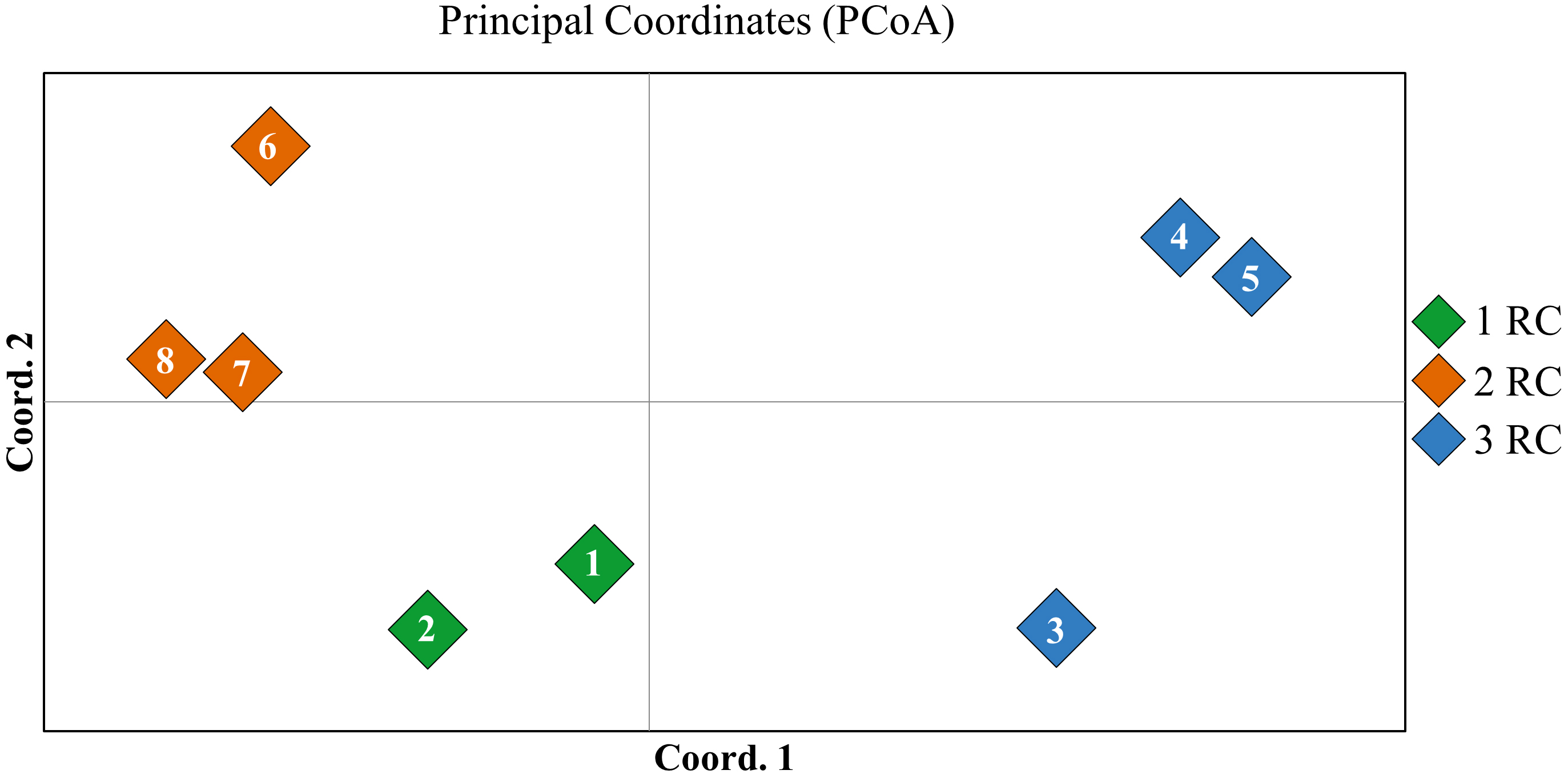 Fig. 6.
Fig. 6. Results of principal coordinate analysis (PCoA) analysis of red clover experimental samples, based on genotyping using REMAP, SRAP, and iPBS markers: 1—RC1 (0); 2—RC1 (50); 3—RC2 (0); 4—RC2 (50); 5—RC2 (100); 6—RC3 (0); 7—RC3 (50); 8—RC3 (100).
According to the results of PCoA, the breeding samples RC2 (100) and RC3 (100) were located at a significant genetic distance from the samples of the standard variety and control initial forms RC2 (0) and RC3 (0). This is an indirect confirmation of the features in the genome structure (increase in size or compression due to insertions/deletions and transpositions) formed during cultivation under selective conditions, and is consistent with the data obtained by other researchers [40, 47].
The results of this study showed that markers based on the polymorphism of terminal repeats of retrotransposons (REMAP and iPBS) can be used successfully for selection and assessment of red clover breeding samples with different tolerance to increased concentrations of toxic Al3+ (50 and 100 mg/L). The most efficient REMAP (PawS6+MS17 and PawS11+MS17) and iPBS (PBS 2257, PBS 2253, PBS 2248, PBS 2232, PBS 2300, PBS 2239) were identified to distinguish genotypes both at the seedlings level and adult plants of F1 and F2. Selected in vitro samples RC2 (100) and RC3 (100) are the promising initial material for development the new varieties with stable inheritability of the aimed trait.
Combination of biotechnological approaches with methods of DNA-typing will contribute to a better understanding of acid tolerance mechanism and to advance red clover breeding programs.
The datasets utilized and/or analyzed within the present study can be obtained from the corresponding author upon reasonable request.
Study conception, IK, LS, MA, LL. In vitro culture experiments, LS, MA, LL. PCR and statistical analysis, VD, AS. Original article preparation, IK, VD, AS. Review and editing, IK, VD, AS, MA. All authors have read and agreed to the final version of the manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work. All authors contributed to editorial changes in the manuscript.
The experimental seeds were obtained by selection from the Kretunovsky variety (https://fanc-sv.ru/products/semena-trav/klever-lugovoj/sort-kretunovskij.html) of the Falenskaya breeding station - Branch of “Federal Agricultural Research Center of the North-East named after N.V. Rudnitsky” (Kirov region, Russia). The seeds of standard variety VIK7 (https://www.vniikormov.ru/sorta/katalog-sortov-kormovykh-kultur-19.php) were obtained from the Federal Williams Research Center of Forage Production and Agroecology.
We wish to thank all the anonymous reviewers for their contributions and critical reviews, which have greatly improved the manuscript.
The research was performed within the framework of the Project No. FGGW-2022-0007 funded by the Ministry of Science and Higher Education of the Russian Federation.
The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/FBE36557.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.






