1 Basic Medical Science Department, Kulliyyah of Medicine, International Islamic University Malaysia, 25200 Kuantan, Pahang, Malaysia
2 Clinical Pharmacology Department, Menoufia Medical School, Menoufia University, 3211 Menoufia, Egypt
3 Department of Neurology, Faculty of Medicine, Delta University for Science and Technology, 11152 Gamasa, Egypt
4 Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA
5 Centre for Alzheimer’s Disease and Related Dementias (CARD), National Institute on Ageing (NIA), National Institutes of Health (NIH), Bethesda, MD 20892, USA
6 Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, WC1N 3BG London, UK
7 Neurology Department, Dr Benbadis University Hospital, 25000 Constantine, Algeria
8 Department of Neurology, School of Medicine, College of Health Sciences, Addis Ababa University, Addis Ababa, Ethiopia
9 Department of Neurology, Beni-Suef University, 62511 Beni-Suef, Egypt
10 King Faisal Specialist Hospital and Research Center, 11564 Riyadh, Saudi Arabia
11 Department of Pharmacy Practice and Pharmacotherapeutics, College of Pharmacy, University of Sharjah, 27272 Sharjah, United Arab Emirates
12 Department of Neurology, Marrakech Faculty of Medicine and Pharmacy, University Hospital Mohamed VI, 5244 Marrakech, Morocco
13 Neuroscience and Geroscience Research Unit, King Fahd Medical Research Center, King Abdulaziz University, 21589 Jeddah, Saudi Arabia
14 National Institute Mongi Ben Hmida of Neurology, 1007 Tunis, Tunisia
§AA-PD-GC: AfrAbia+plus Parkinson Disease Genomic Consortium (AA-PD-GC) members based on the country.
Abstract
Over 80% of genetic studies in the Parkinson’s disease (PD) field have been conducted on individuals of European descent. There is a social and scientific imperative to understand the genetic basis of PD across global populations for therapeutic development and deployment. PD etiology is impacted by genetic and environmental factors that are variable by ancestry and region, emphasising the need for worldwide programs to gather large numbers of patients to identify novel candidate genes and risk loci involved in disease. Only a handful of documented genetic assessments have investigated families with PD in AfrAbia, which comprises the member nations of the Arab League and the African Union, with very limited cohort and case-control studies reported. This review article summarises prior research on PD genetics in AfrAbia, highlighting gaps and challenges. We discuss the etiological risk spectrum in the context of historical interactions, highlighting allele frequencies, penetrance, and the clinical manifestations of known genetic variants in the AfrAbian PD patient community.
Keywords
- AfrAbia
- Parkinson's disease
- genetics
- challenges
- gaps
- prospects
The term “AfrAbia” was first coined by Ali A Mazrui in 1992. The convergence of Arabism, Africanity, and Islam occurred within the context of the Sahara Desert and Red Sea regions in the contemporary global paradigm. The process of Arabization, which aimed to overcome the geographical obstacles posed by the Red Sea many centuries before, occurred concurrently with the spread of Islam and the Arabization of North Africa. The historical and geographical connections between Africa and the Arab world have facilitated cross-cultural interactions, impacting genetic admixture patterns, environmental exposures, and, in turn, disease etiologies [1].
Geographical AfrAbians refer to individuals of Arab and Berber descent who belong to states that are members of the African Union and the Arab League. Countries such as Egypt and Tunisia have a predominantly Arab population, whereas Somalia and the Comoro Islands have a minor Arab presence. Additionally, there exists a group known as genealogical AfrAbians, who trace their ancestry to both Arabs and Black Africans. The Ashkenazi Jewish community is also presumed to have had a substantial and persistent historical presence in Afrabia, hence making significant contributions to the multifaceted cultural and genetic background of the region [2, 3].
Parkinson’s disease (PD)’s etiology lies on a genetic continuum, ranging from monogenic forms to complex sporadic disease. The overwhelming majority of PD cases have no identifiable single genetic cause. Monogenic forms of PD constitute between 5 and 10% of total PD cases [4, 5].
Researchers have linked several well-established genes to monogenic forms of the disease, including autosomal dominant (SNCA, LRRK2, and VPS35) and autosomal recessive inheritance (PRKN, PINK1, and DJ1) [6, 7]. Mutations in GBA1, encoding for glucocerebrosidase, are considered important genetic risk factors for developing PD across different ancestries. A recent study has reported an intronic GBA1 variant as a significant contributor to PD risk in the African population [8]. In recent years, almost 100 genetic risk loci have been associated with PD [9].
Most of our knowledge is based on European population-based studies, and no PD genetic studies aimed at exploring common genetic variation have been conducted in the AfrAbian population [10, 11]. Genetic research in the AfrAbian population represents a crucial opportunity for the study of ancestral differences in allele frequencies and magnitudes of effect of mutations associated with PD risk, the potential risk effect of heterozygous variants in recessive PD genes, and the clinical outcomes for those inheriting mutations in both GBA1 and LRRK2, among others. This initiative can provide significant insights into the complex relationship between hereditary factors and PD in this population [2, 12].
An in-depth understanding of the genetic components involved in the risk and course of the disease has a significant impact on the effectiveness of personalized treatment approaches. It may single out pathogenesis-relevant biological pathways and causal genes and genetic variants. It enables the design of therapeutics that specifically target molecular processes. It also facilitates categorising varying illness manifestations based on common genetic roots. This is crucial for precision medicine, tailoring treatment plans to patients based on a host of factors including their unique molecular backgrounds. Clinical trials involving precision therapeutics are scarce in the AfrAbian population [10, 11]. This highlights the crucial importance of identifying genetic variation associated with the susceptibility and progression of PD, as it may significantly influence the therapeutic strategies being applied.
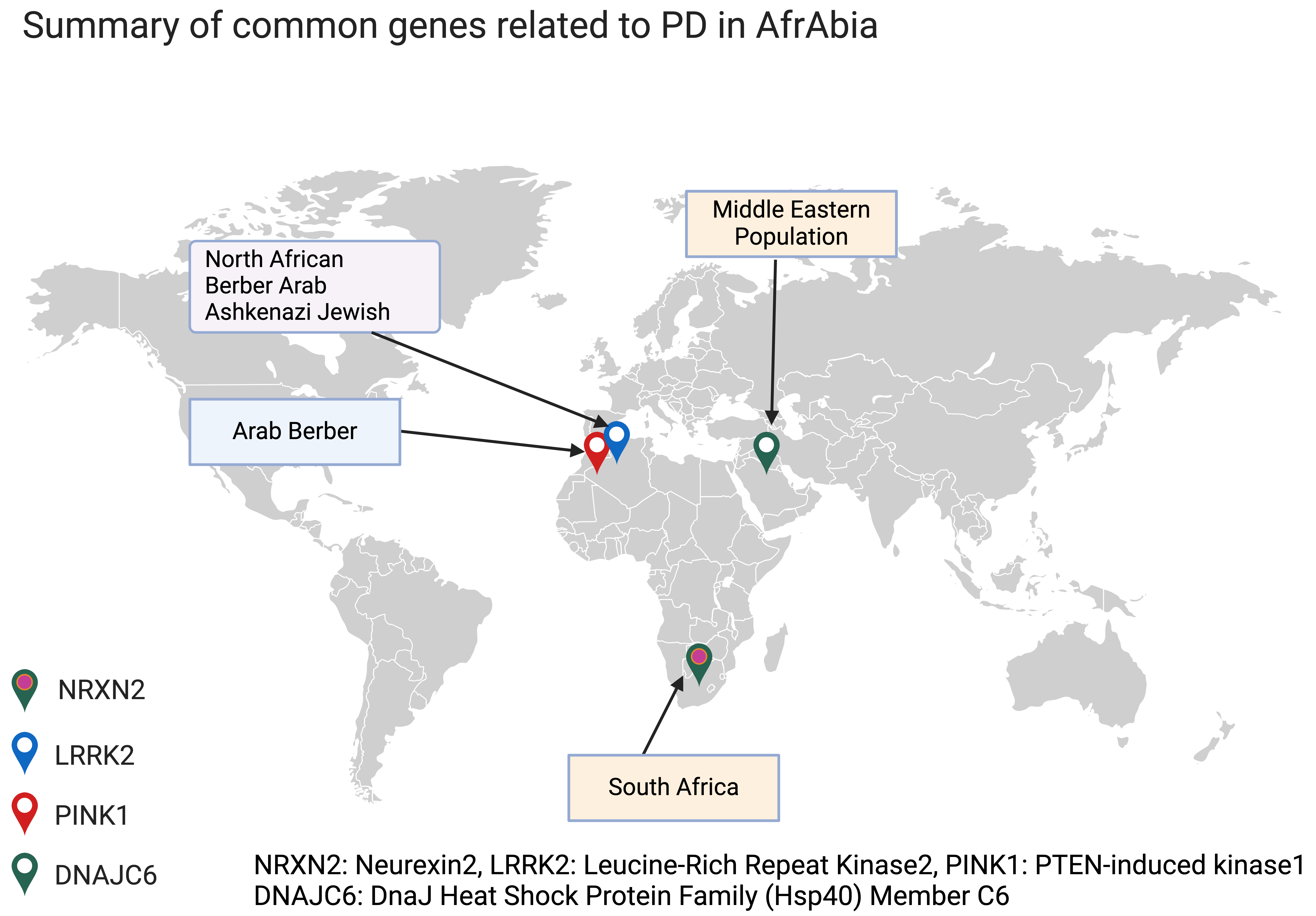
In most groups (including AfrAbia populations), monogenic variants account for less than 10% of PD patients. The necessity to acquire knowledge on the impact of genetic variation among specific populations arises from the observed disparities in disease prevalence, among other ancestral groups [13, 14]. For more details on genotype-phenotype summary for monogenic forms of PD in AfrAbia, see (Fig. 1).
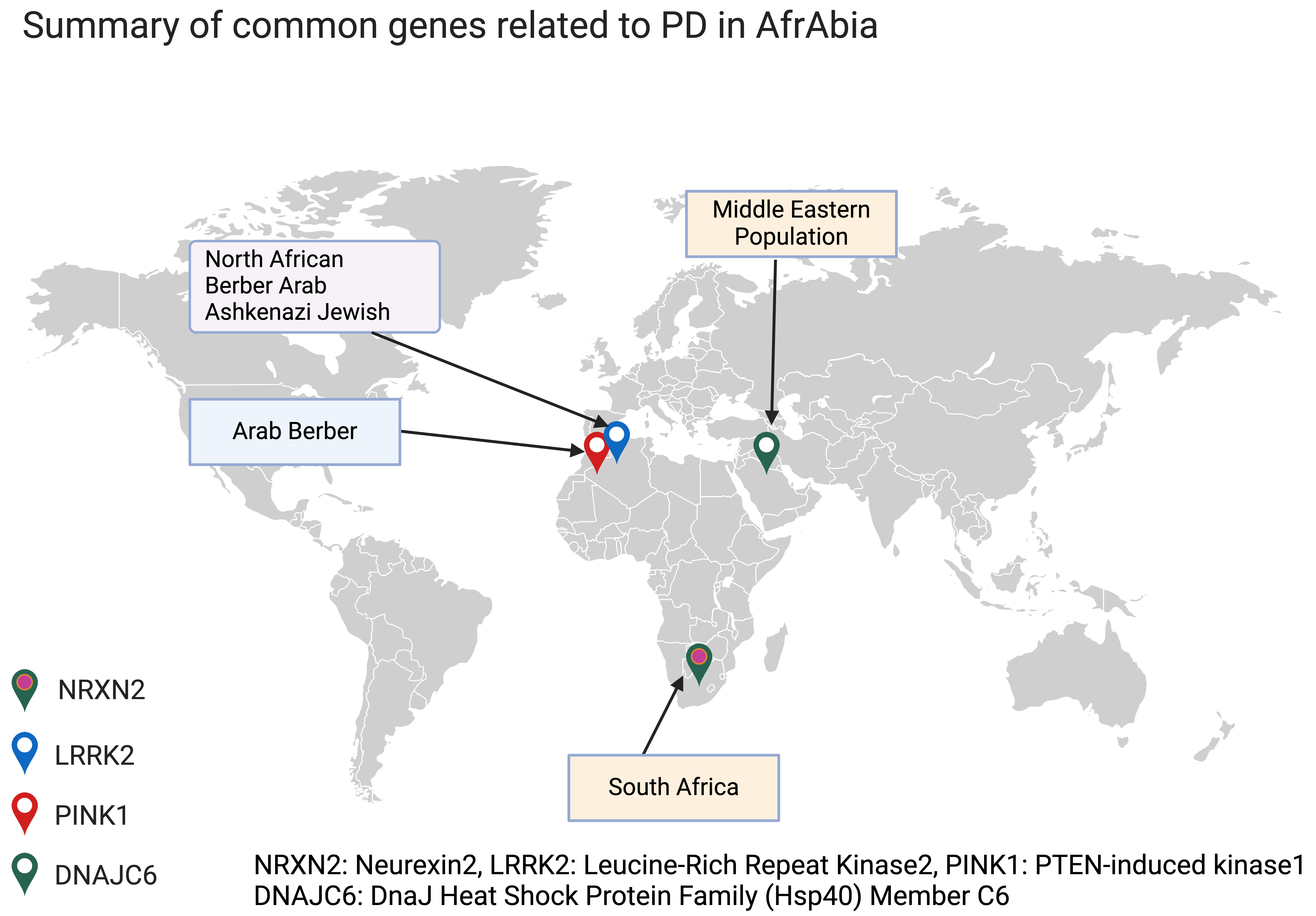 Fig. 1.
Fig. 1.
Common monogenic PD genes in AfrAbia. PD, Parkinson’s disease. The figure was made on Biorender (https://www.biorender.com/).
LRRK2 is a critical genetic risk factor for PD and has been proven to be relevant across various racial and geographical groupings. So far, 80 genetic variants have been uncovered in the LRRK2 gene [15]. Inherited mutations in this gene cause PD with age-dependence penetrance in affected families [16]. Seven of these variants, including p.N1437H, p.R1441C/G/H, p.Y1699C, p.G2019S, and p.I2020T, have been identified as pathogenic, with the p.G2019S mutation being the most frequent. This mutation is responsible for 5–6% of familial and 1–2% of sporadic PD cases [16, 17], and its prevalence varies significantly by location and race. The Ashkenazi Jewish and North African communities have the most significant documented prevalence of mutations in the LRRK2 gene, with up to 20% and 40% of PD patients in these populations being carriers, respectively [18].
The LRRK2 p.G2019S mutation is the most common and well-understood. Three haplotypes have been reported to be linked to the origin of this mutation. People of European ancestry have been shown to carry haplotype 3. These haplotypes have received less attention than haplotype 1. However, they seem more recent in their evolutionary history [8, 19, 20]. Despite its global distribution, haplotype 1 is thought to have originated from a single progenitor [20, 21]. A European cohort study placed the joint founder event at 725 years ago [22]. In contrast, an Ashkenazi Jewish, European, and European-American family study placed it at 2250 years ago [7, 8], coinciding with the Jewish Diaspora between 586 BC and 70 AD. Haplotype 1 has been reported in patients from Tunisia, the United States, Europe, and the Middle East. It is thought that all have a common ancestor who lived about 2600 years ago, according to research by Warren et al. [23].
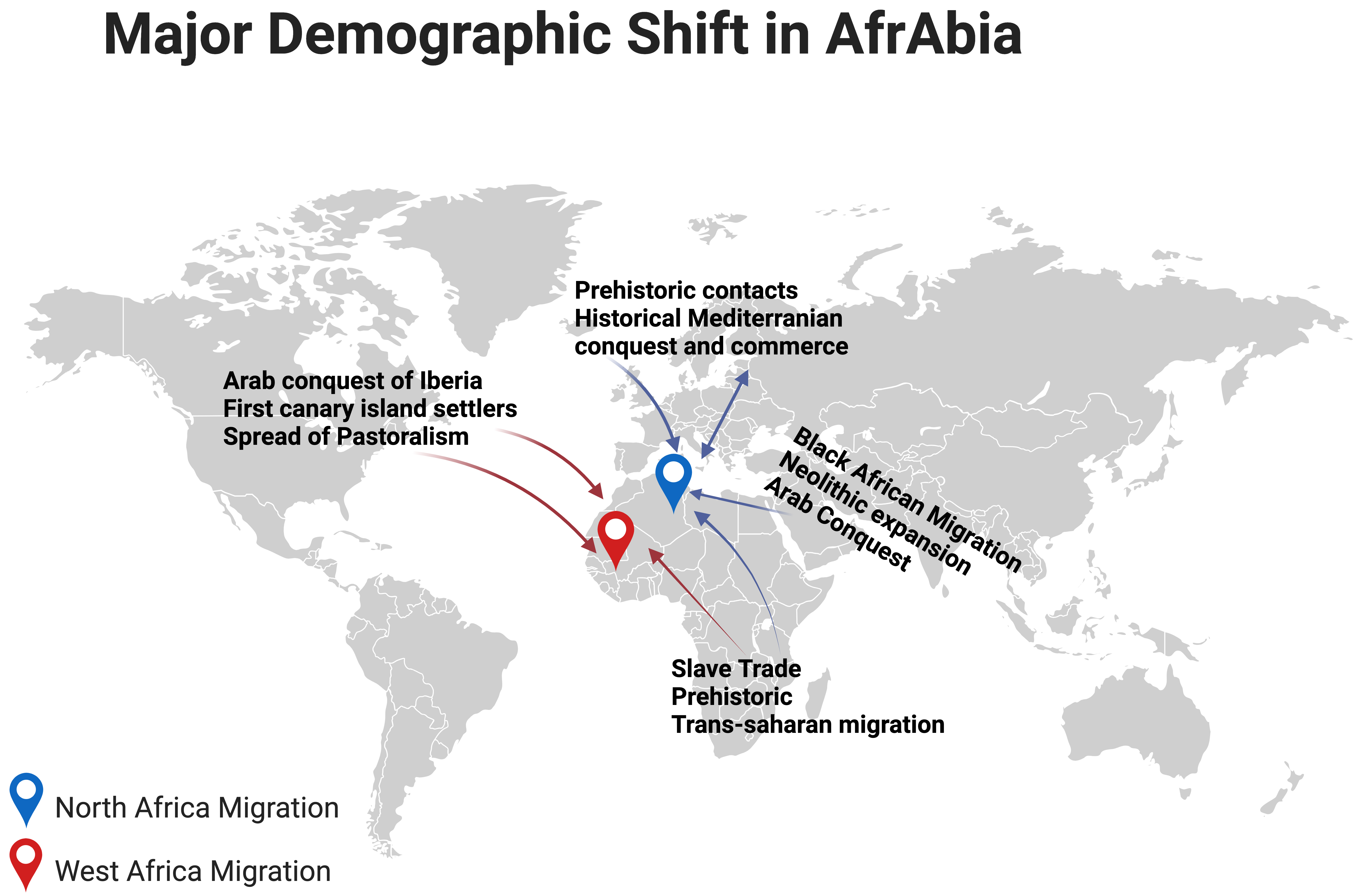
Notably, research conducted on individuals of North African origin has shown that the p.G2019S mutation first appeared in a Moroccan Berber patient, 3840 years ago [24]. Indeed, the LRRK2 p.G2019S mutation is quite common among the Berber and Arab communities that comprise the bulk of North Africa (Fig. 2, Ref. [25]). The ethnic origin of this community has either been assumed to be Arab or Arab-Berber without distinguishing between the two groups in earlier research, likely because this information is simply not recorded [26].
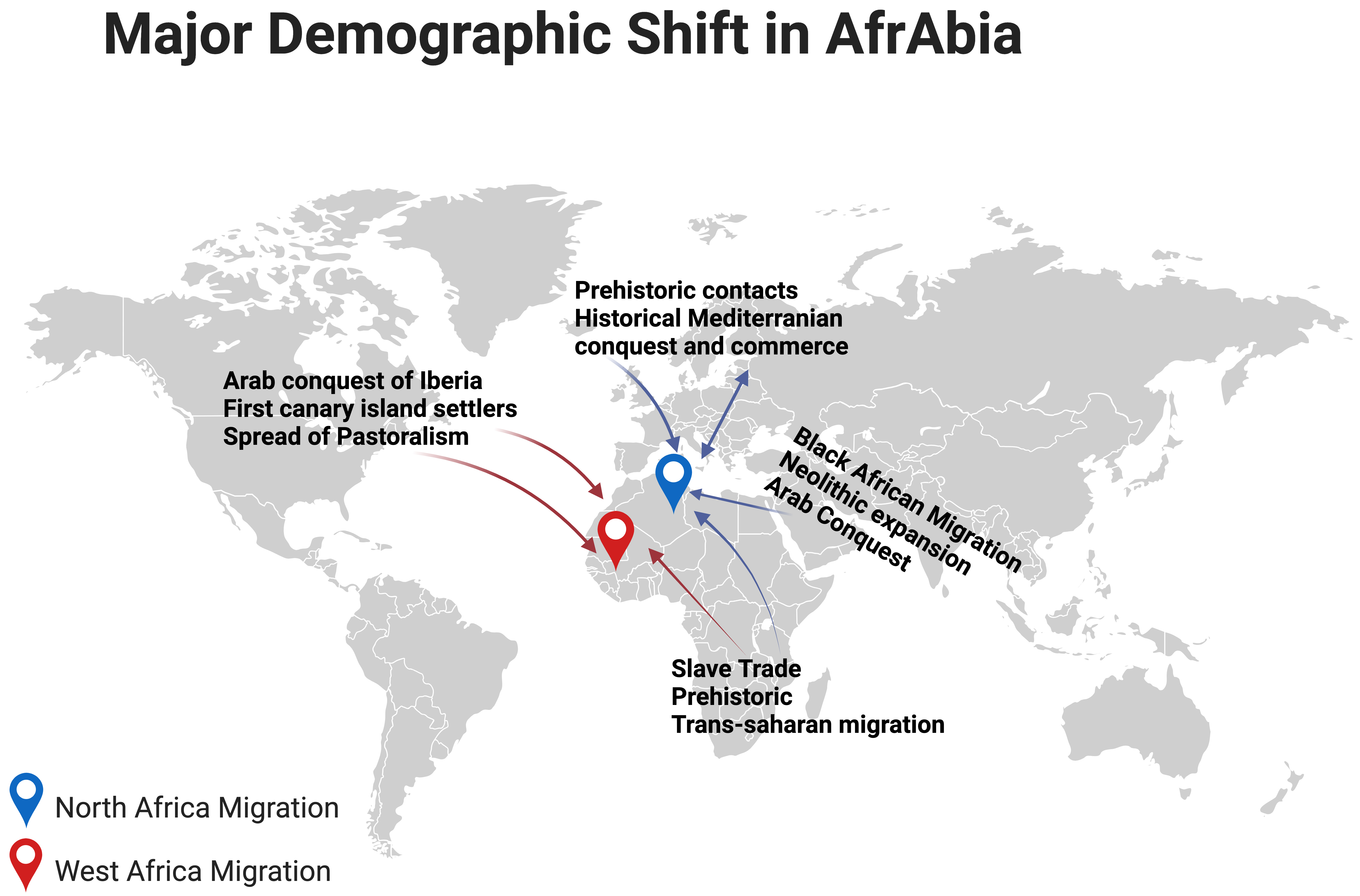 Fig. 2.
Fig. 2.
Major demographic shifts in North and West Africa [25]. The figure was made on Biorender (https://www.biorender.com/).
The Phoenicians, Romans, Vandals, Byzantines, and Arabs have all conquered and influenced the Maghreb and its Berber population, since antiquity. The end of the 15th century also saw a large influx of Moors (Islamic Spaniards) and Jews from Andalusia. A genetics study of PD in Southern Spain found that GBA1 and LRRK2 mutations seem pretty common in this population, which may have resulted from a sizable Jewish population [27]. Despite the influx of diverse populations into the Maghreb region, the native and indigenous people have preserved their genetic lineage for at least 300,000 years [28].
The LRRK2 p.G2019S mutation is most common in Maghreb nations, a region of the Middle East and North Africa. In terms of both frequency and incidence, this mutation is most noticeable in Morocco [29]. According to the golden rule of population genetics, mutations are more prevalent in the geographic area closest to their point of origin [29, 30, 31]. The discovery of uniparental markers may help us determine the most likely way the LRRK2 p.G2019S mutation came into the picture. It is possible that about five thousands years ago, a Maghreb-dwelling Berber progenitor was the carrier of a genetic alteration that would have eventually spread westward, perhaps to Morocco. The mutation may have spread over the northern coast of Africa and into Egypt over several hundred years. It has been hypothesised that during the Muslim expansion from the 18th century to 15th century, there was a flow of genes from Berber-Arab populations (mainly Berbers) into the Iberian Peninsula [32], which accounts for the north-to-south gradient in p.G2019S frequencies observed across Europe [33]. A Genome-Wide Association Study (GWAS) conducted by Campbell and colleagues [34] demonstrated that North Africans of Jewish and non-Jewish backgrounds clustered separately and that this diversity had long existed. Studies comparing Sephardic and Berber LRRK2 p.G2019S carrier frequencies all indicate a Berber origin of this mutation, making its Sephardic Jewish origin very improbable [35].
Berbers have been shown to harbour a higher mutation frequency and a shorter haplotype [24, 32, 36, 37]. Using a multi-ethnic ancestral haplotype, it was estimated that the LRRK2 p.G2019S mutation in Ashkenazim arose 4550 years ago [22]. Our Berber ancestors predate the commencement of the Jewish Diaspora and the emergence of the Jewish people as a distinct ethnic entity. Evidence of Jewish settlement in North Africa dates to at least the Iron Age, as shown by memorial monuments attributing Jewish architecture, customs, and rites [37]. Several waves of immigration bolstered these Judeo-Berbers, notably after the fall of Jerusalem to Titus in 70 AD [38, 39].
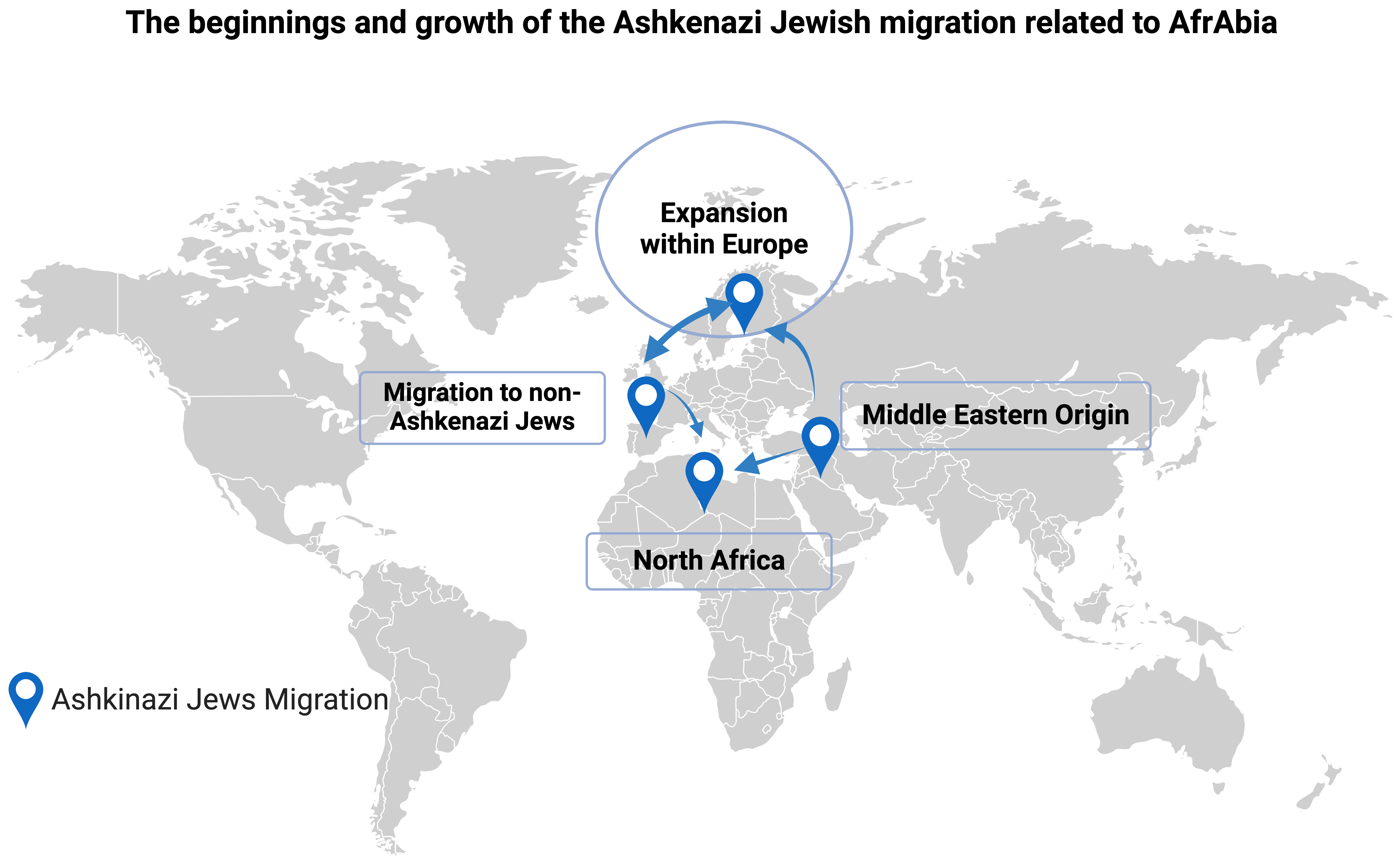
It is believed that Berbers were instrumental in spreading Judaism across North Africa via proselytism, to the point that the Berber Queen Kahina made Judaism the official religion of her country (Fig. 3, Ref. [40]) [1, 2]. Some of these Judeo-Berbers would have not converted to Islam after the Arab invasion of the 7th century, and they would have instead immigrated to the Near East as Jews, where they may have helped create the Ashkenazi branch of Judaism. Thus, it’s possible that isolation and genetic drift are to blame for the LRRK2 p.G2019S mutation’s disproportionate prevalence in the Ashkenazi community [38, 41, 42]. Still, after that, there was a period of tremendous population increase and a high rate of endogamy marriage. Despite their shared ancestry, the modern Jewish population (including Ashkenazi) has been shown to have a relatively diverse genome with no distinct genetic signature because of migrations and diasporas [43]. Lesage et al. [22, 44] conducted a GWAS that lends credence to the idea that indigenous North Africans, from the northwest to the northeast, are closely related populations outside Africa and that their separation from Near Eastern and European people predates the Holocene by more than 12,000 years. Therefore, more phylogenetic research on LRRK2 p.G2019S PD carriers from various ethnicities is warranted to unravel the origins, particularly among Ashkenazi and Sephardic Jews from North Africa and Southern Europe.
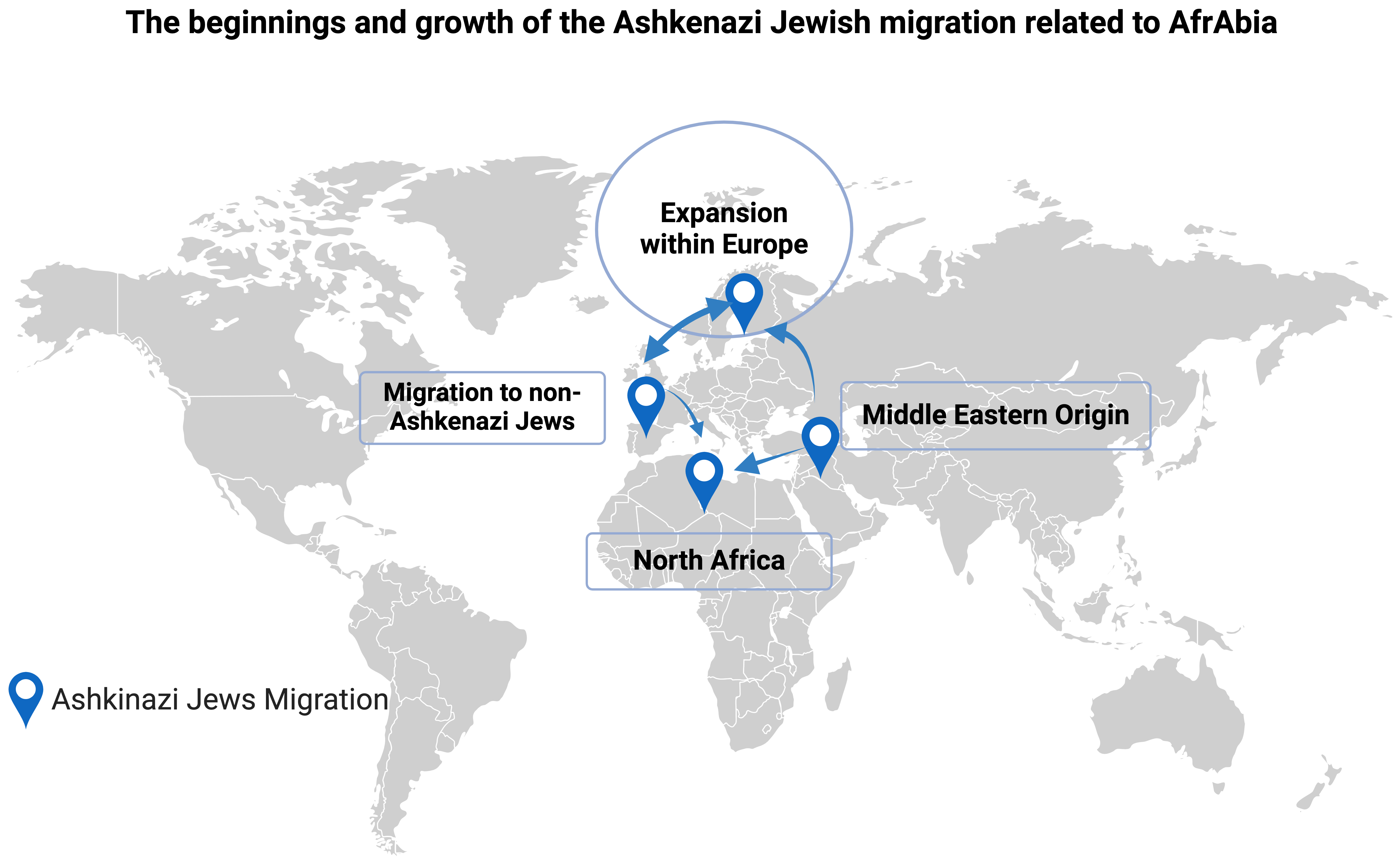 Fig. 3.
Fig. 3.
The beginnings and growth of the Ashkenazi Jewish migration related to AfrAbia [40]. The figure was made on Biorender (https://www.biorender.com/).
Sequencing of the LRRK2 gene in 92 patients with familial PD from Tunisia has revealed that 40% of these patients had a founder mutation, the above-mentioned LRRK2 p.G2019S point mutation [7]. Interestingly, a case-control study involving 238 sporadic Tunisian PD patients and 371 healthy controls showed that up to 30% of sporadic PD patients carried the LRRK2 p.G2019S mutation [45]. The LRRK2 PD phenotype was quite similar to idiopathic PD in this population [3, 46, 47, 48].
Pathogenic GBA1 mutations are the most frequent genetic risk factor for PD in European and Ashkenazi populations. The carrier frequencies of GBA1 coding mutations vary considerably (3–31%) across ethnic groups [46], even when mutations are unique to one ancestral group. Roughly 8.5% of people with PD carry a coding mutation in GBA1 [47, 48]. GBA1 mutations are more prevalent in individuals with early-onset illness (onset around 50 years of age), faster disease progression, and a more severe motor course [49, 50]. The modest, age-dependent penetrance in PD caused by pathogenic mutations in this gene varies between studies, ranging from 8% to 30% by age 80 years [51]. A recent study connected the PD polygenic risk score to penetrance and age of onset in those with a disease associated GBA1 mutation in the European population [52]. In research that examined PD clustering in eight families of non-parkinsonian GBA1 p.N370S homozygous Gaucher patients, the possible involvement of genetic modifiers in PD risk among carriers of GBA1 polymorphisms was further emphasised [53]. Despite these efforts, very little is known about the role of GBA1 variants in the AfrAbia population.
Notably, Rizig et al. [8] have recently nominated a novel intronic variant in the GBA1 gene as a significant population-specific genetic risk factor for PD in people of Nigerian heritage and those with African admixed ancestry [8]. The locus architecture and population-attributable risk differ in this startling finding compared to earlier studies in Northern European and Ashkenazi groups, where genetic GBA1 variation associated with PD risk has been thought to be mostly through coding variants. This finding underscores the need for fair and equitable inclusion of people from different ancestries in clinical trials of treatments for complex diseases like PD, and it comes at a time when the field is shifting towards precision medicine. Given the unique genetics of the Arabian population, replication studies of this novel finding in this population are warranted.
In conclusion, GWAS and other genetic assessments in AfrAbia are hindered by a lack of high-quality prevalence and incidence studies, as PD has not been considered a significant public health issue.
Despite these challenges, genetic diversity, geographical and environmental influences on disease, demographic shift and economic growth, health disparities and their impact on disease outcomes, ethical imperative and virtual learning, and conversations centred around inclusiveness, are promising avenues for further research and development. PD genetics research requires large sample sizes to discover modest effects, making it logistically and financially costly. Data exchange, research, and should be improved via an inclusive research network to empower developing nations. Genetic data from ethnically diverse AfrAbian populations, which contain at least 10% more DNA diversity than human reference genomes and 3 million private alleles, will help us understand the disease not only in these populations but more broadly [8].
Other challenges include distrust between doctors and researchers of different socioeconomic backgrounds and academic assessment systems that may limit regional cooperation. Language and cultural differences might restrict interactions. Middle Eastern and North African cooperation will improve with more global multicenter research and international contributions. The large number of LRRK2 p.G2019S carriers in this area has drawn worldwide attention and encouraged such international cooperation [54, 55, 56]. A diversity road map requires a thorough grasp of PD genetic results by location and ethnicity.
In addition to the rarity of genuine monogenic PD, many variables may make it challenging to identify a disease-causing family PD candidate gene in AfrAbia similar to other underrepresented populations. These include isolated family findings, disease variations in healthy individuals, inaccurate gene-disease relationships, and complicated phenotypes that may indicate various parkinsonisms [57] (Table 1). Robust replication across ancestries must confirm candidate genes as PD genes. This group is crucial for studying novel disease genes that require “proof of pathogenicity” because of high consanguinity frequencies [58, 59].
| Challenges | Opportunities |
| Scarcity of prevalence and incidence studies (Quality and number) | Genetic distinctiveness and diversity |
| Underpowered clinical studies | Geographical and environmental influences on disease |
| Inadequate neurological and neuroscience workforce for research | Demographic shift |
| Immature research infrastructure | Exploration of health disparities and effect on disease outcome |
| Inadequate funding for research | Ethical imperative |
| Insufficient research expertise (basic, clinical, translational) | Global village and virtual learning |
| Under-representation in cohorts included in global studies | Diversity and inclusiveness convo |
| Non-prioritization of PD as a public health issue | |
| Addressing Unmet Needs in AfrAbia PD Genetics Clinical Trials: A holistic approach* | |
| 1. Genetic Diversity: AfrAbia groups may possess distinct genetic characteristics that might impact the development, progression, and response to treatment of diseases. It is essential to consider and acknowledge this variation to create specific and successful solutions. | |
| 2. Socioeconomic Factors: Socioeconomic inequalities among AfrAbia communities might affect their ability to get healthcare, receive diagnoses, and participate in clinical studies. It is crucial to make efforts to understand and address these concerns to guarantee fair and equal representation. | |
| 3. Cultural considerations need the customization of clinical trial designs to accommodate cultural beliefs, habits, and preferences. This encompasses factors such as linguistic concerns, active involvement of the community, and the establishment of research procedures that are culturally sensitive and appropriate. | |
| 4. Healthcare Accessibility: Restricted availability of healthcare facilities and specialised services in some parts of Africa and the Arab world might hinder participation in clinical trials. Collective endeavours are required to enhance infrastructure and guarantee wider accessibility to healthcare resources. | |
| 5. Awareness and Education: Enhancing awareness of PD among AfrAbia populations is crucial. By implementing educational activities, it is possible to debunk misconceptions, diminish social disapproval, and promote engagement in clinical trials as participants get a deeper understanding of the significance of research in improving therapies for PD. | |
* This comprehensive strategy helps create more inclusive, relevant, and successful clinical trials, ultimately improving PD treatment outcomes for AfrAbia populations.
For disadvantaged populations, ethical monitoring by culturally competent agents, a well-thought-out informed consent process, local rules, data protection, and value creation are essential [60, 61]. AfrAbia communities consist of individuals of African and Arab descent. We should extend the same principles and practices we already use for Africans, to AfrAbians. For instance, new rules to protect indigenous people from genetic research abuse include the Human Heredity and Health (H3Africa) Guidelines for Community Engagement (https://h3africa.org/), the San Code of Research Ethics code of ethics (https://www.globalcodeofconduct.org/affiliated-codes/), and the Navajo Nation’s genetic research and data sharing policy [61, 62]. Research objectives, financing, governance, and publishing criteria should also be widely available.
Long-term investigations, independence, and a diversified genetics research pool need capacity development in underrepresented areas [5]. The research process, including objectives, financing, governance, and publishing criteria, should be very accessible. Nations must cooperate to pool resources and samples to recruit enough AfrAbian patients for genetic studies.
The International Parkinson Disease Genomics Consortium [63] and the Global Parkinson’s Disease Genetics Programme (GP2), funded by the Aligning Science Across Parkinson’s (ASAP) initiative [64], aim to rapidly expand our global understanding of PD and address emerging research needs in diverse ancestry populations like AfrArabia. PD is incurable, but genetic studies may illuminate its etiology and help develop personalized treatments.
Ethically sound solutions, less inequality, and more capacity development are all possible outcomes of collaborative efforts on a local and regional scale. These groups may work together to educate researchers in AfrAbia about genomics and bioinformatics, and they may develop collaborations with international groups investigating the genetic etiology of PD (AfraAbia-PD-Genomic Consortium) (https://waelmohamed3.wixsite.com/afrabiaconsortium). Care for patients living in rural areas may be improved by establishing PD support groups and implementing community-based rehabilitation programs.
There has been a scarcity of research conducted on investigating the genetic risk factors associated with PD in the AfrAbia community. There is evidence that modern humans originated in Africa, a region with a complex population structure, making it imperative to thoroughly examine this genetic community. This will aid in enhancing therapy choices and advancing the development of precision medicine specifically tailored to target PD in AfrAbia.
Conceptualization: WM, SC. Literature Search: ME, YM, YZ, WK. Data Analysis and Interpretation: SS, MM, BM, KA, NK, BA, VA. Writing — Original Draft: WM, SC, VA. Writing — Review & Editing: VA, SS, MM. Visualization: WM, BM. Supervision: SC, WM. Funding Acquisition: WM, SS. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work. This review was produced by cooperative efforts among several individuals from several countries within the AfrAbia+-PD-GC. Contributing from a variety of backgrounds and areas of expertise, each contributor helped to create, investigate, and synthesise the material.
Not applicable.
Not applicable.
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging (NIA), National Institutes of Health, Department of Health and Human Services; project number ZIAAG000534, as well as the National Institute of Neurological Disorders and Stroke. The Consortium is funded by the Michael J Fox Program Genetic Diversity in Parkinson’s Disease 2023 (MJFF Grant ID: MJFF-023995) and the Global Parkinson’s Genetic Program (GP2) which is funded by the Aligning Science Across Parkinson’s (ASAP) initiative and implemented by The Michael J. Fox Foundation for Parkinson’s Research (https://gp2.org). For a complete list of GP2 members see https://gp2.org.
Wael Mohamed is serving as one of the Guest editors of this journal. We declare that Wael Mohamed had no involvement in the peer review of this article and has no access to information regarding its peer review. Full responsibility for the editorial process for this article was delegated to Gernot Riedel. The authors declare no conflict of interest.
Supplementary AfrAbia+plus Parkinson Disease Genomic Consortium (AA-PD-GC) members based on the country.
Egypt: Wael Mohamed, MD, PhD, Kulliyah of Medicine, International Islamic University Malaysia, Kuantan, Pahang, Malaysia; Mohamed Abdelhalim Eltantawi, MD, Delta University for Science and Technology, Egypt. Mohammed.tantawy@deltauniv.edu.eg; Walaa A. Kamel, MD, Department of Neurology, Beni-Suef University, Egypt. walaaneuro@yahoo.com; Eman Eltantawy, MD, Assistant lecturer of Neurology Faculty of Medicine, Mansoura University, Egypt. Emantantawy1989@hotmail.com; ElSayed Heiba, MD, Neurology fellow Mansoura international hospital, Egypt. sayedelhussieni@gmail.com; Nehal Abou Bakr Elbeltagy, MD, Neurology specialist Mansoura international hospital, Egypt. nehalelbeltagy92@yahoo.com; Ahmed Abdulatif Hassan Mosa, MD, Lecturer of Neurology, Faculty of Medicine, Delta university for science and technology, Gamasa, Egypt. ahmed.abdulatif@deltauniv.edu.eg; Shady El Rashedy, MD, Lecturer of Neurology, Faculty of Medicine Delta university for science and technology, Gamasa, Egypt. Shady.elrashedy@deltauniv.edu.eg.
Ethiopia: Yared Zenebe Zewde, MD, Department of Neurology, School of Medicine, College of Health Sciences, Addis Ababa University, Ethiopia. yaredzene121@gmail.com; Seblewongel Asmare, MD, Department of Neurology, School of Medicine, College of Health Sciences, Addis Ababa University, Ethiopia. sebleasmare21@gmail.com.
Tunisia: Samia Ben Sassi, MD, National Institute Mongi Ben Hmida of Neurology, Tunis, Tunisia. dr.samiabensassi@gmail.com; Rim Amouri, PhD, Molecular Neurobiology Laboratory. National Institute Mongi Ben Hmida of Neurology, Tunis, Tunisia. rim.amouri@gmail.com; Fatma Nabli, MD, Neurology department. National Institute Mongi Ben Hmida of Neurology, Tunis, Tunisia. fatmafatnassi@gmail.com; Zakaria Saied, MD, Neurology department. National Institute Mongi Ben Hmida of Neurology, Tunis, Tunisia. zakaria.saied@fmt.utm.tn; Rania Zouari, MD, Neurology department. National Institute Mongi Ben Hmida of Neurology, Tunis, Tunisia. zouari.rania12@gmail.com; Amine Rachdi, MD, Neurology department. National Institute Mongi Ben Hmida of Neurology, Tunis, Tunisia. dr.rachdi.amine@gmail.com; Dina Ben Mohamed, MD, Neurology department. National Institute Mongi Ben Hmida of Neurology, Tunis, Tunisia. dinamedicale@gmail.com; Jihene Ketata, MD, Neurology department. National Institute Mongi Ben Hmida of Neurology, Tunis, Tunisia. jiheneketata@gmail.com.
Algeria: Yasser Mecheri, MD, Neurology department, Dr Benbadis University Hospital, Constantine 25000, Algeria. mecheriyasser@outlook.com; Boubekeur Saddik Fekraoui, MD, PhD, Neurology Department, Dr Benbadis University Hospital, Constantine 25000, Algeria. as71fekraoui@gmail.com; Fatima Serradj, MD, PhD, Neurology Department, Dr Benbadis University Hospital, Constantine 25000, Algeria. fatser98@gmail.com; Imene Lemdaoui, MD, Neurology Department, Dr Benbadis University Hospital, Constantine 25000, Algeria. ilemdaoui25@gmail.com.
Morocco: Najib Kissani, MD, Neurology Department, University Hospital Mohamed VI, Marrakech, Morocco. najibkis@gmail.com; Neuroscience Research Laboratory, Faculty of Medicine and Pharmacy, Cadi Ayyad University, Marrakech, Morocco. Mohammed Chraa, MD, Neurology Department, University Hospital Mohamed VI, Marrakech, Morocco. medchraa@gmail.com.
Saudi Arabia: Bashayer R Al-Mubarak, PhD, King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia. bal-mubarak@kfshrc.edu.sa; Badrah S Alghamdi, PhD, King Abdulaziz University, Jeddah, Saudi Arabia. basalghamdi@kau.edu.sa.
Jordan & UAE: Karem H. Alzoubi, PhD, Department of Pharmacy Practice and Pharmacotherapeutics, College of Pharmacy, University of Sharjah, Sharjah, UAE. kelzubi@sharjah.ac.ae; Department of Clinical Pharmacy, Faculty of Pharmacy, Jordan University of Science and Technology, Irbid, Jordan.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.