1 Department of Neurological Surgery, University of Wisconsin, Madison, WI 53792, USA
Academic Editor: Rafael Franco
Abstract
Gliomas are common brain tumors with a variable prognosis based on their tumor grade. With glioblastomas, the prognosis is usually unfavorable. Thus, having accurate and rapid methods for their diagnosis and follow-up are essential for rapid discovery of the tumor and to protect patients from unnecessary procedures. Some glioma cases are challenging since there is a limited ability to differentiate between gliomas, recurrent glioblastomas, and single metastatic lesions. Monitoring treatment responses and follow-ups can also be challenging. While both radiological and serological markers have been identified that can aid diagnosis and assess therapies, a particularly promising new class of serological markers are long non-coding RNAs. Long non-coding RNAs are a relatively recently discovered class of regulatory RNA molecules that play critical roles in many cellular and physiological processes. The potential role that long non-coding RNAs play with glioma pathogenic processes is not fully understood. In this literature review, we highlight the potential for long non-coding RNAs to be used as serum biomarkers in glioblastoma patients, including their potential to serve as non-invasive, easy to use, and rapid diagnostic or prognostic indicators.
Keywords
- non-coding RNA
- lncRNA
- glioblastoma
- glioma
- biomarker
- microRNA
- miRNA
Gliomas originate from multiple glial cell types and can appear anywhere in the
central nervous system [1]. They are the most common primary brain tumors in
adults, comprising 80% of adult malignant brain tumors [1, 2]. Diffuse
histological type glioblastomas (GBM) are the most aggressive and most fatal
primary brain tumor. Fig. 1 shows a simplified algorithm for classification of
diffuse gliomas according to the 2016 WHO Classification of Tumors of the Central
Nervous System [3, 4]. There are two types of GBM, which are defined by the gene
encoding for isocitrate dehydrogenase (IDH). These are: (1) IDH-wild types that
represent
 Fig. 1.
Fig. 1.A simplified algorithm for classification of diffuse gliomas based on histological features and for classification of GBM based on genetic features. Diffuse gliomas are classified histologically into astrocytoma, oligoastrocytoma, oligodendroglioma and glioblastoma. Glioblastomas are classified genetically into IDH wild type and IDH mutant. IDH refers to the gene that encodes isocitrate dehydrogenase.
Due to the severity, high recurrence rate, and poor prognosis of GBM, new and improved diagnostics are highly desired to enable early diagnosis and to monitor tumor progression, regression, and recurrence. Genetic biomarkers are a very promising diagnostic modality. Here we use the term biomarker for a substance that can objectively act as an indicator for a physiological or pathological process, such as a pharmacological response to a particular drug. Biomarkers may be found in tumor tissues or serum, and may include DNA, RNA, enzymes, metabolites, transcription factors, and cell surface receptors [9]. In the present work we will discuss serum long non-coding RNAs (lncRNAs) as promising biomarkers for the diagnosis and prognosis of GBM. We also will elucidate the pathophysiology by which those lncRNAs participate in carcinogenesis and anticancer drugs resistance.
Only a small proportion of our genome is transcribed into mRNAs to produce protein. In contrast, most of our genome consists of ncRNAs that are not translated into proteins [10, 11]. In the past, there was a belief that the most important genetic material product were protein-encoding sequences, and that the remaining genome that does not encode proteins had unknown or obscure less important roles, such as ncRNAs and other gene products that were involved in gene expression and message transfer. But, more recently, it has been determined that ncRNA plays critical roles in diverse cellular and physiological processes including gene regulation, chromatin packaging, cell differentiation and development [10, 11, 12, 13, 14]. NcRNAs can be classified based on the number of nucleotides (nt) into lncRNAs that are greater than 200 nt long, and small ncRNAs that have no more than 200 nt. Small ncRNAs include microRNAs (miRNAs), small nuclear RNAs (snRNAs), small interfering RNAs (siRNAs), small nucleolar RNAs (snoRNAs), and Piwi-interacting RNAs (piRNAs) [15, 16, 17]. These different types of ncRNAs are illustrated in Fig. 2. In contrast to the short-lived miRNAs that have been extensively discussed in the literature, lncRNAs are stable and easily detectable in body fluids including plasma, and usually have higher tissue-specificity, thus these are a promising new class of biomarkers deserving of attention [18, 19, 20].
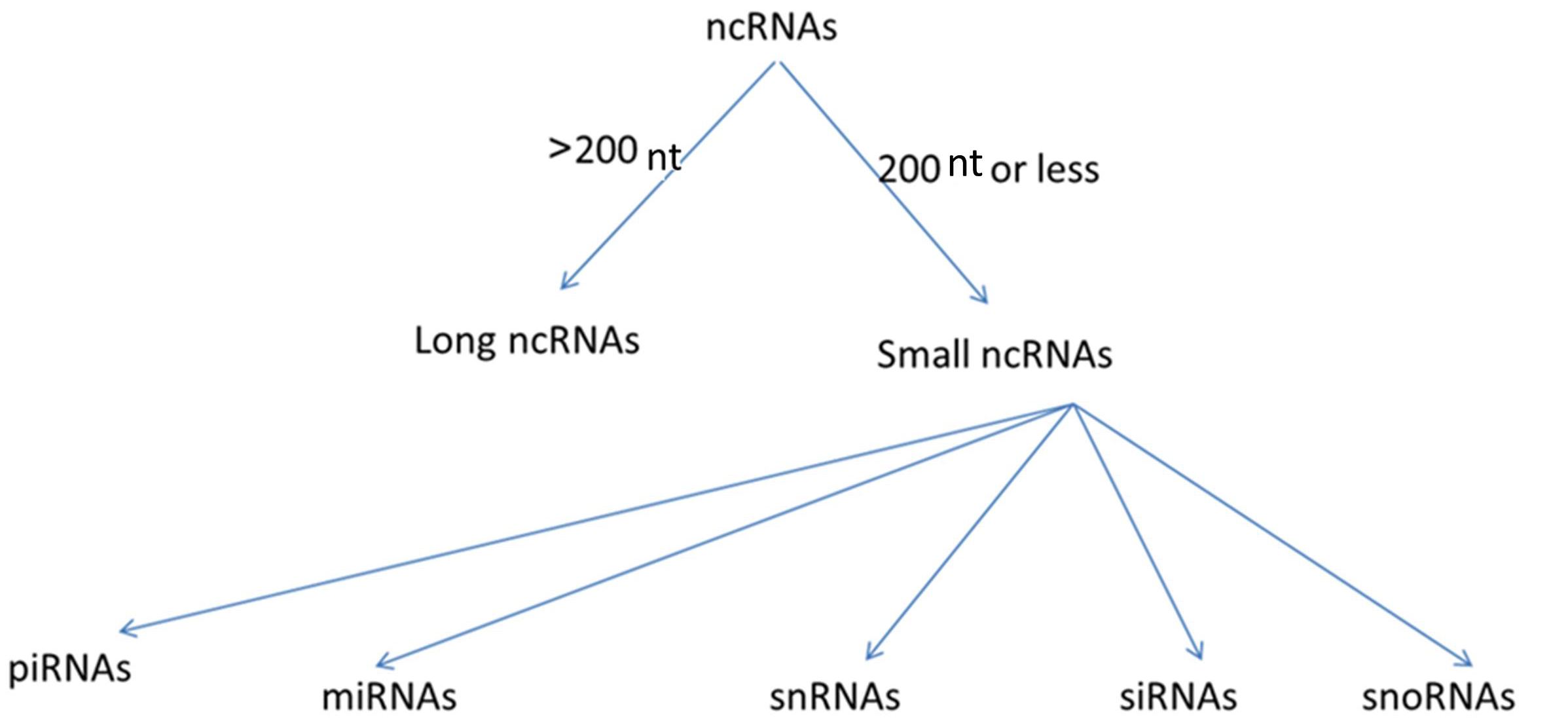 Fig. 2.
Fig. 2.Categorization of types of non-coding RNAs (ncRNAs). NcRNAs are classified based on nucleotide number (nt) into long non-coding RNAs (lncRNAs) and small ncRNAs. Small ncRNAs are further classified based on their functions into many types: miRNAs (microRNAs), snRNAs (small nuclear RNAs), siRNAs (small interfering RNAs), snoRNAs (small nucleolar RNAs), piRNAs (Piwi-interacting RNAs).
LncRNAs are non-protein coding RNA transcripts that are greater than 200 nt in length [21]. These play important roles in tumor proliferation and progression via regulating the expression of oncogenes, tumor suppressor genes, and other cancer-associated genes through epigenetic control, and through transcriptional, posttranscriptional, and translational regulation [22, 23]. Although most research has focused on the intracellular roles of lncRNAs, there is a growing interest in the diagnostic and prognostic role of circulating lncRNAs as potential non-invasive biomarkers for a variety of diseases and tumors. Specific lncRNAs have been shown to be survival predictors in patients with heart failure [24], mortality predictors in patients with acute kidney injury [25], and novel biomarkers for diagnosis of coronary artery disease [26], esophageal squamous cell carcinoma [27], and gastric cancer [28].
In gliomas, most of the reported lncRNAs can cause tumor progression or chemo-resistance, such as HOTAIR (HOX transcript antisense RNA), HOXA11-AS (HOXA11 antisense RNA), lncRNA NEAT1 (nuclear paraspeckle assembly transcript 1), lncRNA SBF2-AS1 (RNA SBF2 antisense RNA1), and lncRNA MALAT1 (metastasis-associated lung adenocarcinoma transcript) [29, 30, 31, 32, 33, 34]. In contrast to those lncRNAs, some lncRNAs have protective roles and their expression decrease may lead to tumor proliferation or chemo-resistance, such as lncRNA HERC2P2 [35].
A review of the literature indicates that there are several lncRNAs associated with GBM, but these levels were measured in biopsies or cell cultures. Using lncRNAs levels that are obtained from biopsies, while useful for research, is problematic for a clinical biomarker since obtaining a biopsy requires surgery, and the biopsy specimen itself, even without assaying for the lncRNA, is already sufficient for diagnosis. It thus would be of interest to determine if there are lncRNAs that might be used as reliable serum biomarkers for clinical diagnosis and prognosis, and as markers for chemoresistance.
The HOTAIR gene is located within a HOXC gene cluster on chromosome 12 between the HOXC11 and HOXC12 genes [36, 37]. This gene encodes a long noncoding RNA (lncRNA) molecule called HOTAIR that stretches for nearly 2200 nucleotides [38]. As illustrated in Fig. 3, the HOTAIR-PRC2-LSD1 complex attaches to the promoters of many tumor suppressor genes enabling HOTAIR to silence those genes through methylation of H3K27me3 and demethylation of H3K4me2 [39, 40]. Because of HOTAIR’s role in carcinogenesis, HOTAIR has an important diagnostic or prognostic role in multiple tumors. For example, it acts as a potential biomarker to diagnose breast cancer [41]. It also correlates with disease progression in bladder cancer [42]. It is considered a novel diagnostic biomarker for esophageal squamous cell carcinoma [36], and generally, its aberrant expression is associated with the metastatic progression of many malignancies [43]. Regarding the level of HOTAIR expression in GBM tissues, in a study that used glioma samples to elucidate the correlation between HOTAIR and glioma, Zhang et al. [44] found a significant increase in HOTAIR transcript levels in GBM, compared with that observed in normal tissues and low-grade gliomas. This study also shows that overall survival was inversely correlated with HOTAIR levels in GBM [44]. In another study, Zhou et al. [45] measured HOTAIR levels in gliomas and normal brain tissues and found a higher expression of HOTAIR in GBM tissues than in low-grade gliomas and normal brain tissues. Thus HOTAIR has potential for use as a serum biomarker for GBM diagnosis and prognosis since it appears to enable a correlation between HOTAIR expression and glioma grade.
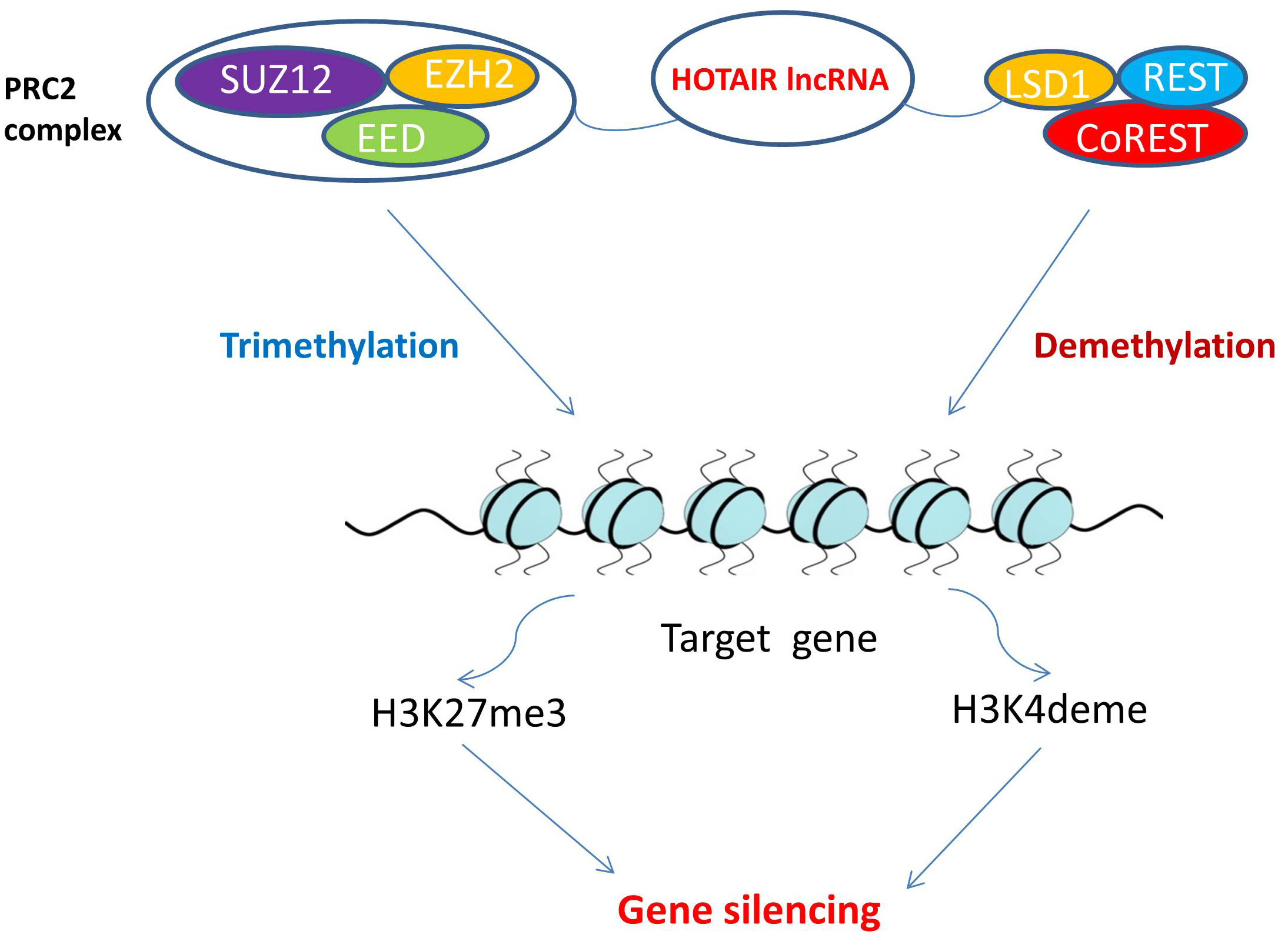 Fig. 3.
Fig. 3.HOTAIR role in epigenetic modifications. PRC2 is a target that HOTAIR can bind to and induce changes. The PRC2 complex then induces lysine methylation on histone H3. This H3K27-methylation is a form of gene silencing and is induced by histone methyltransferase EZH2, a component of PRC2. The LSD1 demethylase creates a repressor complex with REST and CoREST, that together have a pivotal role in gene silencing through mediating enzymatic demethylation of H3K4. HOTAIR interacts with both PRC2 and LSD1. The HOTAIR-PRC2-LSD1 complex attaches to the promoters of many tumor suppressor genes enabling HOTAIR to silence those genes. This histone methylation and demethylation is a dynamic process of epigenetic modification that regulates gene transcription, chromatin packaging, and cellular differentiation. Abbreviations: HOTAIR, HOX transcript antisense RNA; lncRNA, long non-coding RNA; PRC2, polycomb repressive complex 2; SUZ12, suppressor of zeste 12 protein homolog; EED, embryonic ectoderm development; EZH2, enhancer of zeste homolog 2; REST, RE1-silencing transcription factor; LSD1, lysine-specific histone demethylase 1; coREST, REST corepressor 1; H3K27me3, trimtethylated lysine 27 of histone H3; H3K4deme, demethylated lysine 4 of histone H3.
Circulating serum HOTAIR was found to be significantly greater in GBM patients than in normal controls, using real-time quantitative reverse transcription PCR (qRT-PCR), as reported by Tan et al. [46]. This finding thus indicates the potential for using HOTAIR as a biomarker to diagnose GBM. Further supporting this potential is the finding by Shen et al. [47] that high levels of HOTAIR were associated with a 2.04-fold decrease in survival.
Another mechanism by which lncRNAs can indirectly alter gene expression is by acting as miRNA sponges or as competitive endogenous RNAs (ceRNA) by binding to miRNAs [48]. A single miRNA molecule can directly affect the expression of hundreds of genes by targeting hundreds of mRNAs so miRNAs can repress protein synthesis [49, 50]. As shown in Fig. 4, lncRNA binding to miRNAs can de-repress the expression of all target genes that were acted upon tumor suppressor genes or oncogenes [51].
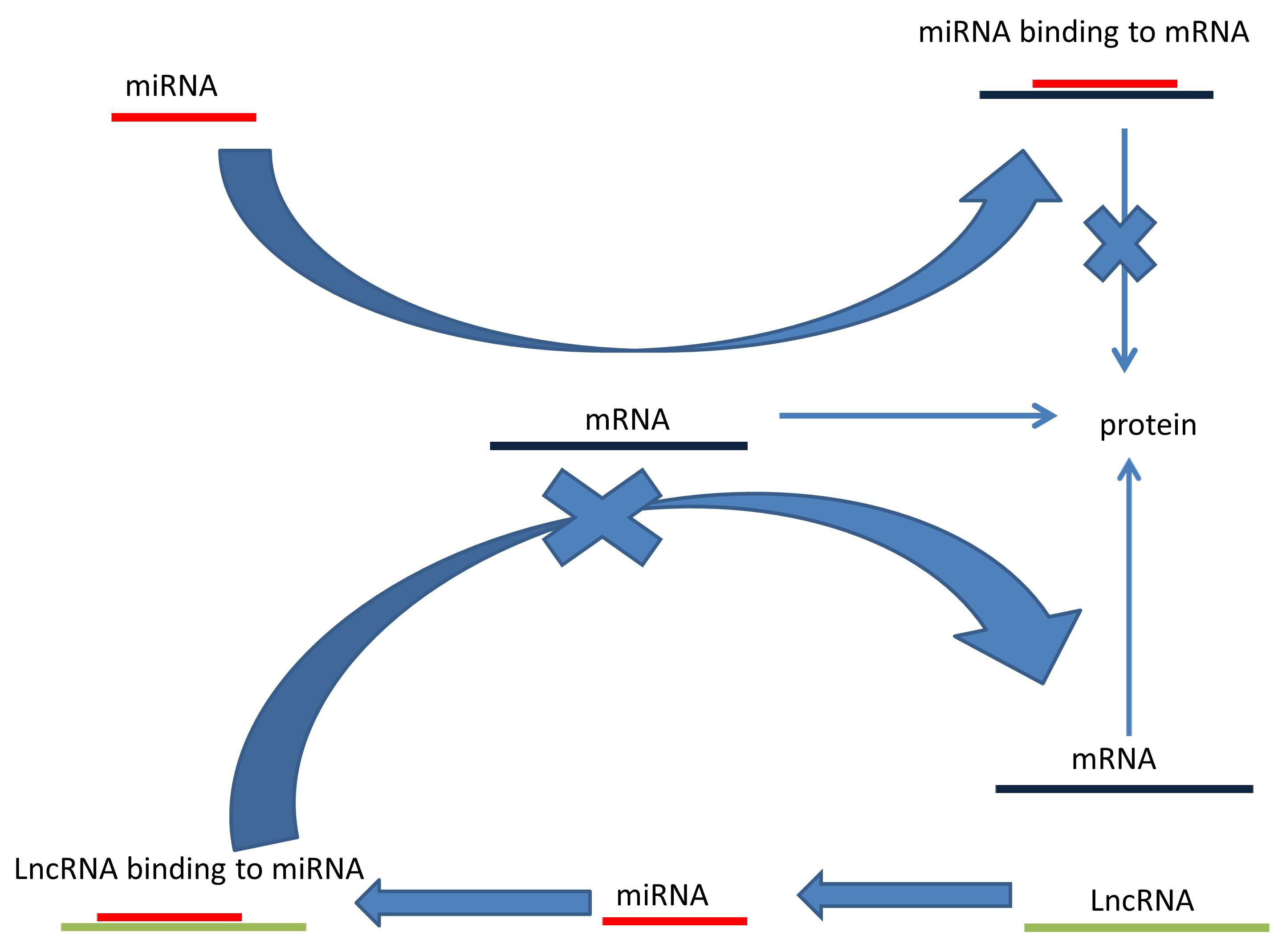 Fig. 4.
Fig. 4.Long noncoding RNA acts as a competitive endogenous RNA. LncRNAs (1) bind to miRNAs. This prevents miRNAs (2) from binding to mRNA (3) thus allowing mRNA translation (4).
LncRNA NEAT1 acts as a ceRNA, as discussed above. LncRNA NEAT1 binds to many miRNAs that have a suppressive effect on glioma cells growth and proliferation, including miR-139-5p, miR-132, miR-128-3p, miR-98-5p, and miR-107 [52, 53, 54, 55, 56, 57]. Subsequently, this leads to tumor cell proliferation and metastasis. Using qRT-PCR to measure NEAT1 expression in cultured tumor tissues from patients with gliomas and non-tumorous tissues, He et al. [58] found significantly higher NEAT 1 expression in glioma tissues. They also reported larger tumor sizes, higher WHO grade, and recurrence were strongly correlated with high levels of NEAT1 upregulation. Further, unfavorable prognoses were associated with patients receiving postoperative chemoradiotherapy with high NEAT1 expression [58]. This suggests a role for lncRNA NEAT1 expression in high-grade gliomas cell proliferation and metastasis, and hence the potential for novel treatments by targeting lncRNA NEAT1. Clinically, Wu et al. [59] found high expression of lncRNA NEAT1 in the plasma of GBM patients.
LncRNA GAS5 gene is located on chromosome 1q12.1 [60]. LncRNA GAS5 is considered an antioncogenic lncRNA and is downregulated in many tumors including ovarian and breast cancers, non-small cell lung cancers, large B‑cell lymphoma, and gastric and laryngeal cancers [60, 61, 62, 63, 64, 65]. In contrast, lncRNA GAS5 acts as an oncogenic lncRNA and is overexpressed in other tumors such as hepatocellular carcinoma and cholangiocarcinoma [66, 67]. LncRNA GAS5 suppresses glioma proliferation and growth by acting as a ceRNA through targeting multiple miRNAs including miRNA-222, miRNA-10b, and miR-196a-5p [68, 69, 70].
In GBM tissues, Toraih et al. [71] found that lncRNA GAS5 was significantly under-expressed. Clinically, Shen et al. [47] found that high serum levels of GAS5 were associated with a 56% decrease in the likelihood of death in GBM patients. This indicates the possibility of using LncRNA GAS5 as a prognostic and diagnostic biomarker for GBM patients.
LncRNA SAMMSON gene is located on chromosome 3p13–3p14 and is known to act as an oncogene in different tumors, such as melanoma, liver tumor, gastric cancer, papillary thyroid carcinoma, and oral squamous cell carcinoma [72, 73, 74, 75, 76]. In GBM tissues and cells, Ni et al. [77] detected high expression of lncRNA SAMMSOM. SAMMSON acts as a ceRNA through binding to miRNA-622. Through this mechanism, it increases GBM cell proliferation [78]. Clinically, Xie et al. [78] found that lncRNA SAMMSON was overexpressed in GBM patient plasma comparing to normal controls.
We recognize that many lncRNAs have been examined in GBM biopsy specimens and
cell cultures that show a significant correlation between tumor progression and
patient prognosis. However, clinical evidence for these lncRNAs as serum
biomarkers is lacking. For instance, HOXA11-AS is an example of a potentially
promising lncRNA biomarker. HOXA11-AS is another HOX family member related to
GBM. Like other HOX genes that cause tumor formation when
inappropriately expressed or dysregulated. HOXA11-AS expression plays a role in
tumorigeneses including non-small cell lung cancers, osteosarcoma, and
hepatocellular carcinoma [79, 80, 81]. Li et al. [82] found that miR-140-5p
has a role in inhibition proliferation and induction apoptosis of Wilms’ tumor.
Others have also reported that glioma cell proliferation and invasion are
inhibited effectively by overexpressed miR-140-5p levels [83]. This indicates a
protective role for miR-140-5p against tumorigenesis. Cui et al. [84]
suggested that HOXA11-AS attaches to miR-140-5p and prevents its protective role.
Subsequently, this leads to glioma cells proliferation and invasion. Wang
et al. [85] found that HOXA11-AS is overexpressed in GBM tissues and
high levels of HOXA11-AS are inversely associated with overall survival of GBM
patients, thus suggesting that HOXA11-AS could be a prognostic biomarker.
Bountali et al. [86] showed that the long-term survival of GBM cells is
decreased when lncRNA MIAT is downregulated. This shows a role for lncRNA MIAT in
GBM growth and progression. Through binding to miRNA-182, LncRNA UCA1 promotes
GBM cells proliferation and invasion by de-repressing expression of PFKFB2 which
is responsible of increasing glycolysis in GBM cells [87]. Another lncRNA that
plays an important role in GBM growth through increasing glycolysis in GBM cells
is lncRNA SNHG9. LncRNA SNHG9 increases Wnt2 expression by sponging
miRNA-199a-5p. De-repressed Wnt2 increases aerobic glycolysis in GBM cells which
thus increases GBM cell growth and proliferation capacity [88]. By producing
miR-22-3p and miR-22-5p, lncRNA MIR22HG promotes GBM cell progression and
self-renewal. Han et al. [89] found that MIR22HG silencing leads to
loses in miR-22-3p and miR-22-5p, which subsequently leads to the inactivation of
Wnt/
| LncRNA | Expression in GBM | Role | Targets | Measured in GBM patient serum | References |
| HOTAIR | Upregulated | Oncogenic | Interacts with the PRC2 complex | Yes | [44, 45, 46, 92] |
| NEAT1 | Upregulated | Oncogenic | MiR-139-5p, miR-132, miR-128-3p, miR-98-5p, and miR-107 | Yes | [52, 53, 54, 55, 56, 57, 58] |
| GAS5 | Downregulated | Anti-oncogenic | MiRNA-222, miRNA-10b, and miR-196a-5p | Yes | [68, 69, 70] |
| SAMMSON | Upregulated | Oncogenic | MiRNA-622 | Yes | [78] |
| HOXA11-AS | Upregulated | Oncogenic | MicroRNA-140-5p | No | [83, 84] |
| MIAT | Upregulated | Oncogenic | Acting as ceRNA | No | [86] |
| UCA1 | Upregulated | Oncogenic | MiRNA-182 | No | [87] |
| SNHG9 | Upregulated | Oncogenic | MiRNA-199a-5p | No | [88] |
| MIR22HG | Upregulated | Oncogenic | MiR-22-3p and miR-22-5p | No | [89] |
| SNHG4 | Upregulated | Oncogenic | MiR-138 | No | [90] |
| LINC01579 | Upregulated | Oncogenic | MiR-139-5p | No | [91] |
| SBF2-AS1 | Upregulated | Oncogenic | MiR-151a-3p | Yes | [33] |
| MALAT 1 | Upregulated | Oncogenic | MiRNA‐101 and miRNA 203 | Yes | [93, 94] |
| TP73-AS1 | Upregulated | Oncogenic | Regulation of the expression of metabolism- related genes and ALDH1A1 | No | [95] |
| OIP5-AS1 | Upregulated | Oncogenic | MiRNA-129-5p | No | [96] |
| CRNDE | Upregulated | Oncogenic | PI3K/Akt/mTOR pathway and ABCG2 expression | No | [97] |
| LINC00511 | Upregulated | Oncogenic | MiRNA-126-5p | No | [98] |
| TUSC7 | Downregulated | Anti-oncogenic | MiRNA-10a | No | [99] |
| AC003092.1 | Downregulated | Anti-oncogenic | MiRNA-195 | No | [100] |
A major barrier to chemotherapy efficacy with GBM is that tumors become metastatic and/or chemoresistant in a large portion of patients following chemotherapy [101, 102]. The absence of effective therapies for GBM makes it crucial to understand the pathophysiology of chemoresistance. Of interest, there are specific lncRNA and miRNA associations with GBM chemoresistance. The lncRNA mechanism for chemoresistance development is largely unknown, hence its study could provide new therapeutic targets that can reverse GBM chemoresistance. Additional clinical research could also confirm the association between high levels of identified lncRNAs and GBM chemoresistance so these lncRNAs can also be examined as potential biomarkers for GBM chemoresistance, thus protecting those patients from ineffective and thus unnecessary and harmful chemotherapies.
By binding to the HOTAIR promoter, Bromodomain Containing 4 (BRD4) protein controls HOTAIR levels and increases its expression. Pastori et al. [92] found that by using small molecule BET inhibitors, which are promising epigenetic modulators currently used in clinical trials, BRD4 activity can be inhibited to decrease BRD4 binding at the HOTAIR promoter. HOTAIR levels have been shown to decrease both in vitro and in vivo [92]. This inhibition of HOTAIR transcription is crucial to induce cell cycle arrest in GBM cells. Pastori et al. [92] suggested that HOTAIR can be used as a biomarker for the responsiveness of GBM cells to BET inhibitors. More research to examine the correlation between serum HOTAIR and GBM resistance to BET inhibitors is thus warranted.
TMZ is an alkylating agent that works by attaching an alkyl group (CnH2n+1) to the purine bases of DNA [103]. TMZ is metabolized finally to the electrophilic alkylating methyldiazonium cation that acts as a donor of a methyl group on the O6 position of guanine. Although the majority of DNA-methyl adducts are on the N7 of guanine and the N3 of adenine, the O6 position methylations accounts for about 5% of DNA adducts and are primarily responsible for the cytotoxic effects of TMZ [104]. The subsequent methyl group transferring to the O6 position of guanine forms O6-methylguanine lesion leading to DNA double-strand breaks. As a result, apoptosis and/or autophagy occur that cause cell death [105, 106]. TMZ was approved by the U.S. Food and Drug Administration for the treatment of glioblastoma [107]. There are many mechanisms of TMZ resistance for GBM [108], but here we will focus on lncRNAs role in developing TMZ resistance, their potential use as a biomarker for TMZ resistance, and their use as a novel therapeutic agent.
In contrast to bacteria that develop antibiotic resistance by acquiring protein-coding DNA pieces from other bacteria, non-coding genomic pieces make GBM resistant to TMZ. Of interest, by transferring those lncRNAs from chemoresistance GBM tissue or culture to a chemo-sensitive one, the chemo-sensitive tissue becomes chemoresistant [33].
The lncRNA SBF2-AS1 gene is located on chromosome 11p15.1. The proliferation or invasion of many tumor types are associated with altered lncRNA SBF2-AS1 serum levels; Diffuse large B-cell lymphoma, colorectal cancer, hepatocellular carcinoma, serous ovarian carcinoma, clear cell renal cell carcinoma, esophageal squamous cell carcinoma, and lung cancer are associated with increased SBF2-AS1 serum levels [109, 110, 111, 112, 113, 114, 115], while laryngeal squamous cell carcinoma [116] is associated with decreased levels. LncRNA SBF2-AS1 can indirectly alter gene expression by acting as a ceRNA. LncRNA SBF2-AS1 binds to protective miRNAs, thus inducing tumor proliferation and invasion [109, 110, 111]. In contrast, lncRNA SBF2-AS1 acts as an anti-oncogene in laryngeal squamous cell carcinoma by targeting a different miRNA associated with laryngeal cancer invasion and migration [116].
In regard to GBM, lncRNA SBF2-AS1 binds to miR-151a-3p which promotes TMZ sensitivity in GBM cells by targeting the XRCC4 gene [33]. Zhang et al. [33] found that overexpression of long non-coding RNA SBF2-AS1 occurs in TMZ-resistant GBM tissues. This overexpression changes the tumor microenvironment and promotes TMZ resistance. Of interest, in lncRNA SBF2-AS1-depleted Rec GBM cells, the cells became more sensitive to TMZ. Thus, exosomal lncSBF2-AS1 in human serum may serve as a possible diagnostic biomarker for TMZ-resistant GBM [33]. This assumption has been confirmed clinically; Zhang et al. [33] found a poor response to TMZ in GBM patients having high levels of lncSBF2-AS1 in serum exosomes.
LncRNA MALAT1, also called nuclear-enriched abundant transcript 2, is overexpressed in tumor tissues and correlates with prognosis for many tumors, including non-small cell lung cancer, hepatocellular carcinoma, bladder carcinoma, breast cancer, and prostate cancer [117, 118, 119, 120, 121, 122]. With respect to gliomas, lncRNA MALAT1 promotes growth and progression by acting as a ceRNA sponge for miRNA-613 and miRNA-124 [123, 124].
With respect to chemoresistance, lncRNA MALAT1 has been found to have a major role in developing chemoresistance for osteosarcomas, chronic myeloid leukemia, ovarian cancer, and hepatocellular carcinoma [125, 126, 127, 128]. MALAT1 promoted the TMZ resistance through suppressing miRNA-101 and miRNA 203 signaling pathways in GBM cells [93, 94]. Clinically, Chen et al. [94] found that high serum MALAT1 levels are correlated with TMZ chemoresistance. This indicates the possibility for lncRNA MALAT1 as a biomarker for GBM chemoresistance.
Mazor et al. [95] found that lncRNA TP73-AS1 promotes TMZ resistance in GBM cancer stem cells and that there is a significant overexpression of lncRNA TP73-AS1 in GBM biopsies. This suggests a potential for this lncRNA to be used as a biomarker to predict TMZ chemoresistance. LncRNA OIP5-AS1 acts as a ceRNA by binding to miRNA-129-5p. Since miRNA-129-5p targets and downregulates IGF2BP2, lncRNA OIP5-AS1 can cause TMZ sensitivity if it is inhibited [96]. Thus, targeting this lncRNA could provide a novel therapeutic pathway for treating GBM patients with TMZ resistance. Another lncRNA that can cause TMZ sensitivity when it is knocked down is lncRNA CRNDE [97]. In GBM cells, LncRNA LINC00511 can promote TMZ chemoresistance by sponging miRNA-126-5p [98]. In contrast to those lncRNAs, there are other lncRNAs which have a protective role and induce chemosensitivity. For instance, lncRNA TUSC7 targets miRNA-10a in GBM cells which inhibits TMZ resistance [99]. LncRNA AC003092.1 is another lncRNA that promotes TMZ chemosensitivity in GBM cells which it does through binding to miRNA-195 [100].
In contrast to other invasive modalities of GBM diagnosis, circulating genetic biomarkers are easier, faster, and less costly. These represent an interesting potential method for diagnosis, treatment, prognosis, and avoiding chemoresistance when chemotherapy is unneeded. LncRNAs have shown a potential strong correlation with GBM diagnosis, prognosis, and grading. Furthermore, this suggests other mechanisms for chemoresistance that require further elucidation. There is thus great hope that these biomarkers can be used clinically in the future to decrease the need for invasive diagnostic procedures, to help patients with chemoresistance in their diagnosis and treatment, and to target those lncRNA that cause chemoresistance. LncRNAs have been proven to have a significant value in diagnostic and prognostic purposes. Robust experimental and clinical research should be utilized to determine the most sensitive and highly specific markers for correlating between high plasma levels with GBM type and virulence and to examine the possibility of using lncRNAs as easy, rapid, and noninvasive methods for GBM diagnosis and treatment.
AME performed the research and wrote the manuscript. SLG and AK provided reviewing, editing. MKB provided help, advice, and editing. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Not applicable.
Not applicable.
This research received no external funding.
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.




