1 Department of Stomatology, Sichuan Provincial People's Hospital, School of Medicine, University of Electronic Science and Technology of China, 611731 Chengdu, Sichuan, China
2 Department of Prosthodontics, The Affiliated Stomatology Hospital, Southwest Medical University, 646000 Luzhou, Sichuan, China
3 Oral & Maxillofacial Reconstruction and Regeneration of Luzhou Key Laboratory, 646000 Luzhou, Sichuan, China
4 Department of Periodontics & Oral Mucosal Diseases, The Affiliated Stomatology Hospital, Southwest Medical University, 646000 Luzhou, Sichuan, China
Abstract
Osteoradionecrosis of the jaw (ORNJ) is a common complication following radiotherapy for head and neck cancer. Thus, this study aimed to explore the effects of active components in Lycium barbarum on ORNJ through network pharmacology and to conduct experimental verification to identify potential therapeutic targets.
The main active ingredients in Lycium barbarum (Gouqi), a traditional Chinese herbal medicine, was used in this study. After identifying ferroptosis-related genes associated with ORNJ, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses and constructed an interaction network. A molecular interaction force analysis was then performed on eight binding conformations to identify the optimal conformation with the lowest binding free energy. We established a model of ORNJ in Sprague-Dawley (SD) rats and administered oral Lycium barbarum glycopeptide (LbGP) to the experimental group. Moreover, reverse transcription quantitative polymerase chain reaction (RT-qPCR), enzyme-linked immunosorbent assay (ELISA), and immunohistochemical staining techniques were then employed to detect the mRNA and protein levels of various relevant cytokines.
Based on network pharmacology predictions, this study identified three potential active components in Lycium barbarum, namely β-sitosterol, glycitein, and quercetin. The possible effects of these components in the treatment of ORNJ were analyzed. Seven hub genes related to ferroptosis and ORNJ were identified, and LbGP was selected for in vivo verification. The expression levels of TP53, EGFR, IL-6, and TNF were significantly altered in the LbGP group compared to the untreated group, as revealed by the qPCR, ELISA, and immunohistochemistry assays.
The use of Lycium barbarum extract may exert therapeutic effects on mandible injury following ORNJ by regulating the expression of TP53, EGFR, IL-6, and TNF.
Keywords
- osteoradionecrosis
- lycium
- network pharmacology
- ferroptosis
Osteoradionecrosis of the jaw (ORNJ) is a critical complication related to the treatment of head and neck cancer, with an incidence ranging from 5.4% to 13% [1]. Indeed, ORNJ is considered to be one of the particularly severe complications among the potential adverse reactions of extensive high-dose radiation therapy in the oral cavity, maxilla, mandible, and salivary glands. Moreover, the primary feature of ORNJ is a non-healing, exposed bone area that can occur without affecting the mucosa or skin [2]. The etiology and pathogenesis of ORNJ remain uncertain and are associated with factors such as radiation trauma and infection, bone injury, and the three-low theory [3]. Presently, no universally accepted treatment protocol exists for ORNJ, with current treatment options ranging from conservative approaches (such as antibiotics, antifibrotic agents, and antioxidants) to radical surgical interventions and/or hyperbaric oxygen therapy (HBO). The selection of treatment is based on the severity of clinical presentations.
Lycium barbarum (Gouqi) is a traditional Chinese medicinal herb containing various chemical components, including polysaccharides, flavonoids, anthocyanins, and alkaloids. Notably, Lycium barbarum has multiple pharmacological effects, including immune regulation, anti-aging, blood sugar and lipid level regulation, blood pressure reduction, anti-tumor properties, and the potential to reduce inflammatory responses.
Notably, ferroptosis, a form of regulated cell death distinct from apoptosis, necrosis, and autophagy, has recently emerged as a critical mechanism in various diseases [4]. Growing evidence highlights the role of ferroptosis in mediating cellular damage and inflammatory responses, suggesting its potential relevance in the progression of ORNJ. Moreover, primitive radioresistant cancer cells can become radiosensitive through the inhibition of ferroptosis using Solute Larrier Camily 7 member 11 (SLC7A11) or Glutathione Peroxidase 4 (GPX4) inhibitors [4].
Lycium barbarum polysaccharide (LBP) is a key biologically active component of Lycium barbarum, functioning as an antioxidant and preventing cell death due to oxidative stress [5]. Lycium barbarum glycopeptide (LbGP) is derived and purified from LBP and consists of five glycopeptides with strong immunomodulatory and anti-aging properties. Our previous study demonstrated that LbGP regulates oxidative stress and ferroptosis through the Nrf2 pathway and ameliorates epithelial injury induced by ionizing radiation [6]. Nonetheless, the precise molecules in Lycium barbarum that are associated with ferroptosis in ORNJ remain unknown. Therefore, this study aimed to investigate the interplay between disease targets, active compounds of Lycium barbarum, and the relevant pathways by leveraging network pharmacology. Correlations between this herbal medicine and ferroptosis-linked radiogenic mandibular osteomyelitis should enhance our understanding of the mechanism of action through which Lycium barbarum functions at the molecular level.
The Traditional Chinese Medicine Systems Pharmacology Database and analysis platform (TCMSP) (https://tcmsp-e.com/tcmsp.php) [7] is based on the traditional Chinese Medicine Systems pharmacology framework. The TCMSP includes 837 related disorders, 29,384 constituents, 3311 targets, and 499 varieties of Chinese therapeutic plants from the Chinese Pharmacopeia. The TCMSP also contains 12 crucial Absorption, Distribution, Metabolism, Excretion (ADME)-associated attributes of drugs, including drug similarity, human oral bioavailability (OB), Caco-2 permeability, half-life, blood–brain barrier integrity, and compliance with Lipinski’s rule-of-five for drug screening and assessment. Compositional information for Lycium barbarum was sourced from the TCMSP database.
ADME [8] refers to the Absorption, Distribution, Metabolism, Excretion, and Toxicity of drugs. Thus, the ADME content is applied to contemporary drug design and screening in pharmacokinetics research. This research is based on the characteristics of the ADME TCMSP databases and provides relevant data with a selected oral exploitation degree OB of
We used a two-step procedure to predict and screen the target proteins of effective Chinese medicine ingredients. First, the TCMSP database was utilized to predict protein targets for the active components of Lycium barbarum. Subsequently, the predicted targets in the TCMSP database were transformed into gene names using UniProtKB (http://www.uniprot.org) [9].
The GeneCards database (https://www.genecards.org/) offers extensive information on genes for individuals [10]. Therefore, we identified a total of 445 ferroptosis-related genes (FRGs) in the GeneCards database using the keyword “ferroptosis” and a relevance score of
The search term “osteoradionecrosis of the jaw” was used to examine the GeneCards database. A total of 35 genes linked to ORNJ targets, with a relevance score
The intersection of targeted proteins for screened Chinese herbal ingredients and screening genes associated with ORNJ (ORNJ-related genes, or ORNJRGs) yielded Lycium barbarum.
The screened genes relating to ORNJ and ferroptosis were intersected to identify any FRGs in Lycium barbarum therapy. These were designated as hub genes. Individual proteins that interact with one another form protein–protein interaction networks. The STRING database (https://cn.string-db.org) [13] can be employed to search for connections between anticipated and existing proteins. Using this database, the biological species was set to human with a confidence level of
Gene Ontology (GO) analysis is a general method for large-scale functional enrichment, including both molecular functions (MFs) and biological processes (BPs). The Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.kegg.jp) is an extensive database that contains information on genomes, illnesses, biological pathways, and drugs [14]. The R package clusterProfiler [15] was employed to perform GO annotation analysis of hub genes, with entry screening standards of p.adjust
The ENCORI database (https://rnasysu.com/encori/) [16] is version 3.0 of the starBase database. Interactions in ENCORI, including RNA-binding protein (RBP)–mRNA, RNA–RNA, ncRNA–RNA, RBP–ncRNA, miRNA–mRNA, and miRNA–ncRNA, are formulated based on CLIP-seq and degradome sequencing data specific to plants. The interplay between hub genes and miRNAs was predicted using the ENCORI database. Subsequently, the mRNA–miRNA data were screened. Three or more databases supported the establishment of this predicted mRNA–miRNA interplay network.
The CHIPBase database [17] (version 3.0) (https://rna.sysu.edu.cn/chipbase/) recognizes hundreds of thousands of binding motifs and binding sites from DNA-binding protein ChIP-seq information, while predicting transcriptional regulatory relationships between millions of genes and transcription factors (TFs). Meanwhile, TFs that bind to hub genes were identified using the CHIPBase database. Subsequently, to build an mRNA–TF interplay network, we filtered mRNA–TF data supported by four or more databases.
Additionally, the Comparative Toxicogenomics Database (CTD) [18] (http://ctdbase.org/) was used to predict underlying drugs or small-molecule compounds that interact with hub genes. Subsequently, mRNA–drug data supported by four or more databases were filtered to build an mRNA–drug interplay network.
The RCSB PDB database (https://www.rcsb.org) was used to extract crystal structures for the hub genes in pdb format [19]. Core small-molecule structures were obtained in mol2 format from the TCMSP database. For crystal structures downloaded from the RCSB PDB database, we used PyMOL (https://pymol.org/2/) to remove ligands, select chains, and extract them into PDB format. Protein molecule active docking sites were predicted using Discovery Studio 2.5.5 (Dassault Systèmes BIOVIA, San Diego, CA, USA). For the extracted protein structures mentioned above, the AutoDockTool-1.5.7 program (The Scripps Research Institute, La Jolla, CA, USA) [20] was utilized to eliminate water molecules, supplement hydrogen atoms, compute charges, and export files in both pdb and pdbqt formats. For the core small-molecule structures in mol2 format, we used the AutoDockTool-1.5.7 program to add hydrogen atoms, calculate charges, and determine rotatable bonds for the ligands. These formats were then exported as pdbqt format files.
Autodock Vina (1.2.0, The Scripps Research Institute, La Jolla, CA, USA) [21] was utilized for docking proteins and small molecules, generating nine complex conformations for each protein–molecule docking. The docking score Affinity in AutoDock Vina indicates the strength of binding: Affinity
Sprague-Dawley (SD) rats aged 6–8 weeks were randomly divided into the normal control (NC), ORNJ, and ORNJ + LbGP groups. To establish the ORNJ animal model, rats from the ORNJ and ORNJ + LbGP groups were irradiated once daily at doses of 7 Gy per fraction for a total of five fractions [23]. The NC rats did not receive radiation treatment. The ORNJ + LbGP rats were also administered oral LbGP at a dose of 400 mg/kg/d, whereas the NC and ORNJ rats were administered the same concentration of normal saline. After 3 weeks of radiotherapy, the first molar of the mandible was extracted under isoflurane local anesthesia. The rats were euthanized with excessive anesthesia via inhalation using isoflurane (2% concentration) delivered at 0.41 mL/min with a fresh gas flow rate of 4 L/min. The mandibular tissues were extracted, and jaw bone marrow mesenchymal stem cells (JBMMSCs) were isolated and cultured for 3–5 generations for the follow-up experiments. The identity of these cells was confirmed through morphology, surface marker expression, and differentiation ability. All the cultures were subjected to routine tests, and mycoplasma contamination was found to be negative. RT-qPCR was performed to evaluate the expression of the hub genes in the JBMMSCs. First, cDNA synthesis was performed under the following conditions: reverse transcription at 42 °C for 60 minutes, followed by enzyme inactivation at 70 °C for 5 minutes. The used primers are listed in Supplementary Table 3. The RT-qPCR reaction mixture consisted of 10 µL of 2
Statistical analysis was performed using R software (v4.3.0, R Foundation for Statistical Computing, Vienna, Austria) for bioinformatics and GraphPad Prism (v9.0.0.121, GraphPad Software (Dotmatics) Boston, MA, USA) for experimental data. Enrichment analyses (GO and KEGG) were conducted using the clusterProfiler package 4.2 (Guangzhou Medical University, Guangzhou, Guangdong, China), with significance defined as p.adjust
Data from at least three independent experiments were expressed as the mean
The flowchart for this study is shown in Fig. 1.
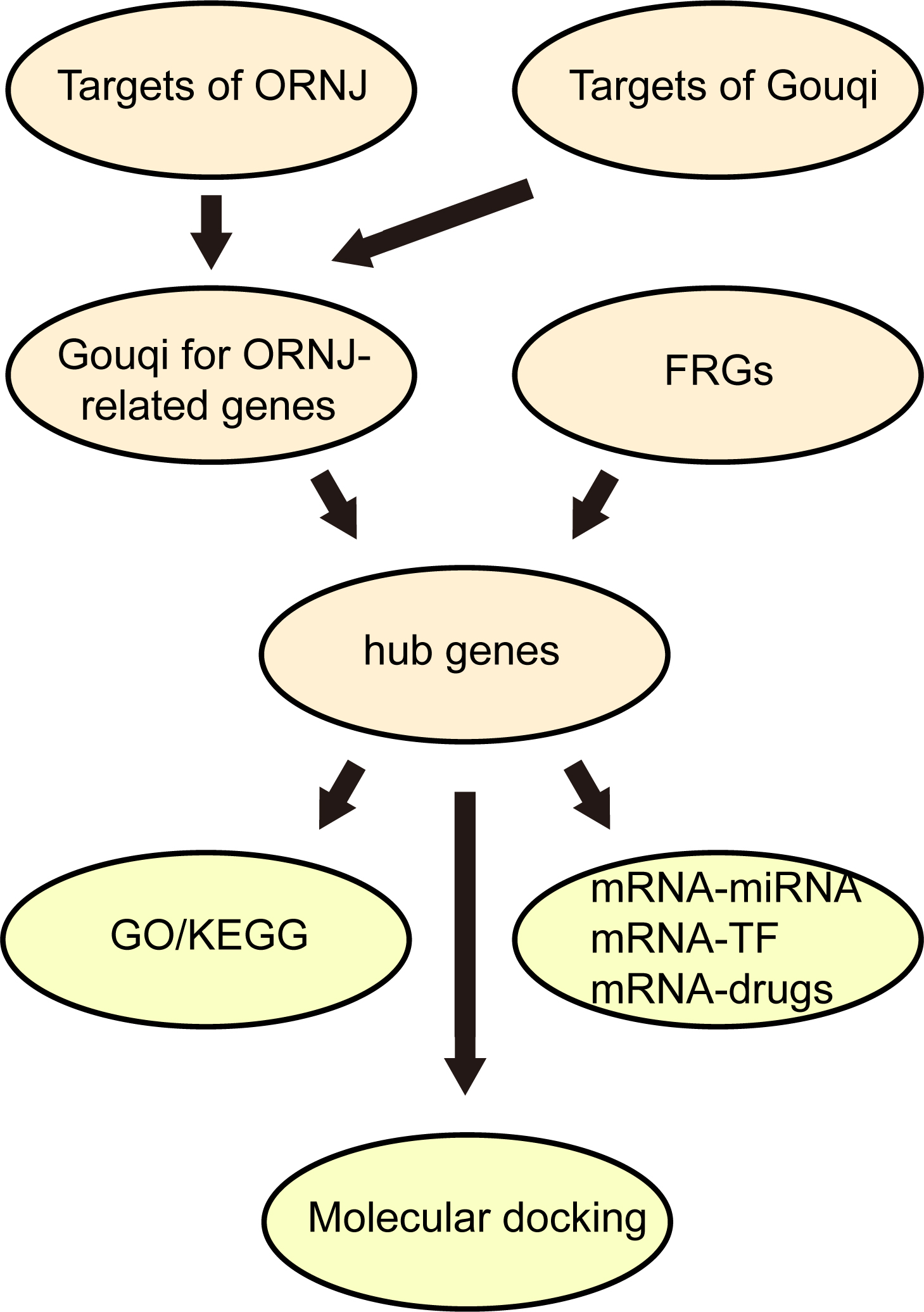 Fig. 1.
Fig. 1. Technology roadmap. ORNJ, osteoradionecrosis of the jaw; FRGs, ferroptosis-related genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; TF, transcription factors.
A total of 188 chemical components from Lycium barbarum were extracted from the TCMSP database. Among these, 45 active compounds were selected through screening for values of OB
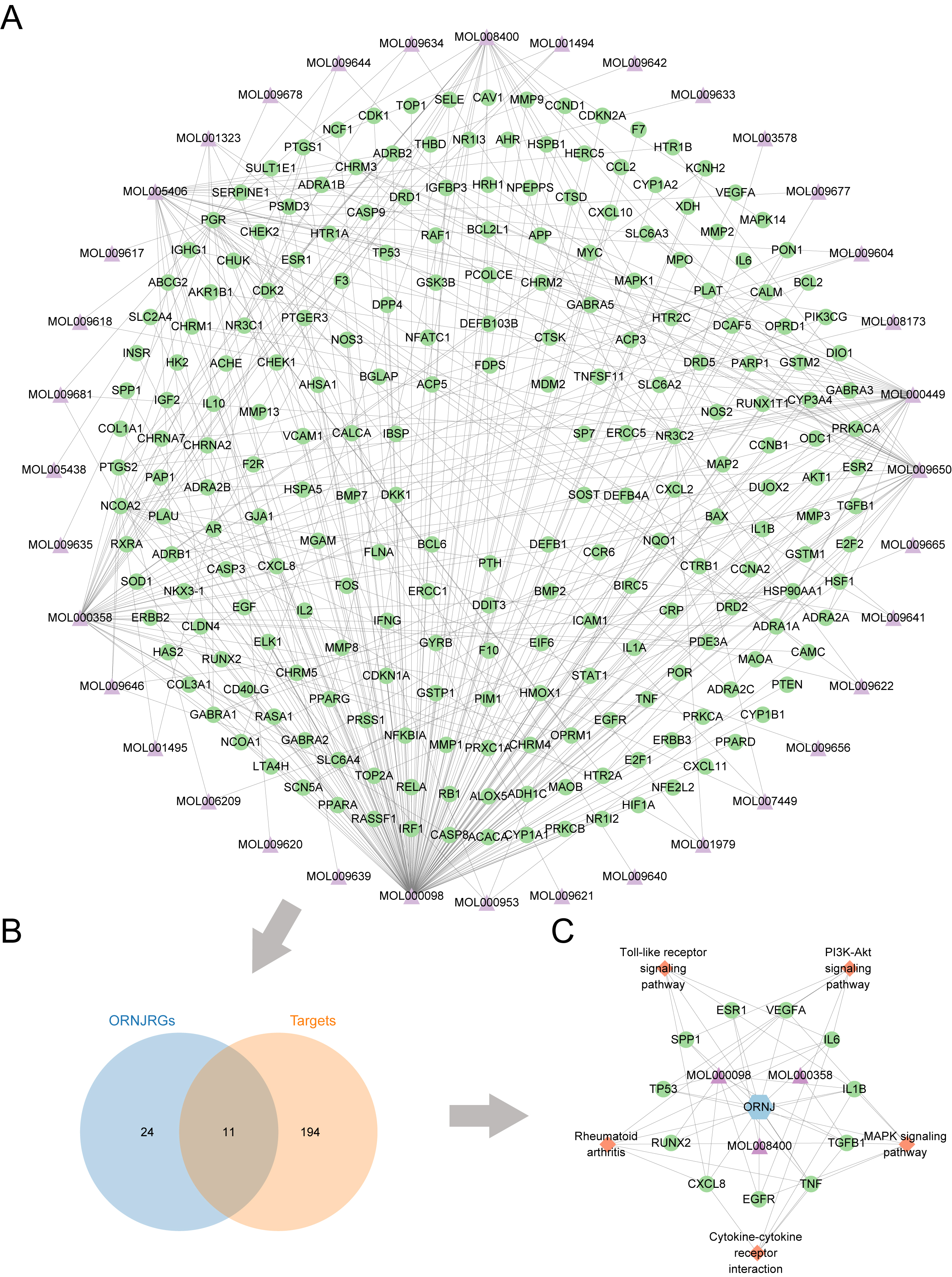 Fig. 2.
Fig. 2. Target screening for Lycium barbarum and ORNJ. (A) Network diagram of predicted targets of active chemical constituents in Lycium barbarum. (B) Venn diagram of the predicted targets of active components in Lycium barbarum and ORNJ-related genes. (C) Active chemical components, ORNJ-related genes, and the KEGG pathway network map for Lycium barbarum. ORNJ, osteoradionecrosis of the jaw; ORNJRGs, osteoradionecrosis of the jaw related genes; KEGG, Kyoto Encyclopedia of Genes and Genomes. In panels (A) and (C), the green circles represent mRNAs, purple triangles are chemical components, blue hexagons are ORNJ, and orange quadrangles are the KEGG pathways.
| MOL_ID | Compound | OB (%) | DL |
| MOL001323 | Sitosterol alpha1 | 43.28127042 | 0.78354 |
| MOL003578 | Cycloartenol | 38.68565906 | 0.78093 |
| MOL001494 | Mandenol | 41.99620045 | 0.19321 |
| MOL001495 | Ethyl linolenate | 46.10096327 | 0.19716 |
| MOL001979 | LAN | 42.11918897 | 0.74787 |
| MOL000449 | Stigmasterol | 43.82985158 | 0.75665 |
| MOL000358 | Beta-sitosterol | 36.91390583 | 0.75123 |
| MOL005406 | Atropine | 45.97058178 | 0.19328 |
| MOL005438 | Campesterol | 37.57681789 | 0.71488 |
| MOL006209 | Cyanin | 47.42092269 | 0.75918 |
| MOL007449 | 24-methylidenelophenol | 44.19264545 | 0.75330 |
| MOL008173 | Daucosterol_qt | 36.91390583 | 0.75316 |
| MOL008400 | Glycitein | 50.47891366 | 0.23826 |
| MOL010234 | Delta-carotene | 31.80094312 | 0.54639 |
| MOL000953 | CLR | 37.87389754 | 0.67677 |
| MOL009604 | 14b-pregnane | 34.77923299 | 0.33723 |
| MOL009612 | Alpha (r)-4-methyl-24-ethylcholesta-7,25-dien-3beta-ylacetate | 46.35749925 | 0.83980 |
| MOL009615 | 24-methylenecycloartan-3beta,21-diol | 37.31728162 | 0.79751 |
| MOL009617 | 24-ethylcholest-22-enol | 37.09454086 | 0.75110 |
| MOL009618 | 24-ethylcholesta- 5,22-dienol | 43.82985158 | 0.75636 |
| MOL009620 | 24-methyl-31-norlanost-9(11)-enol | 37.99968530 | 0.75092 |
| MOL009621 | 24-methylenelanost-8-enol | 42.36819868 | 0.76769 |
| MOL009622 | Fucosterol | 43.77639556 | 0.75668 |
| MOL009631 | 31-norcyclolaudenol | 38.68209614 | 0.81391 |
| MOL009633 | 31-norlanost-9(11)-enol | 38.35394137 | 0.72490 |
| MOL009634 | 31-norlanosterol | 42.20462055 | 0.73012 |
| MOL009635 | 4,24-methyllophenol | 37.83467433 | 0.74999 |
| MOL009639 | Lophenol | 38.12940252 | 0.71400 |
| MOL009640 | 4alpha,14 alpha,24-trimethylcholesta-8,24-dienol | 38.90988973 | 0.75772 |
| MOL009641 | 4alpha,24-dimethylcholesta-7,24-dienol | 42.65304098 | 0.75297 |
| MOL009642 | Alpha4-methyl-24-ethylcholesta-7,24-dienol | 42.29509453 | 0.78304 |
| MOL009644 | 6-fluoroindole-7-dehydrocholesterol | 43.72602513 | 0.72224 |
| MOL009646 | 7-O-methylluteolin-6-C-beta-glucoside_qt | 40.77368843 | 0.30497 |
| MOL009650 | Atropine | 42.15897078 | 0.19299 |
| MOL009651 | Cryptoxanthin monoepoxide | 46.95371937 | 0.56103 |
| MOL009653 | Cycloeucalenol | 39.72647216 | 0.79446 |
| MOL009656 | (E,E)-1-ethyl octadeca-3,13-dienoate | 41.99620045 | 0.19364 |
| MOL009660 | Methyl (1R,4aS,7R,7aS)-4a,7-dihydroxy-7-methyl-1-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy-1,5,6,7a-tetrahydrocyclopenta[d]pyran-4-carboxylate | 39.42847682 | 0.46558 |
| MOL009662 | Lantadene A | 38.67942417 | 0.57405 |
| MOL009664 | Physalin A | 91.70647491 | 0.27207 |
| MOL009665 | Physcion-8-O-beta-D-gentiobioside | 43.90358656 | 0.62426 |
| MOL009677 | lanost-8-en-3beta-ol | 34.22630373 | 0.74036 |
| MOL009678 | Lanost-8-enol | 34.22630373 | 0.74167 |
| MOL009681 | Obtusifoliol | 42.55200222 | 0.75650 |
| MOL000098 | Quercetin | 46.43334812 | 0.27525 |
ADME, Absorption, Distribution, Metabolism, Excretion; OB, oral bioavailability; DL, drug-likeness; LAN, Number of Liked Approved drug Analoques; CLR, Clearance.
The intersection of 205 potential target points for Lycium barbarum components and 35 disease target points was plotted in a Venn diagram (Fig. 2B). This yielded 11 potential Lycium barbarum, in traditional Chinese medicine formulation, treatment targets for ORNJ: RUNX2 (Runt-Related Transcription Factor 2), TGFB1 (Transforming Growth Factor Beta 1), TP53 (Tumor Protein p53), TNF (Tumor Necrosis Factor), IL-6 (Interleukin-6), VEGFA (Vascular Endothelial Growth Factor A), ESR1 (Estrogen Receptor 1), SPP1 (Secreted Phosphoprotein 1), IL-1
Seven intersection genes (TP53, EGFR, TGFB1, IL-6, IL1B, TNF, ESR1) were obtained from the intersection of FRGs and Lycium barbarum treatment ORNJRGs. These intersection genes were deemed as hub genes in the Venn diagram (Fig. 3A). Protein–protein interactions were examined in terms of the seven hub genes, resulting in the creation of a protein–protein interaction network (Fig. 3B), visualized through Cytoscape (Fig. 3C). Furthermore, for each of the seven hub genes, an interaction network of functionally related genes was predicted and built using the GeneMANIA website (Fig. 3D). This allowed the physical interactions, shared protein domains, gene interactions, and other relevant information for the hub genes to be observed.
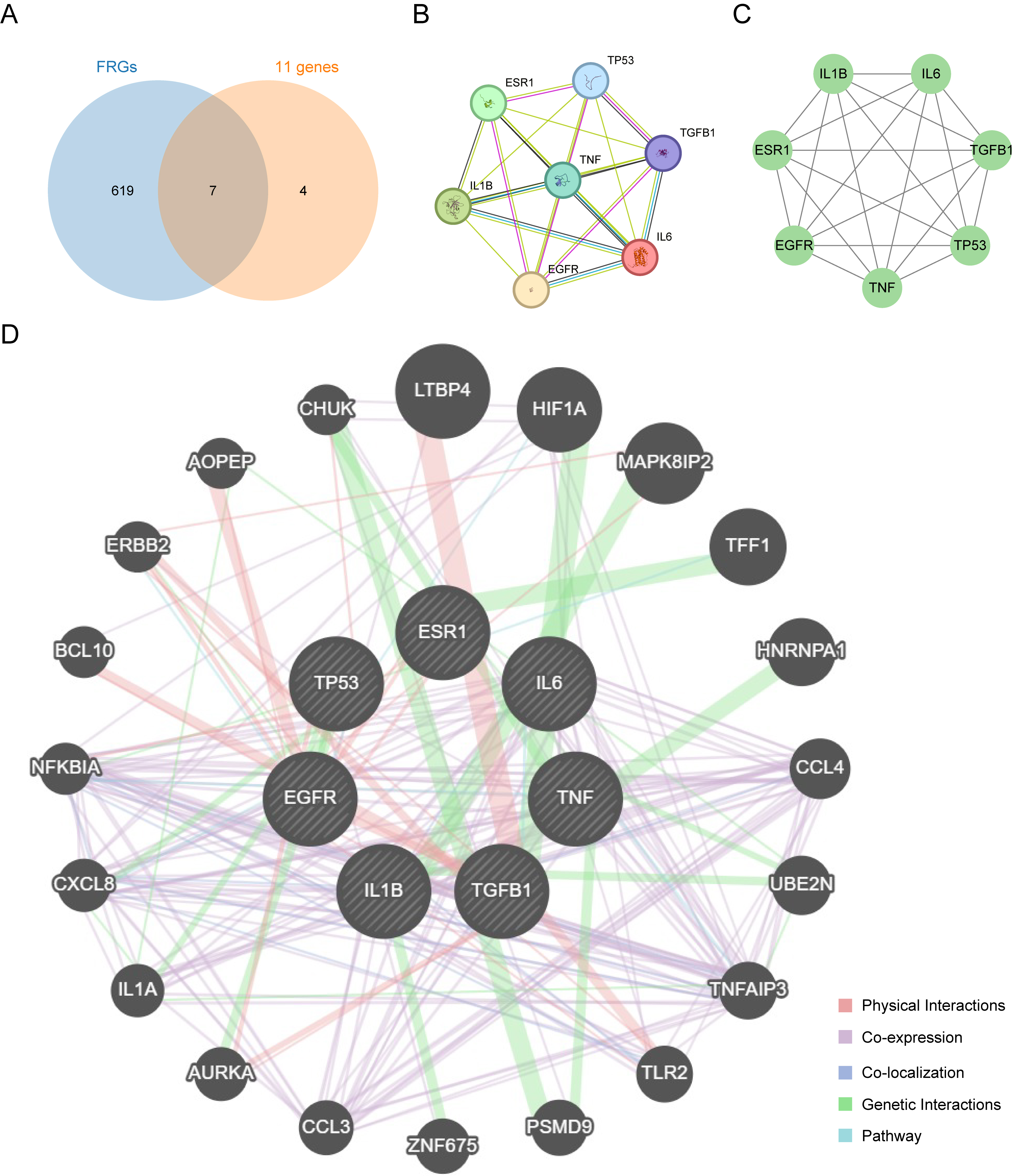 Fig. 3.
Fig. 3. Protein–protein interaction network. (A) Intersection Venn diagram of ferroptosis-related genes and 11 genes related to Lycium barbarum treatment of radiation-induced osteonecrosis of the jaw. (B) Protein interaction network diagram obtained from the STRING database. (C) Protein interaction network drawn by Cytoscape. (D) Interaction network of functionally similar genes of hub genes on the GeneMANIA website. In this network, black circles with white slash lines represent input hub genes, while other black circles without white slash lines represent predicted functionally similar genes.
The BP, MF, CC (Cellular Component), and biological pathways were analyzed for the seven hub genes (TP53, EGFR, TGFB1, IL-6, IL1B, TNF, ESR1). The results of the enrichment analysis are presented as bar graphs (Fig. 4A–C). Furthermore, the correlations between hub genes, GO results (Fig. 4D,E), and KEGG (Fig. 4F) enrichment analyses are presented as a circular network diagram.
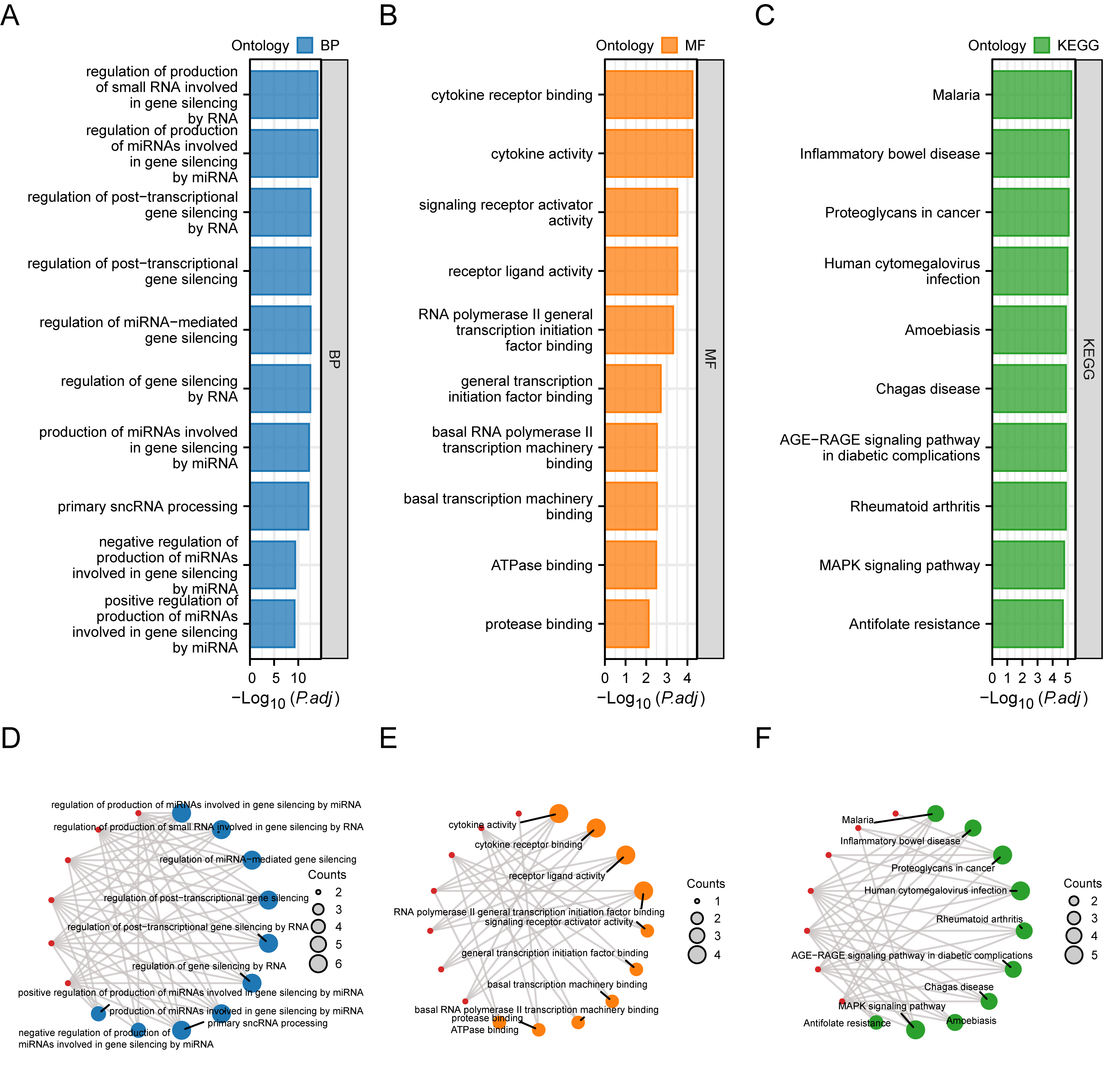 Fig. 4.
Fig. 4. Functional enrichment analysis (GO) and pathway enrichment (KEGG) analysis of hub genes. (A) Bar graph of the results of BP functional enrichment analysis of hub genes. (B) Ring bar chart of MF functional enrichment analysis results of hub genes. (C) Bar chart of KEGG pathway enrichment analysis results of hub genes. (D) Network diagram of BP functional enrichment analysis results of hub genes. (E) Ring network diagram of MF functional enrichment analysis results of hub genes. (F) Network diagram of KEGG pathway enrichment analysis results of hub genes. GO, Gene Ontology; BP, biological process; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes. (D–F): the blue circles are used to denote the BP entries, orange circles for MF entries, green circles for KEGG entries, and red circles for genes.
As shown in Fig. 4A–F, the hub genes were mainly enriched in regulating the generation of miRNAs involved in gene silencing through microRNAs, small RNA generation involved in gene silencing through RNA, miRNA-mediated gene silencing, gene silencing through RNA, post-transcriptional gene silencing, post-transcriptional gene silencing through RNA, miRNA generation involved in gene silencing through miRNA, primary sncRNA processing, and the negative and positive regulation of the generation of miRNAs involved in gene silencing through miRNA. The regulation of other BPs included cytokine receptor binding, signaling receptor activator activity, cytokine activity, receptor ligand activity, RNA polymerase II common transcription initiation factor binding, common transcription initiation factor binding, basal transcription machinery binding, basal RNA polymerase II transcription machinery binding, and ATPase binding, as well as MFs such as protease binding. The hub genes were mainly enriched in malaria, inflammatory bowel disease, cancer proteoglycans, cytomegalovirus infection, rheumatoid arthritis, the Advanced Glycation End-products-Receptor for Advanced Glycation End-products (AGE-RAGE) signaling pathway in diabetic complications, Chagas disease, Amoebiasis, the MAPK signaling pathway, and antifolate resistance pathway (Table 2).
| ID | Description | GeneRatio | BgRatio | p-value | p.adjust | q-value |
| GO:1903798 | Regulation of the production of miRNAs involved in gene silencing by miRNA | 6/7 | 22/18,800 | 8.51780 | 9.56455 | 1.49200 |
| GO:0070920 | Regulation of the production of small RNAs involved in gene silencing by RNA | 6/7 | 23/18,800 | 1.15236 | 9.56455 | 1.49200 |
| GO:0060964 | Regulation of miRNA-mediated gene silencing | 6/7 | 42/18,800 | 5.98311 | 2.67277 | 4.16931 |
| GO:0060147 | Regulation of post-transcriptional gene silencing | 6/7 | 44/18,800 | 8.05050 | 2.67277 | 4.16931 |
| GO:1900368 | Regulation of post-transcriptional gene silencing by RNA | 6/7 | 44/18,800 | 8.05050 | 2.67277 | 4.16931 |
| GO:0060966 | Regulation of gene silencing by RNA | 6/7 | 46/18,800 | 1.06814 | 2.95519 | 4.60988 |
| GO:0035196 | Production of miRNAs involved in gene silencing by miRNA | 6/7 | 52/18,800 | 2.32094 | 5.50395 | 8.58574 |
| GO:0070918 | Primary sncRNA processing | 6/7 | 56/18,800 | 3.70084 | 7.67924 | 1.19790 |
| GO:1903799 | Negative regulation of the production of miRNAs involved in gene silencing by miRNA | 4/7 | 11/18,800 | 2.21775 | 4.09051 | 6.38089 |
| GO:1903800 | Positive regulation of the production of miRNAs involved in gene silencing by miRNA | 4/7 | 12/18,800 | 3.32620 | 5.52149 | 8.61310 |
| GO:0005125 | Cytokine activity | 4/7 | 235/18,410 | 8.78981 | 5.67143 | 1.16082 |
| GO:0005126 | Cytokine receptor binding | 4/7 | 272/18,410 | 1.57540 | 5.67143 | 1.16082 |
| GO:0048018 | Receptor ligand activity | 4/7 | 489/18,410 | 1.61493 | 0.00030746 | 6.29304 |
| GO:0030546 | Signaling receptor activator activity | 4/7 | 496/18,410 | 1.70811 | 0.00030746 | 6.29304 |
| GO:0001091 | RNA polymerase II general transcription initiation factor binding | 2/7 | 24/18,410 | 3.40678 | 0.000490576 | 0.00010041 |
| GO:0140296 | General transcription initiation factor binding | 2/7 | 52/18,410 | 0.000162845 | 0.001954141 | 0.00039997 |
| GO:0001098 | Basal transcription machinery binding | 2/7 | 74/18,410 | 0.000330387 | 0.002973487 | 0.000608608 |
| GO:0001099 | Basal RNA polymerase II transcription machinery binding | 2/7 | 74/18,410 | 0.000330387 | 0.002973487 | 0.000608608 |
| GO:0051117 | ATPase binding | 2/7 | 82/18,410 | 0.000405637 | 0.003245097 | 0.000664201 |
| GO:0002020 | Protease binding | 2/7 | 136/18,410 | 0.001110341 | 0.007588935 | 0.001553291 |
| hsa05144 | Malaria | 4/7 | 50/8164 | 4.29924 | 5.84697 | 1.94597 |
| hsa05321 | Inflammatory bowel disease | 4/7 | 65/8164 | 1.25831 | 8.55649 | 2.84775 |
| hsa05205 | Proteoglycans in cancer | 5/7 | 205/8164 | 1.91753 | 8.69279 | 2.89311 |
| hsa05163 | Human cytomegalovirus infection | 5/7 | 225/8164 | 3.05480 | 1.03863 | 3.45675 |
| hsa05323 | Rheumatoid arthritis | 4/7 | 93/8164 | 5.38164 | 1.32803 | 4.41990 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 4/7 | 100/8164 | 7.21259 | 1.32803 | 4.41990 |
| hsa05142 | Chagas disease | 4/7 | 102/8164 | 7.81191 | 1.32803 | 4.41990 |
| hsa05146 | Amoebiasis | 4/7 | 102/8164 | 7.81191 | 1.32803 | 4.41990 |
| hsa04010 | MAPK signaling pathway | 5/7 | 294/8164 | 1.15907 | 1.75149 | 5.82925 |
| hsa01523 | Antifolate resistance | 3/7 | 30/8164 | 1.55196 | 2.11067 | 7.02466 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, Mitogen-Activated Protein Kinase.
The mRNA–miRNA data identified miRNAs that interact with the seven hub genes (TP53, EGFR, TGFB1, IL-6, IL1B, TNF, and ESR1). A visual representation of the mRNA–miRNA interaction network was created using the Cytoscape software (Fig. 5A). Six hub genes (TP53, EGFR, TGFB1, IL-6, IL1B, ESR1) and 73 miRNA molecules were included in this network, yielding 129 pairs of interactions between mRNA and miRNA. Detailed information on these interactions is shown in Supplementary Table 5.
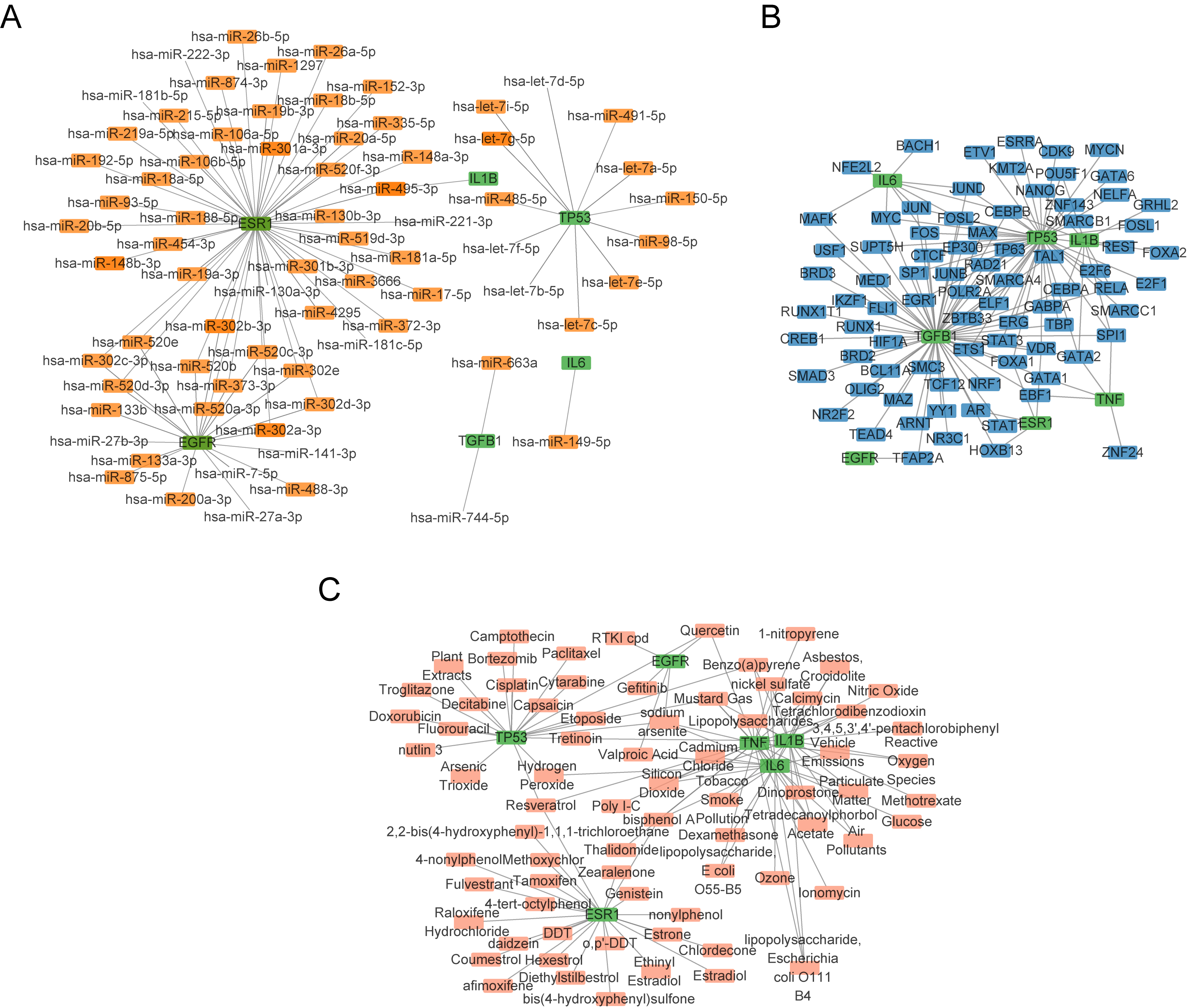 Fig. 5.
Fig. 5. Construction of mRNA–miRNA, mRNA–TF, and mRNA–drug interaction networks. (A–C) mRNA–miRNA (A), mRNA–TF (B), mRNA–drug (C) interaction networks of hub genes. Green squares are mRNAs; orange squares are miRNAs; blue squares are TFs; pink squares are small-molecule drugs. TFs, transcription factors.
We next searched the CHIPBase database for TFs that interact with the hub genes. Interaction information was extracted from the two databases and intersected with the seven hub genes, thereby demonstrating interactions between the seven hub genes and 81 TFs, as visualized using Cytoscape (Fig. 5B). Details of the mRNA–TF interactions are shown in Supplementary Table 6.
Possible medications or chemical compounds for the seven hub genes were identified using the CTD database. The mRNA–drug interaction network (Fig. 5C) revealed 73 possible pharmaceuticals or chemical substances that interact with six hub genes (TP53, TGFB1, IL-6, IL1B, TNF, ESR1) (Supplementary Table 7).
Based on the disease target–Lycium active ingredient–pathway network shown in Fig. 2C, three active ingredients related to the seven hub genes were identified:
| Gene | Compound | Binding energy |
| TGFB1 | Beta-sitosterol | 6.0 |
| ESR1 | Glycitein | 7.1 |
| EGFR | Quercetin | 8.6 |
| TNF | Quercetin | 6.8 |
| IL-6 | Quercetin | 7.0 |
| TP53 | Quercetin | 5.8 |
| IL1B | Quercetin | 6.3 |
| TGFB1 | Quercetin | 6.5 |
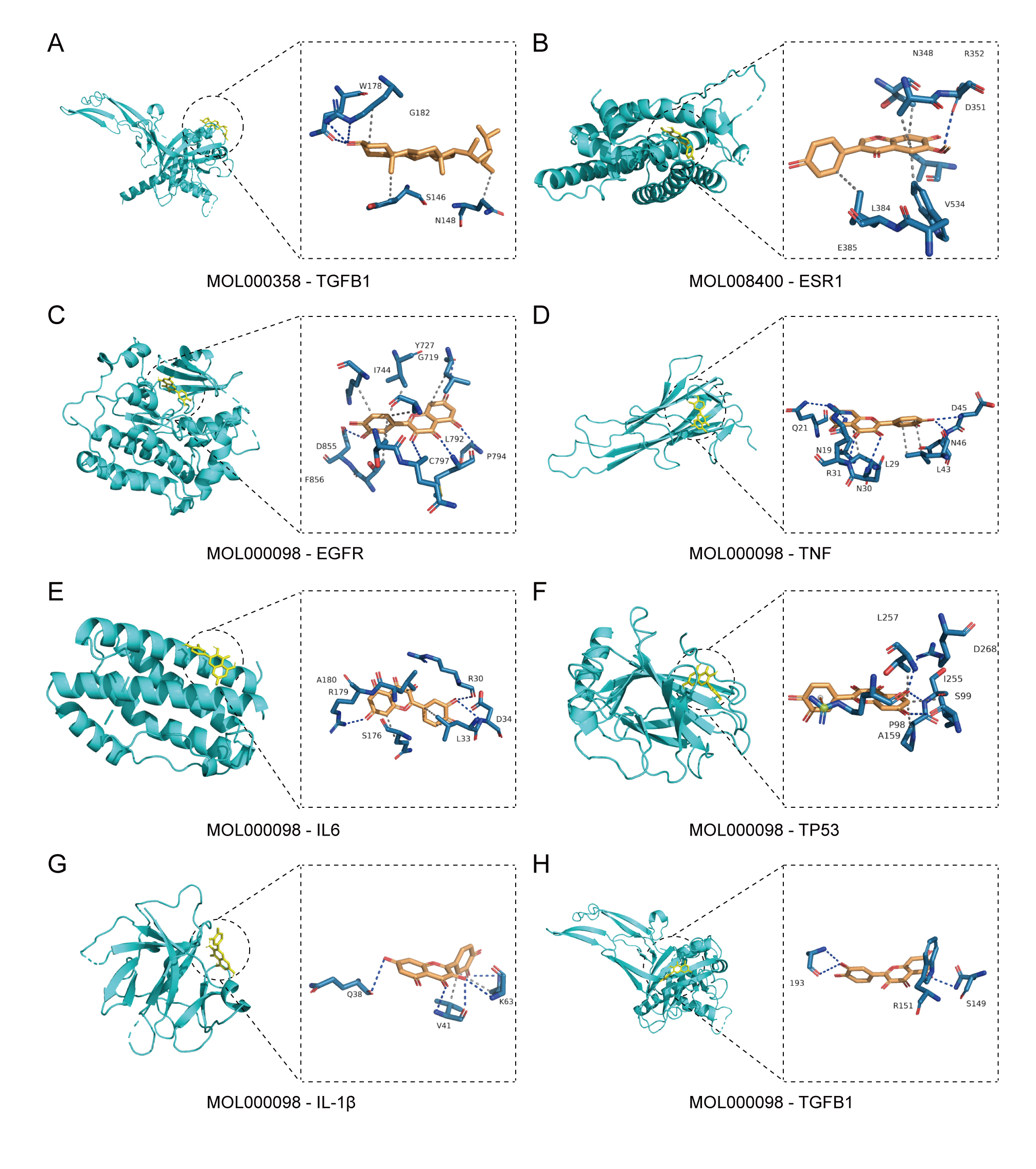 Fig. 6.
Fig. 6. Docking analysis of hub genes with core active components. (A) Mode of binding of Transforming Growth Factor Beta 1 (TGFB1) to
We next investigated the therapeutic effects of LbGP on ORNJ. The RT-qPCR data showed that the mRNA levels of IL1B and IL-6 were significantly reduced in rat JBMMSCs treated with LbGP compared to the ORNJ group; however, the levels of these mRNAs remained higher than in the NC group. The mRNA levels of TP53 and TNF were also lower in the LbGP group, whereas those of EGFR and TGFB1 increased. The mRNA level of ESR1 showed no significant change (Fig. 7A). The ELISA results exhibited significantly decreased protein levels of TNF-
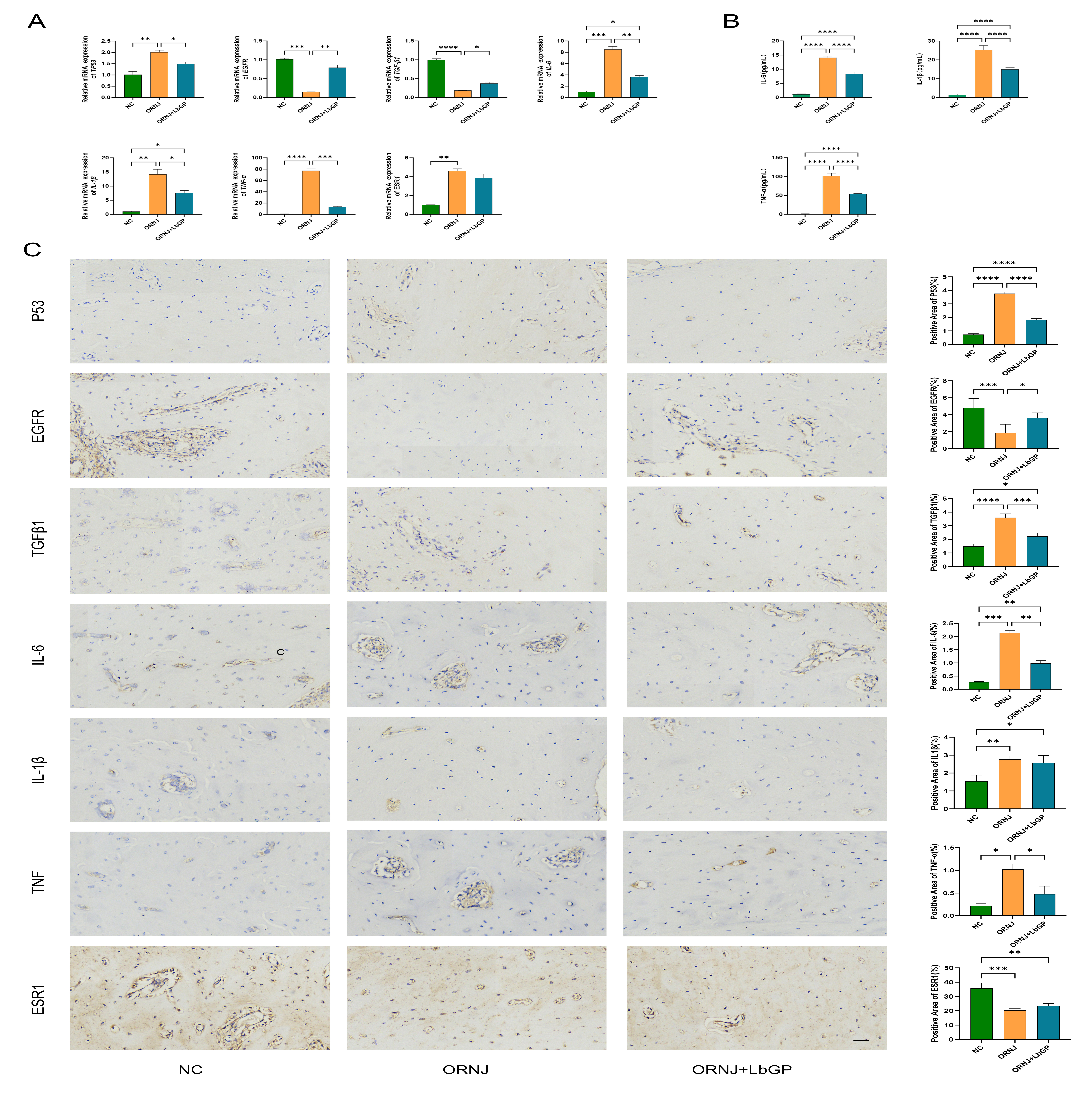 Fig. 7.
Fig. 7. RT-qPCR validation, ELISA validation, and immunohistochemical validation of paraffin-embedded sections of rat mandible tissue. (A) Relative mRNA expressions of hub genes in the normal, ORNJ, and treatment groups. (B) Protein expressions of tumor necrosis factor-alpha (TNF-
The incidence of osteoradionecrosis among patients receiving radiotherapy for head and neck malignancies ranges from 0% to 23%, representing a serious complication of the applied treatment. Meanwhile, a higher prevalence is observed among older and male patients [1]. Following radiotherapy, most patients experience reduced salivary secretion, increased susceptibility to rampant caries, secondary odontogenic infections, and prolonged non-healing wounds resulting from extractions or other injuries. The latter occasionally leads to fistula formation with minimal purulent discharge, accompanied by persistent pain and halitosis. Occasionally, soft tissue may undergo necrosis and ulceration, exposing necrotic bone that is non-mobile, leading to a chronic inflammatory process that significantly impacts the quality of life and oral health of the patients. Currently, treatment modalities for osteoradionecrosis mainly comprise oral antibiotics, oral hygiene measures, surgical interventions, and hyperbaric oxygen therapy. However, existing treatment modalities have limitations, such as relying solely on medications and local wound care. Although partially effective, this may overestimate clinical resolution due to the inclusion of so-called “mild” cases of osteoradionecrosis. Additionally, surgical interventions entail inherent risks, while hyperbaric oxygen therapy is both expensive and laborious.
Thus, the investigation of the potential therapeutic effects of Lycium barbarum is of significant importance and urgency. Lycium barbarum possesses numerous medicinal features, including anti-inflammatory, immunomodulatory, and antioxidative properties, which can exert beneficial effects in treating osteoradionecrosis. Nevertheless, thorough studies on the effectiveness of Lycium barbarum and its mode of action in osteoradionecrosis remain limited. Further screening of the bioactive components of Lycium barbarum is needed to evaluate its feasibility and efficacy as a treatment for osteoradionecrosis. This will provide a scientific basis for clinical practice and should eventually enhance treatment results and patient quality of life.
We first obtained the compositional information of Lycium barbarum from the TCMSP database and utilized its ADME-related characteristic data. Active constituents were screened based on the OB and DL values. Subsequently, the active ingredients were converted into gene names to predict the corresponding protein targets in Lycium barbarum. To assess the involvement of ferroptosis in ORNJ, we identified 626 unique FRGs from the GeneCards, MSigDB, and FerrDb databases. Additionally, we identified Lycium barbarum treatment-related genes for ORNJ by intersecting these genes with the previously screened ORNJRGs. Subsequently, seven hub genes (TP53, EGFR, TGFB1, IL-6, IL1B, TNF, ESR1) were identified, allowing a corresponding protein–protein interaction analysis to establish a network.
Utilizing the disease target–Lycium barbarum active ingredient–pathway network, we identified three active ingredients in Lycium barbarum associated with the seven hub genes; notably,
GO and KEGG enrichment analyses were performed to identify the regulatory pathways and biological processes of key genes. Our findings suggest that Lycium barbarum may be involved in biological processes, including cytokine receptor binding and receptor–ligand interactions. Immune cells release a variety of cytokines in response to infection, injury, or damage, including TNF and IL-6. These cytokines trigger inflammatory responses that lead to vasodilation and increased vascular permeability, containing the spread of infection. However, excessive or prolonged inflammatory responses can be detrimental to the body. Gene expression can be modulated through the inhibition of target gene translation or through mRNA degradation. This can occur via the regulation of miRNAs, by mediating gene silencing through miRNAs, and by post-transcriptional gene silencing. The MAPK signaling pathway can be stimulated by diverse inflammatory factors [29], exerting a crucial regulatory function in the development and spread of inflammation. Investigating the influence of Lycium barbarum on inflammatory factors holds great potential for preventing and managing infections and inflammatory reactions. Finally, we investigated the underlying treatment effects of LbGP extract on ORNJ. Using LbGP as the intervention drug, mesenchymal stem cells from the mandible of ORNJ rats were cultured and examined by qPCR. The mRNA expression levels of TP53, IL-6, and TNF were observed to decrease, while the expression of EGFR increased. ELISA confirmed the reduced expression of IL-6 and TNF-
Although this study identified some bioactive compounds in Lycium barbarum, time constraints prevented us from carrying out functional experiments to demonstrate their therapeutic effects on osteoradionecrosis. The animal experiments in this study primarily focused on the expression of inflammatory factors and certain key genes. Specific indicators of ferroptosis, such as GPX4, ACSL4, and lipid peroxides, were not included in the analysis. Therefore, the role of ferroptosis is currently based on the extrapolation of bioinformatics data and indirect evidence. Secondly, the core small molecule components identified in the computational analysis were
This study initially identified seven key hub genes (TP53, EGFR, TGFB1, IL-6, IL1B, TNF, and ESR1) that interact with active components of Lycium barbarum and may play a role in the treatment of ORNJ. The active components, identified as
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. Data are available in a public, open access repository, Data are available on reasonable request. This study also utilized data from the following publicly available resources: TCMSP (https://tcmsp-e.com/tcmsp.php), GeneCards (https://www.genecards.org/), MSigDB (https://www.gsea-msigdb.org/gsea/msigdb/), FerrDb (http://www.zhounan.org/ferrdb/current/), STRING (https://cn.string-db.org), KEGG (https://www.kegg.jp), ENCORI (https://rnasysu.com/encori/), CHIPBase (https://rna.sysu.edu.cn/chipbase/), CTD (https://ctdbase.org/), and RCSB PDB (https://www.rcsb.org).
Conceptualization and design of the study were conducted by LF and YM. Data analysis and interpretation were performed by LF, JW, ZW, LS and LZ. LF, JW and LZ wrote manuscript. YM, JW, LS and LZ revised the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
This study complies with international and national ethical guidelines, and follows the ARRIVE (Animal Research: Reporting In Vivo Experiments) guidelines. Approval for the animal experiments was obtained from the appropriate Institutional Animal Care and Use Committee (Ethics Committee of Sichuan Provincial People’s Hospital), with approval number 2021 No. 336.
We extend our heartfelt gratitude to all participants, researchers, technicians, and reviewers who contributed to this study. Their invaluable support and contributions have been instrumental in the successful completion of this research.
This work was supported by the Chengdu Science and Technology Program (Grant Number: 2024-YF09-00026-SN).
The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/IJP44912.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.







