- Academic Editor
-
-
-
†These authors contributed equally.
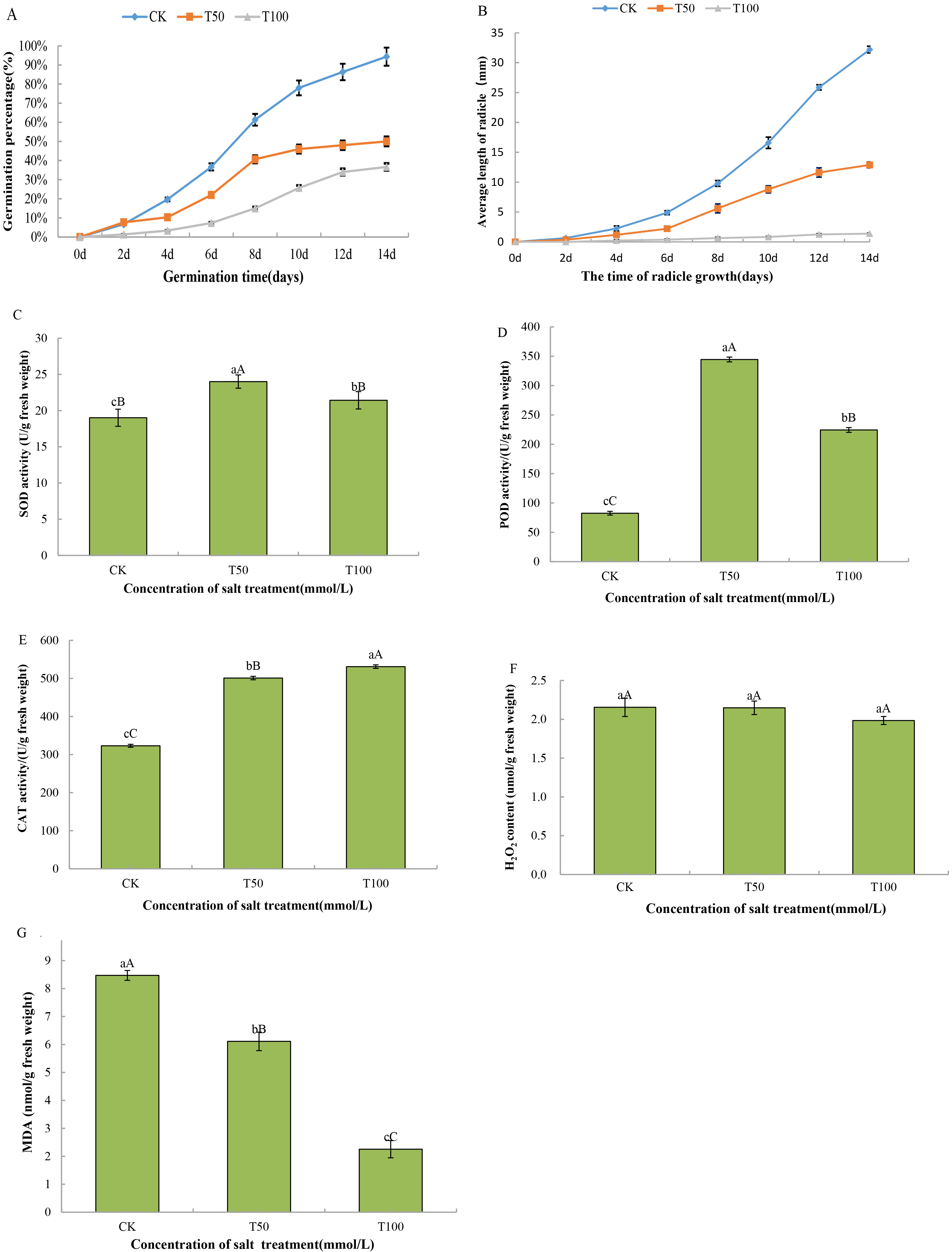
Background: Salinity is the main abiotic stress that affects seed germination, plant growth and crop production. Plant growth begins with seed germination, which is closely linked to crop development and final yields. Morus alba L. is a well-known saline-alkaline tree with economic value in China, and the most prominent method of expanding mulberry tree populations is seed propagation. Understanding the molecular mechanism of Morus alba L. salt tolerance is crucial for identifying salt-tolerant proteins in seed germination. Here, we explored the response mechanism of mulberry seed germination to salt stress at physiological and protein omics levels. Methods: Tandem mass tag (TMT)-based proteomic profiling of Morus alba L. seeds germinated under 50 mM and 100 mM NaCl treatment for 14 days was performed, and the proteomic findings were validated through parallel reaction monitoring (PRM). Results: Physiological data showed that salt stress inhibited the germination rate and radicle length of mulberry seeds, decreased the malondialdehyde (MDA) content and significantly increased superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) activities. Then, a TMT marker technique was used to analyze the protein groups in mulberry seeds with two salt treatment stages, and 76,544 unique peptides were detected. After removing duplicate proteins, 7717 proteins were identified according to TMT data, and 143 (50 mM NaCl) and 540 (100 mM NaCl) differentially abundant proteins (DAPs) were screened out. Compared with the control, in the 50 mM NaCl solution, 61 and 82 DAPs were upregulated and downregulated, respectively, and in the 100 mM NaCl solution, 222 and 318 DAPs were upregulated and downregulated, respectively. Furthermore, 113 DAPs were copresent in the 50 mM and 100 mM NaCl treatments, of which 43 were upregulated and 70 were downregulated. Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed that the DAPs induced by salt stress during mulberry seed germination were mainly involved in photosynthesis, carotenoid biosynthesis and phytohormone signaling. Finally, PRM verified five differentially expressed proteins, which demonstrated the reliability of TMT in analyzing protein groups. Conclusions: Our research provides valuable insights to further study the overall mechanism of salt stress responses and salt tolerance of mulberry and other plants.