1 Department of Microbiology and Immunology, Basic Medicine College, Jinan University, 510632 Guangzhou, Guangdong, China
2 School of Chemistry & Pharmaceutical Sciences, Guangxi Normal University, 541001 Guilin, Guangxi, China
3 School of Pharmacy, Guangdong Lingnan Institute of Technology, 510663 Guangzhou, Guangdong, China
4 Key Laboratory of Tropical Disease Control (Sun Yat-Sen University), Ministry of Education, 510632 Guangzhou, Guangdong, China
5 Department of Blood Transfusion, The Sixth Affiliated Hospital, Sun Yat-Sen University, 510632 Guangzhou, Guangdong, China
†These authors contributed equally.
Abstract
Background: Stability of intestinal flora is not only important for
maintaining stable immune functions; it is also a key immune channel
communicating the interaction between lung and intestine. In this study,
probiotics and fecal microbiota transplantation (FMT) were used to regulate
influenza-infected mice with antibiotic-induced intestinal dysbiosis and the
effects of intestinal microorganisms on these mice were subsequently observed and
evaluated. Methods: Mice are housed in a normal environment with
intranasal infection with influenza virus (FM1). Real-time quantitative
polymerase chain reaction (RT-qPCR) was used to determine messenger RNA
expression and lung viral replication of toll-like receptor 7 (TLR7), myeloid
differentiation primary reaction 88 (MyD88) and nuclear factor
Keywords
- intestinal flora
- probiotics
- influenza A virus FM1 mouse lung adaptation strain
- fecal microbiota transplantation
- TLR7
The complex relationship between intestinal flora and the human body spans various physiological activities and pathological processes, and consequently, is indispensable to the organism. The imbalance of the intestinal flora not only affects the functional state of the intestinal tract but also extensively influences the immune status and host function. Long-term antibiotic use can inhibit sensitive intestinal bacteria, allowing an excessive proliferation of drug-resistant bacteria, which is the primary reason for imbalances in intestinal flora [1]. Common methods for regulating imbalances in intestinal flora include the consumption of probiotics and fecal microbiota transplantation (FMT) from healthy donors [2]. Probiotics are active, beneficial microorganisms, commonly Bifidobacterium and Lactobacillus, colonizing the host and through the regulation of host mucosal and systemic immune functions or balancing of intestinal flora, promote nutrient absorption and maintenance of intestinal health. Therefore, promoting microorganisms or microbiota is beneficial to host health. FMT has been successfully used for the treatment of several diseases because FMT can regulate imbalances and restore homeostasis in the intestinal flora [3].
Influenza viruses belong to the Orthomyxoviridae family and can classified into the four following types: A, B, C, and D [4]. Influenza A viruses can easily cause an influenza pandemic owing to antigenic variation [5]. Studies in antibiotic-treated mice have shown that mice with disorders of intestinal flora experience aggravated mucosal destruction, lymphoid tissue damage, disturbances in immune cell homeostasis, and changes in intestinal composition, and are susceptible to severe infectious and inflammatory disease outcomes [6].
Both the respiratory tract and the gastrointestinal tract belong to the mucosal
immune system, have the same embryonic origin, and have similar structural
similarities. Interactions between the intestine and the lung are mediated by
microorganisms, metabolites, and free immune cells. Throughout the
lung–intestinal axis, the intestinal flora remains stable, crucial to in
maintaining the immune state and respiratory tract balance [7]. Recognition of
microbial pathogens through pattern recognition receptors (PRRs) is required to
initiate the natural immune response. PRRs recognize several conserved pathogenic
molecules, or pathogen-associated molecular pattern (PAMP). As important PRRs,
toll-like receptors (TLRs) can recognize pathogen molecules [8]. TLR7, highly
expressed in human plasmacytoid dendritic cells [9], is a member of the TLR
family that can recognizes single-stranded RNA viruses [10] including the
influenza virus. After PAMP recognition, TLR7 recruits specific linker molecules
that bind to Toll-IL-1 receptor (TIR) domains, including myeloid differentiation
factor 88 (MyD88), and contain TIR structures, which can induce the
interferon-
In this study, we hypothesized that gut microbes could be used for treating antibiotic-induced intestinal dysbiotic influenza-infected mice through the TLR7 signaling pathway. To verify our hypothesis, we first established an animal model for influenza-infected mice with antibiotic-induced intestinal dysbiosis. After infecting with the influenza virus FM1 strain, these mice were treated with probiotics or FMT to restore intestinal flora. Finally, we assessed the effect of an imbalance in intestinal flora on the TLR7 signaling pathway in the lungs of influenza-infected mice. TLR7 signaling pathway was found to play an important role in reducing the inflammatory process in antibiotic-induced influenza-infected mice with intestinal dysbiosis.
Animal experiments were performed with the approval and supervision of the
Experimental Animal Ethics Committee of Jinan University (Guangdong, China).
SPF-grade normal C57BL/6 wild-type mice were purchased from the Experimental
Animal Center of Guangdong Province (production license SCXK [Guangdong]
2019-0056). Specific-pathogen-free (SPF), 6–8-week-old female C57BL/6J wild type
(WT) mice, weighing 20
Thirty WT and 30 Tlr7-/- mice were randomly divided into five groups:
normal control (untreated), virus control (Virus), antibiotic treatment + virus
(Model), model + probiotic recovery (Probiotics), and model + fecal microbiota
transplantation (FMT). Model, Probiotics, and FMT groups were treated with
antibiotics mixed in drinking water for 25 days. All mice were fed sterilized rat
food. On day 26, feces from each group of mice were collected from their sterile
containers; mice were then anesthetized using a 1% pentobarbital sodium
intraperitoneal injection. The untreated group was treated with a 50
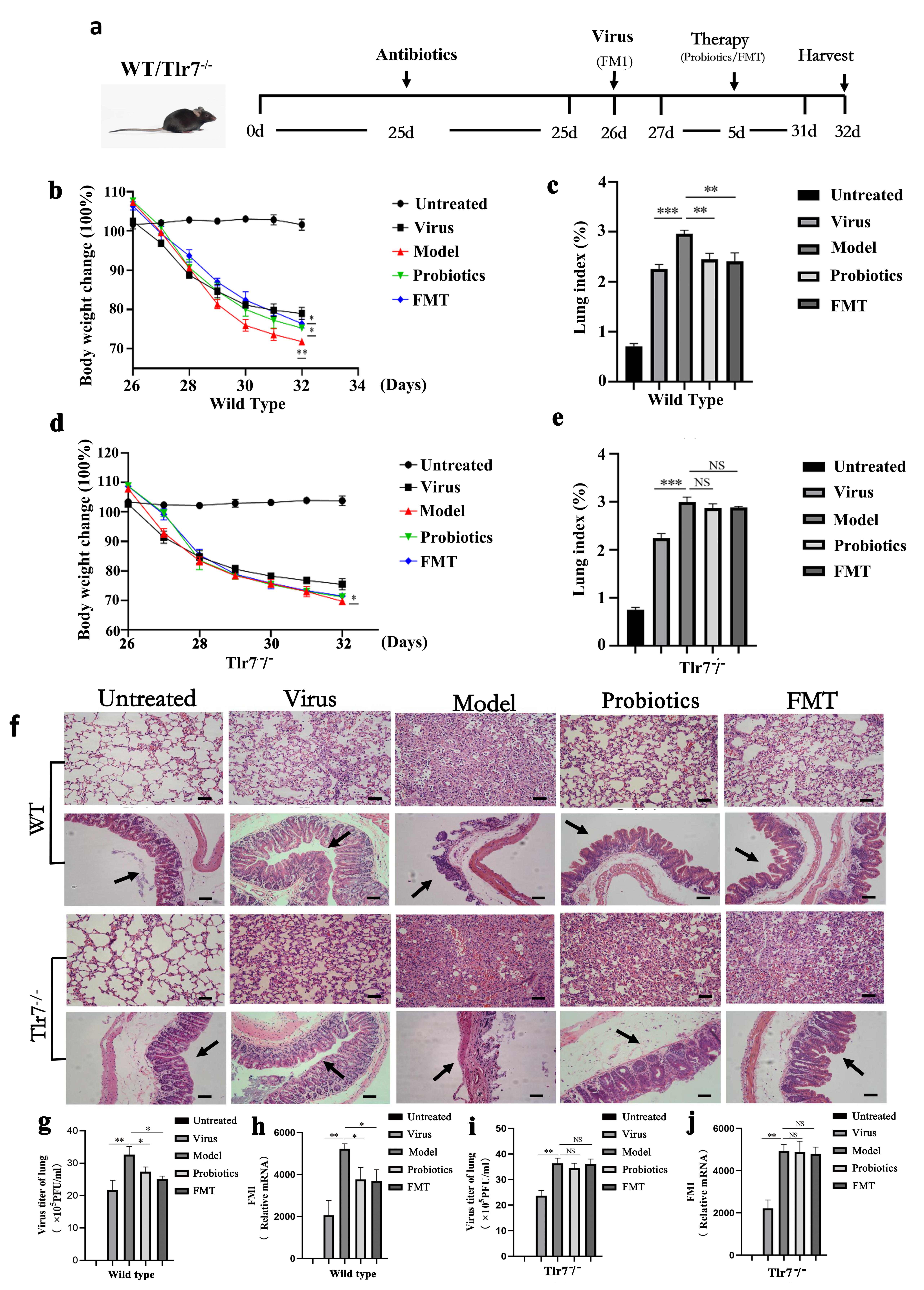
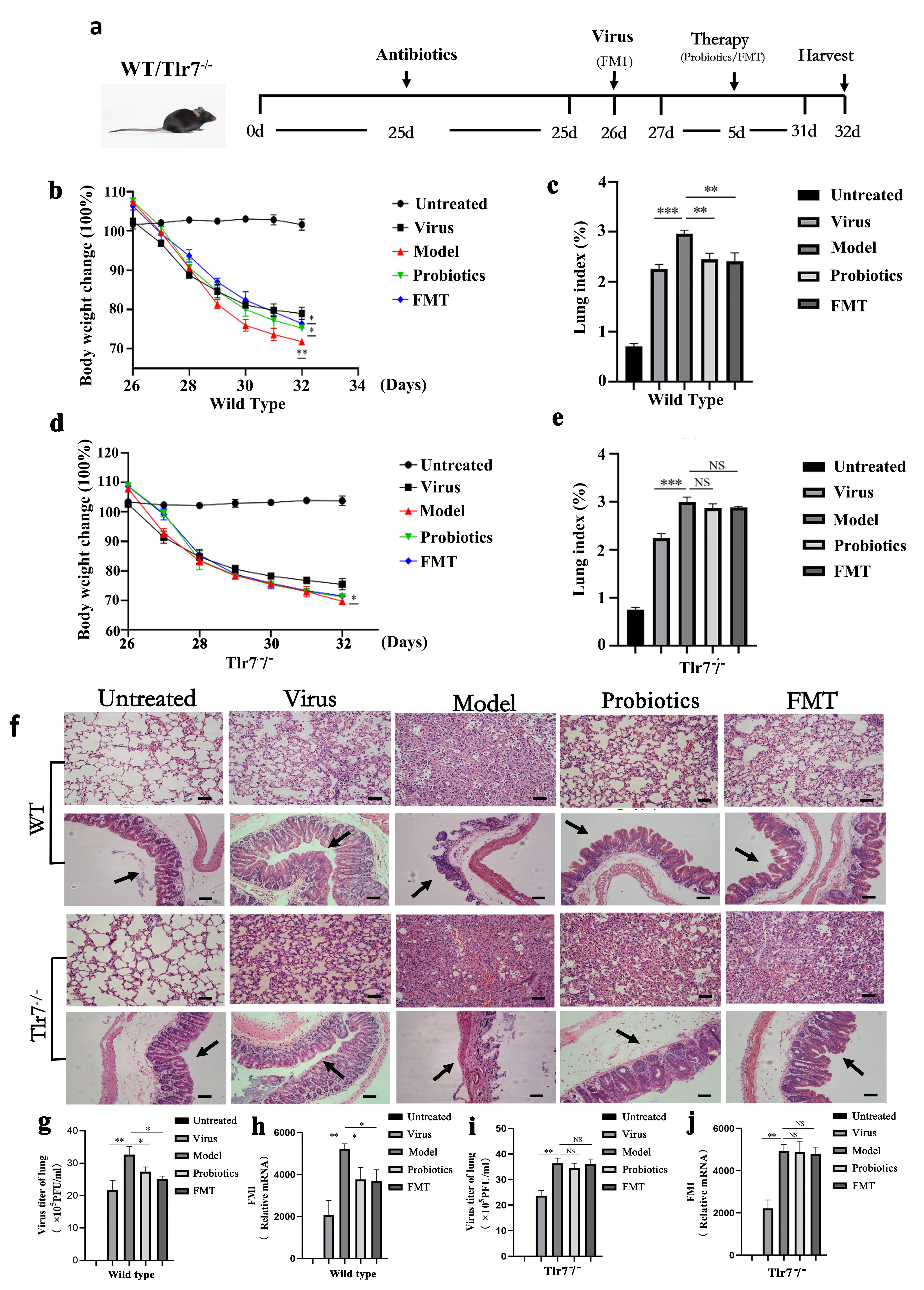 Fig. 1.
Fig. 1.The changes of body weight and lung index, pathological
analysis and virus replication were observed. (a) is experimental flow graph.
(b) and (c) represent changes to body weight and lung index, respectively, in
each group of WT mice (n = 6). (d) and (e) represent changes to body weight and
lung index, respectively, in Tlr7
The antibiotic drinking water mix was prepared by adding ampicillin (1 mg/mL, Lot: 7918010110), neomycin (1mg/mL, Lot: 9311010150), metronidazole (1mg/mL, Lot: c10473109), gentamicin (0.5 mg/mL, Lot: H04J10Z89946), and vancomycin (0.5 mg/mL, Lot: H26M10Z84068) (all from Guangzhou Dingguo Biotechnology Co., Ltd., Guangzhou, China) to sterilized pure drinking water; this was replaced every three days.
Bifidobacterium and Lactobacillus triple live bacteria tablets (Inner Mongolia Shuangqi Pharmaceutical Co., Ltd., Inner Mongolia,China , Chinese medicine S19980004) were used strictly according to the manufacturer’s instructions; the daily dosage for each mouse was 0.78 g/kg. Probiotics were dissolved in sterilized saline and administered intragastrically at 0.2 mL/d [12, 13].
Twenty-six days after antibiotic treatment, before being infected with virus, anus of each mouse was wiped with cotton swabs to stimulate defecation on a sterile container collecting feces. On day 32, following euthanasia, fecal samples were collected from the rectum of mice by opening the abdominal cavity using sterilized equipment in a biosafety cabinet. Collected feces were placed into sterile EP tubes and frozen at –80 °C. All procedures were completed within 30 min. The samples were sent to Guangdong Magigene technology Co., Ltd (Guangzhou, China) for testing.
The 16S rRNA gene was sequenced by Guangdong Magigene Biotechnology Co., Ltd (HiSeq2500/Miseq, PE250/PE300, Guangdong, China). Genomic DNA was extracted using the ALFA-SEQ Advanced Stool DNA Kit; the concentration and purity of DNA were detected using NanoDrop gel One. The hypervariable region of 16S rDNA V4–V5 was amplified using polymerase chain reaction (PCR), with barcode-tagged primers and TaKaRa Premix Taq® Version 2.0 (RR901A, TaKaRa, Guangzhou, China). Mixed PCR products were recovered using an E.Z.N.A.® Gel Extraction Kit (Omega, Nocross, Georgia, USA); target DNA fragments were recovered via elution with TE buffer. The Illumina Nova 6000 platform was used for PE250 sequencing of the expanded sublibrary. Head and tail primers were simultaneously removed using Cutadapt software, thus obtaining quality-controlled, paired-end clean reads. Fastp was used to tailor raw tag data with sliding window quality (– W4–M20), thus obtaining an effective splicing segment (clean tags). Clean tags of all samples were clustered (the similarity was 97% by default) to generate operational taxonomic units (OTUs); singleton OTUs and chimeras were simultaneously removed. To obtain annotated species information, each OTUs representative sequence was compared against SILVA (16s), RDP (16s), and Greengenes (16s) databases using usearch-sintax(version v11, San Francisco, USA).
Fresh feces from mice in the untreated group were collected and immediately
re-suspended in aseptic saline; insoluble particles were filtered out. The
filtrate was centrifuged at 4 °C for 15 min at 3000
Influenza A virus (A/FM1/1/47) was provided by the Department of Immunology and
Microbiology at the Jinan University and stored at –80 °C. After mice
were anesthetized via intraperitoneal injection of 1% pentobarbital sodium, each
was given 50
The status, coat color, vitality, drinking water, and survival status of mice in
each group were observed daily. The body weight of each mouse was recorded at the
same time each day, and the percentage of weight change was calculated according
to the formula: percentage of weight change = (nth celestial body weight/day 0
body weight). Lung tissue sample was obtained and rinsed with aseptic phosphate
buffer (PBS); the surface water was dried with filter paper before weighing. Lung
index was calculated as (lung weight/body weight)
Lung tissue and cecum of mice were removed, fixed with 4% paraformaldehyde,
embedded in paraffin, and sliced into 4
On day 6 after infection (day 32), lung tissue from mice was homogenized in 1 mL
of Dulbecco’s Modified Eagle’s Medium (DMEM), and the supernatant obtained using
centrifugation. Supernatant of lung tissue was diluted 10 times and to different
concentrations. The Madin–Darby canine kidney (MDCK) cell plaque assay was used
to determine viral titer in the supernatant of lung tissue. MDCK cells were
cultured in a monolayer on a cell culture plate. Following full growth, after
each well was washed with PBS, the supernatant of lung tissue with different
dilution times was added to each well and incubated at 37 °C for 1 hr. A
mixture of 2% carboxymethyl fiber and medium was added and incubated at 37
°C and 5% CO
Total RNA was extracted from lung tissue homogenate using RNAiso plus (Takara,
Kusatsu, Japan). After testing RNA quality, the PrimeScriptTM RT kit and SYBR
Green PCR Master Mix (Takara, Kusatsu, Japan) were used for cDNA synthesis and
qPCR detection. Reverse transcription was performed in a Bio-Rad S1000 thermal
cycler (Bio-Rad, Berkeley, CA, USA). qPCR was performed using the CFX Connect
real-time PCR detection system (Bio-Rad, Berkeley, CA, USA). Experimental
procedures were as follows: 95 °C, 30 s, 5 s, 60 °C, 30 s,
cycle 40 amplification cycles, 95 °C, 10s. All primers (Table 1) were
designed and synthesized by Shanghai Shenggong Bioengineering Co., Ltd
(Guangzhou, China). All samples were equipped with 3 multiple holes, and the
experiment was repeated three times. GAPDH was used as the internal reference and
expression was calculated using the cycle threshold method
(2
| Gene | Forward (5′ to 3′) | Reverse (5′ to 3′) |
|---|---|---|
| Gapdh | CTGAGCAAGAGAGGCCCTATCC | CTCCCTAGGCCCCTCCTGTT |
| FM1 | GACCAATCCTGTCACCTCTGAC | AGGGCATTNTGGACAAAGCGTCTA |
| Tlr7 | GGGTCCAAAGCCAATGTG | TGTTAGATTCTCCTTCGTGATG |
| MyD88 | CGATTATCTACAGAGCAAGGAATG | ATAGTGATGAACCGCAGGATAC |
| NF‐κB p65 | ATTCTGACCTTGCCTATCTAC | TCCAGTCTCCGAGTGAAG |
Mice spleen tissues were washed and homogenized in 1640 medium without serum.
Peripheral blood mononuclear cells (PBMCs) were collected using a mouse
lymphocyte separation solution (Multi Sciences, Hangzhou, China), according to
the manufacturer’s instructions. PBMCs were then suspended in 1640 culture
containing 10% fetal bovine serum (FBS), and the cell concentration adjusted to
1
| Antibody | Labels used | Company |
|---|---|---|
| CD4 | FITC | eBioscience |
| IFN- |
APC | eBioscience |
| IL-4 | PE | eBioscience |
| IL-17A | PE-cyanine7 | eBioscience |
| CD25 | APC | eBioscience |
| Foxp3 | PE | eBioscience |
Mouse lung tissue was extracted using RIPA lysis buffer (LOT: 6171803, Multi
Sciences, Hangzhou, China), with total protein extracted after homogenization.
Total protein was quantified using a BCA protein quantitative kit (LOT: A91041,
Multi Sciences, Hangzhou, China). Protein was separated using 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a
polyvinylidene fluoride membrane (PVDF) membrane sealed with 5% skim milk
powder. The membrane was then incubated with rabbit anti-mouse monoclonal
antibodies against RIG-I (CST, 4200S, 1:1000), MA VS (CST, 4983S, 1:1000),
NF-
Statistical analysis was performed using GraphPad Prism 8.0 software (Beijing,
China). Results were expressed as mean
The Venn diagram for Operational Taxonomic Units (OTU) shows the number of
common and endemic species between different groups. Total OTUs for WT and
Tlr7
Following the FM1 influenza virus infection, the body weight of mice decreased
significantly, and they experienced loss of appetite, fluffy and rough hair, and
decreased activity. Six days after infection, weight loss in the WT Model group
was more severe than in the WT Virus group (p
Microscopic observation showed that alveoli structure in the untreated group of
WT and Tlr7
Intestinal microflora in mice of each group was analyzed 16S sequencing
analysis,
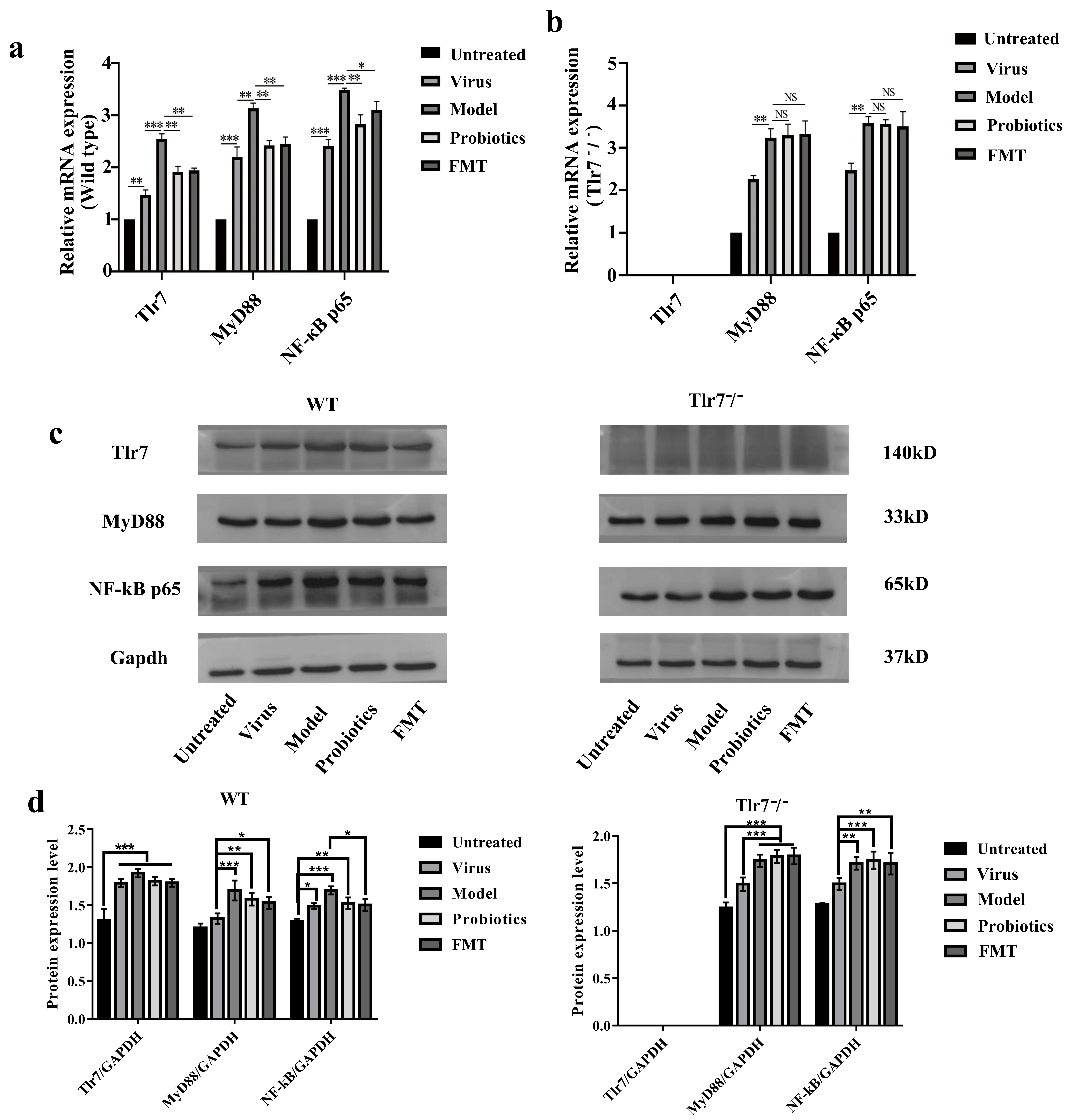
To further verify the relationship between intestinal flora and the TLR
signaling pathway, we analyzed the expressions of TLR7 and downstream MyD88 NF
and NF-
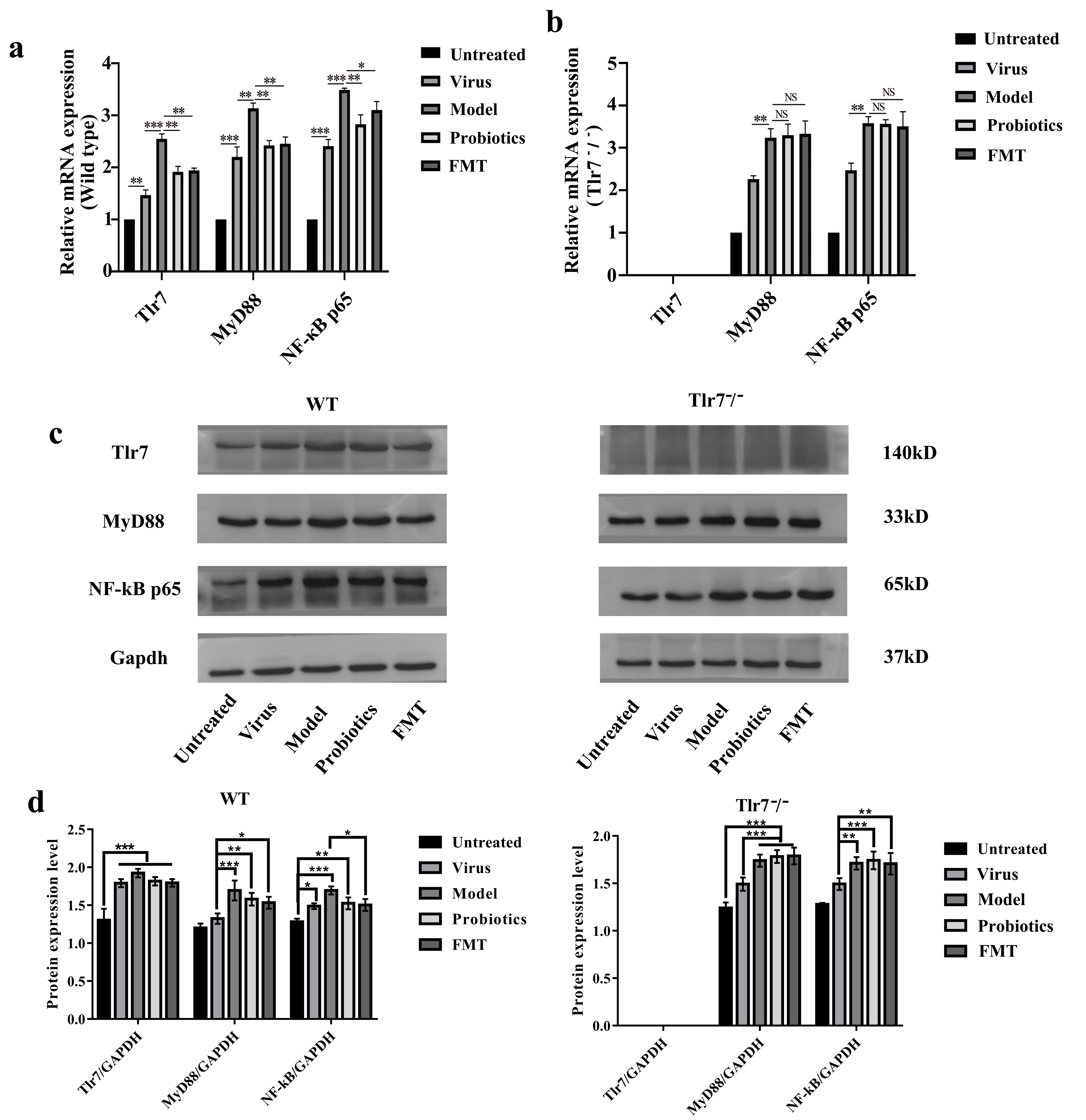 Fig. 2.
Fig. 2.mRNA and proteins expressions of TLR7, MyD88, and NF-
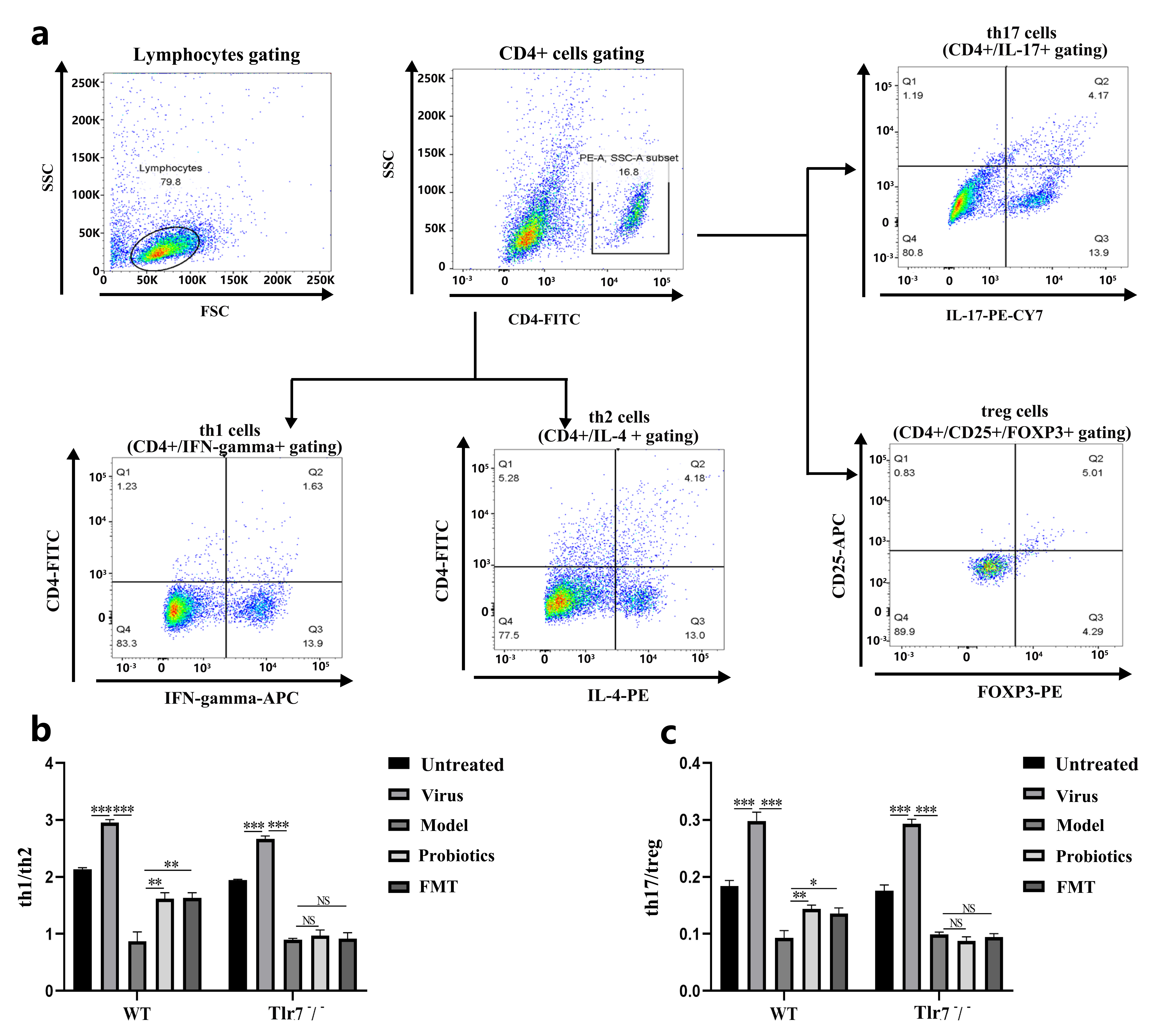
To further assess inflammation in mice, Th1/Th2 and Th17/Treg were detected in
the spleen tissues of mice by flow cytometry (Fig. 3a). In WT mice, the balance
between Th1/Th2 and Th17/Treg in the Model group was significantly disrupted
(Fig. 3b,c); however, the ratio was different compared to the Virus group,
showing a decrease when bacteria were destroyed using mixed antibiotics (Fig. 3b,
c). The ratio in the Probiotics and FMT groups was significantly adjusted
relative to the Model group (Fig. 3b,c). However, in Tlr7
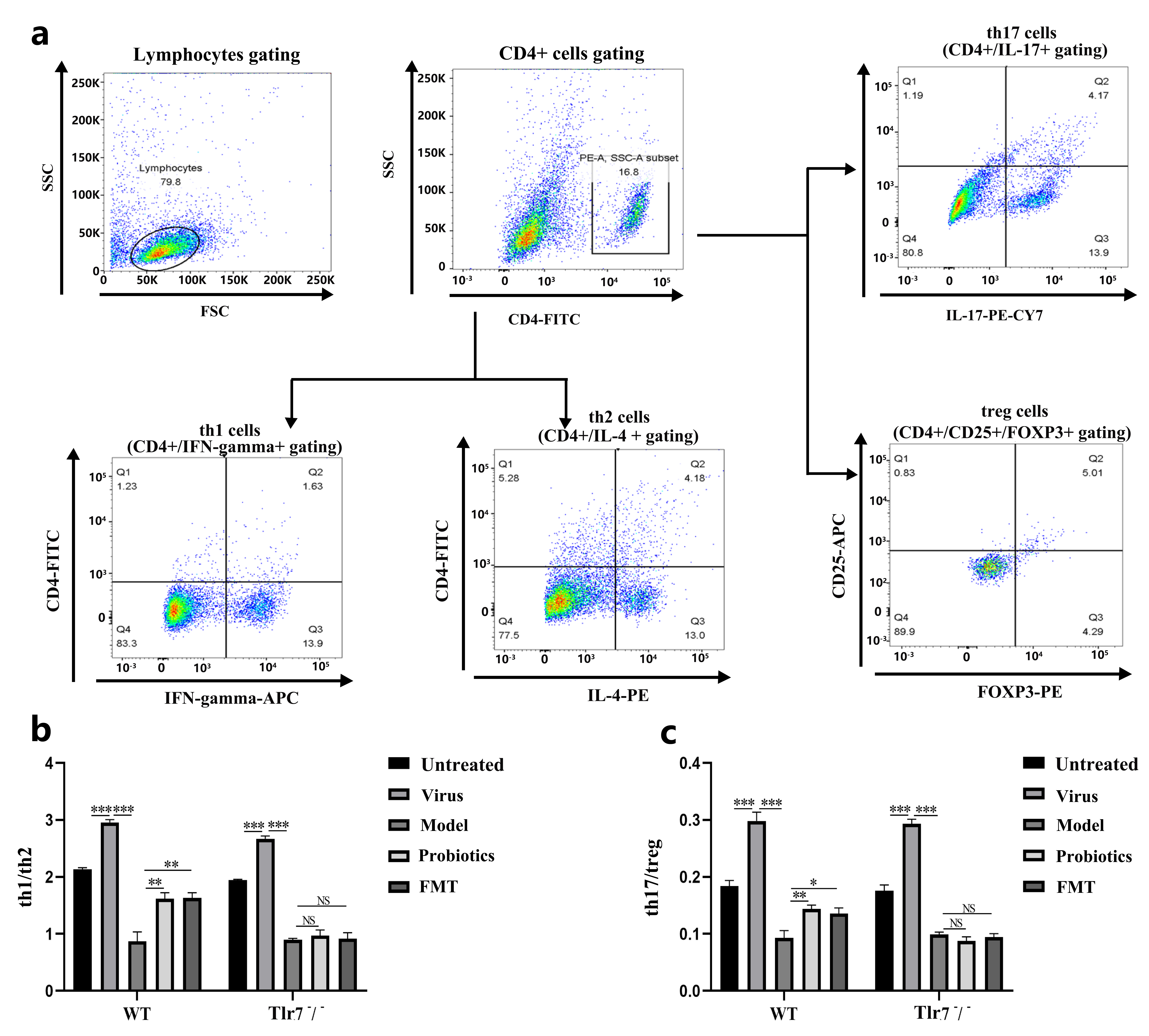 Fig. 3.
Fig. 3.Results of Th1/Th2 and Th17/Treg in mice. (a) Th1, Th2, Th17,
and Treg cells were analyzed using flow cytometry. (b) Results of Th1/Th2 in WT
and TLR7
In this study, we focused on the important effects of regulation of intestinal
flora through the TLR7 signaling pathway in influenza-infected mice with
antibiotic-induced intestinal dysbiosis. Inflammation of the lungs and intestines
in Tlr7
First, it was necessary to establish an influenza mouse model with antibiotic-induced intestinal dysbiosis. Under normal conditions, the intestinal flora is in a dynamic balance, with rich and diverse species showing relatively stable proportions. The four main phyla—Firmicutes Bacteroidetes Actinobacteria and Proteobacteria account for more than 90% of total intestinal bacteria [18, 19]. Many factors can cause an imbalance in the intestinal flora, including environmental, nutritional, drug use, and antibiotic abuse [20]. In this study, five different antibiotics were administered to mice for destroying the intestinal flora, resulting in serious damage to species abundance [21, 22]. Mice were infected with the FM1 influenza virus to establish an influenza-infected mouse model with antibiotic-induced intestinal dysbiosis and probiotics and FMT were used to regulate and rebuild intestinal microbial balance and prevent diseases. Common probiotics include Lactobacillus, Bifidobacterium, Streptococcus, Escherichia, and Bacillus [23]. The Bifidobacterium and Lactobacillus triple live bacteria tablets used in this study are common probiotic preparations comprising B. longum, L. bulgaricus, and S. thermophilus. FMT is also an effective method to restore and regulate destroyed intestinal flora, often playing an important role in many diseases, including diabetes, liver cirrhosis, and Clostridium difficile infections [24].
McDermott and others propose that mucosal immune cells distributed across the
body can interact with different mucosal tissues and organs [25]. In the Virus
group, infection with the influenza virus in the lungs led to intestinal
inflammatory cell infiltration. After the regulation of intestinal flora in the
Probiotics and FMT groups, lung tissues followed the recovery trend of the
intestinal mucosa, to alleviate inflammation associated with viral infections. A
study by Takeshi Ichinohe reported that balanced intestinal flora could regulate
respiratory mucosal immunity through the activation of inflammatory bodies, thus
potentially regulating the respiratory mucosa via PRRs and releasing interleukin
along with Treg cells regulation [26]. Furthermore, probiotics or FMT repaired
intestinal flora and improved intestinal mucosa in Tlr7
TLR7 recognizes the ssRNA genome of the influenza viruses, which acts by
activating MyD88, inducing NF-
According to phenotypic and functional characteristics, CD4+ T cells can be
classified as Th1, Th2, Th17, Th22, T follicular helper cells (TfH), and Treg.
The ratio of Th1/lr7
In summary, damage to lung tissue and intestinal mucosa in influenza-infected mice with antibiotic-induced intestinal dysbiosis is more serious compared to simple virus-infected mice. Improving intestinal flora using probiotics or FMT can alleviate intestinal inflammation and improve pulmonary inflammation through the TLR7 signaling pathway. Therefore, further understanding of the relationship between the molecular mechanism underlying the TLR7 signaling pathway and lung and intestinal mucosa may provide a new strategy for the development of antiviral therapy.
The data and material for the current study are available from the corresponding author upon reasonable request.
Conceptualization—JG, HC, LX, SL, HY, LJ, WC, ZJ. Data curation—JG, HC, LX, SL. Funding acquisition—ZJ. Investigation—JG, HY, LJ. Methodology—LX, SL, HY, LJ, WC. Software—JG and SL. Visualization—JG and HC. Writing – original draft—JG. Writing – review & editing—JG, HC, LX, SL, HY, LJ, WC, ZJ. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
This study was approved by the Experimental Animal Ethics Committee of Jinan University (Guangzhou, China) (Approval No. IACUC-20191120-06).
Not applicable.
This study was financially supported by the Science and Technology Projects of Guangdong Province (2019A1515011071), the National Natural Science Foundation of China (no. 82074133), National Science and Technology Major Infectious Diseases Project during the 12th 5-Year Plan Period (2014ZX10003002-003–002). The Fundamental Research Funds for the Central Universities (NO: 21621101).
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.