Academic Editor: Ranieri Bizzarr
Background: Drosophila
Phosphatase of Regenerating Liver-1 (PRL-1) is the only homolog of the mammalian
PRLs with which it shares high sequence and structural similarities. Whilst PRLs
are most notable for their high expression in malignant cancers and related
promotion of cancer progression, the specific biological functions of the PRLs
remain largely elusive. Methods: Here, using a gain-of-function
approach, we found that PRL-1 functions during wing vein
development in Drosophila melanogaster (Drosophila). Overexpression of Drosophila PRL-1
caused dose-dependent wing vein proliferation. Results: Genetic
screening of the main TGF-
Drosophila PRL-1 is the only homolog for the three human PRL proteins, PRL1, PRL2, and PRL3. The PRLs belong to a family of dual specificity phosphatases (DSP) classified as a subgroup of protein tyrosine phosphatases (PTP). They were originally discovered as immediate-early gene products in the regenerating liver [1, 2, 3]. Mammalian PRLs commonly possess a tyrosine phosphatase domain and a CAAX prenylation motif (where C is the cysteine, A is an aliphatic amino acid and X is any amino acid) which is located next to a polybasic region in the C-terminal for membrane anchoring [3]. Many studies on clinical samples have demonstrated the connection between PRL overexpression and the malignancy of various types of cancers such as colorectal, breast, and gastric carcinomas [4], and the implantation of cultured cancer cells, PRL1 or PRL3-overexpressed cells into nude mice, resulted in metastatic tumors [5].
After the recognition of its intrinsic involvement in metastatic cancers and in
actively driving malignant progression, various animal models such as
Drosophila [6], Mus musculus [7, 8], and Rattus
norvegicus [9] were then used to gain insight into the biological function and
the underling molecular mechanisms of the PRLs. Drosophila PRL-1 was
identified to be a growth inhibitor that could counteract the oncogenic activity
of Src. Drosophila PRL-1 localizes to the lateral membrane of the wing
discs and has also been reported as ubiquitously expressed and localized to both
the cytoplasm and the plasma membrane in various tissues including embryo
epidermis, larval midgut, gastric caecum, developing eye and in the wing discs
themselves. The ability of Drosophila PRL-1 to inhibit growth was
confirmed to be dependent on the CAAX motif that is required to localize
Drosophila PRL-1 to the apical edge of the lateral membrane in the wing
discs [6]. Several other recent studies have showed that Drosophila PRL-1 functions in the central nervous system. The absence of Drosophila
PRL-1, for example, specifically reduces synapse organization in the terminal
arbor of one target area of mechanosensory neurons. It has also been reported
that untranslated Drosophila PRL-1 mRNA (the long UTRs sequences of
Drosophila PRL-1 mRNA) mediate the local translation of
Drosophila PRL-1 in local synaptic arbor formation and that
Drosophila PRL-1 provides a specificity factor to restrict InR-Akt
signaling and synapse formation in a subcellular compartment of neurons [10].
Notably, knockdown of Drosophila PRL-1 in PDF clock neurons dramatically
lengthens the circadian period. Drosophila PRL-1 could also mediate the
dephosphorylation of TIM to set period length and behavioral phase in darkness,
enabling behavioral adjustment to changes in day-night cycles [11]. Our previous
study found that Drosophila PRL-1 is involved in protecting the nervous
system against olfactory CO
Despite there being a rapidly growing amount of literature on PRLs, our
knowledge of the specific biological function of the PRL family of proteins
remains limited. Our study used the Drosophila wing to examine the
overexpression of PRL-1 in genetically controlled models. This system reveals
that Drosophila PRL-1 functions as a newly discovered regulator
of the transforming growth factor-
The adult wing of Drosophila has a stereotypical pattern of five longitudinal veins (LVs, L1–L5) and two crossveins (ACV, PCV) to act as the rigid supports necessary for flight [13]. Wing veins consist of rows of cells that differentiate into heavily pigmented cuticles and smaller apical sized cells. These are formed through specification of cell fate, not cell migration or cell shape change [14, 15]. During metamorphosis several signaling pathways, including those mediated by EGF, BMP/Dpp, Hedgehog, Notch, and Wnt, define the expression of many transcription factors involved in the partition of the wing disc epithelium into proveins and interveins [14]. For example, in the third instar larvae imaginal wing disc, EGFR signaling is activated within the proveins to direct the subdivision of each provein in a central region that will then differentiate as a vein [16, 17]; Notch signaling is involved in establishing the correct width of various wing veins [18]; and BMP signaling is required for the specification of both longitudinal proveins and intervein regions in the imaginal disc at the larval stage, also later required for crossvein formation during pupariation [14, 17].
BMP (Bone Morphogenetic Protein) is another member of the TGF-
In this study, we used a gain-of-function approach to investigate
Drosophila PRL-1 functions during wing development. Overexpression of
Drosophila PRL-1 caused the formation of extra vein tissue at the
posterior end of the PCV along the longitudinal vein 5 (L5), with a dose
dependent effect. Genetic screening found that RNAi-mediated knockdown of Mad
could alleviate the extra vein phenotype caused by overexpressed
Drosophila PRL-1 and lead to loss of the posterior of longitudinal vein
4. Knockdown of Smox on the background of overexpressing Drosophila
PRL-1 in the wing disc showed a phenotype similar to the knockdown of Smox
alone. Clonal analyses revealed that
overexpression of Drosophila PRL-1 resulted in lower expression of the
activated phospho-Mad protein as measured by immunostaining. Real-time PCR showed
that the transcriptional levels of
Smox were significantly increased upon the overexpression of
Drosophila PRL-1 in the wing disc, with a dose dependent effect. We
propose that the main function of PRL-1 in Drosophila wing development
is to affect the phospho-Mad levels and Smox transcriptional levels, therefore
potentially influencing the competitive balance for Medea between Mad and Smox.
Our study demonstrates a novel role for PRL-1 in regulating TGF-
The Drosophila PRL-1 amino acid sequence was downloaded from Flybase (http://flybase.org/) and human PRL1-3 amino acid sequences were downloaded from the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/). The Clustal X 2.0 software (European Bioinformatics Institute, Hinxton, Cambridgeshire, CB10 1SD, UK) was used for the multiple amino acid sequence alignments of Drosophila PRL-1 and human PRL1-3. Results were presented using the DNAman software (version 9.0, Lynnon Corporation, 116 RUE DU MILICIEN, VAUDREUIL-DORION QC J7V 9M5, Canada).
Flies were reared on a standard cornmeal medium at 25 °C. The following
strains were used: w
We used the flip-out system to generate overexpression clones in the brain. To
induce ectopic expression of Drosophila PRL-1, we crossed
hsFlp;Act5C
Adult female or male wings (N = 20 for each genotype, repeated three times) were removed and mounted in 80% glycerol mounting medium. Images of wings were obtained on an Eclipse 80i microscope (Nikon, Tokyo, Japan). Wing posterior and total areas were measured using ImageJ (version 1.51, LOCI, University of Wisconsin, Madison, Wisconsin, USA). Statistical analysis was performed using a student’s t-test.
Drosophila brains from third instar larvae were dissected in ice cold
1
Total RNA was isolated from dissected disc cells using Trizol®
Reagent (Invitrogen, Carlsbad, CA, USA, #15596018). A 1
Rp49 was used as an endogenous control. The following PCR primers
(5
The Drosophila PRL-1 gene (CG4993), encoding the single Drosophila PRL protein (Drosophila PRL-1), is located on the left arm of the second chromosome. It consists of 176 amino acids in approximately 20-kDa protein (http://flybase.org/). To evaluate the conservative properties of PRL phosphatases between humans and Drosophila, we aligned their amino acid sequences using public databases. Results showed that Drosophila PRL-1 shares an overall 75.7% similarity in comparison to all three human PRLs (Fig. 1A). More specifically, the amino acid similarity of Drosophila PRL-1 shows a high positive comparison with each of the human PRLs individually, with a 73% similarity between Drosophila PRL-1 and human PRL-1, 75% for human PRL-2, and 74% for human PRL-3. We then performed bioinformatics analysis on the structural characteristics of Drosophila PRL-1. Drosophila PRL-1 contains several domains also characteristic of mammals including the putative ‘CXnE’ motif [3], the conserved catalytic PTP domain that is critical to phosphatase activity [28], the polybasic region, and the CAAX prenylation motif for targeting to the plasma membrane [29] (Fig. 1A,B, red rectangles), showing that that Drosophila PRL-1 shares high sequence and structural similarities with human PRLs.
 Fig. 1.
Fig. 1.Structural characteristics of PRLs. (A) Multiple sequence alignments of PRLs from Homo sapiens and PRL-1 from Drosophila melanogaster. Identical residues are marked with a black background, conserved residues with a blue background, and similar residues with a salmon background. The putative ‘CXnE’ motif, WPD-loop, P-Loop, a polybasic region and a CAAX prenylation motif are indicated with red squares. (B) Schematic diagrams of Drosophila PRL-1. The putative ‘CXnE’ motif begins at C54.
We had previously generated a Drosophila PRL-1 deletion mutant which
showed a wing hold-up phenotype when stimulated under a high concentration of
CO
 Fig. 2.
Fig. 2.Overexpression of Drosophila PRL-1 caused
dose-dependent wing vein proliferation. (A) Adult wings from flies expressing
the following transgenes under the control of En-Gal4; Left panes are wings from
males; Right panes are wings from females. The lines divide wings into anterior
(A) and posterior (P) parts. (B,B’) The ratio of posterior compartment (P) to the
entire wing (A+P) (n = 20 in each genotype, and with experiments repeated three
times). (B) Male, (B’) female. (C) The percentage of adult wings with ectopic
vein branches; n = 20, repeated three times. (D) Wing vein pattern in the
PRL-1 mutants. Data are presented as means
Interestingly, we observed that the posterior wing compartment of both male and
female showed an extra branch at the posterior end of the PCV along the
longitudinal vein 5 (L5) in En-Gal4
To further investigate the functions and molecular mechanism of Drosophila PRL-1 in wing vein development, we obtained several RNA interference (RNAi) transgenic fly lines that were known to be involved in the pathways that control vein formation. We performed screening experiments using the flies with a background of PRL-1 overexpression driven by En-Gal4. When the antisense RNA of the target gene is expressed by the En-Gal4 driver, the RNA transcribed from the target DNA forms double-stranded RNA with the antisense RNA that is disrupted by the RNAi machinery and depleted from the target gene product at the posterior of wing. Using RNAi, we could therefore compare the phenotype resulting from a specific gene knockdown in the posterior of wing tissue and attempt to clarify the relationship between the specific gene and PRL-1.
We found that RNAi-mediated knockdown of Mad
could alleviate the extra vein phenotype
caused by overexpressed PRL-1 (Fig. 3A–D, yellow arrows), but this also led to a
loss of the posterior portion of longitudinal vein 4 (Fig. 3D, red arrows), which
was an identical phenotype seen in some Dpp partial loss-of-function mutants [26, 30]. Notably, downregulating Mad only in the posterior portion of wing tissue did
not result in any obvious phenotype (Fig. 3C), while knock down of Mad together
with simultaneous overexpression of PRL-1 could induce vein loss. This
demonstrates that the efficiency of the knockdown of Mad alone is not sufficient
to lead vein loss, and that this only occurs in combination with PRL-1. In this
way the data clearly indicates a Drosophila PRL-1 genetic interaction
with Mad. We then wondered whether overexpressing PRL-1 has any effect on Mad
levels. We carried out a flip-out method using heat shock induced FLP (hsFLP);
Act5C
 Fig. 3.
Fig. 3.Adult wings from flies expressing the following transgenes under
the control of En-Gal4. (A) En-Gal4
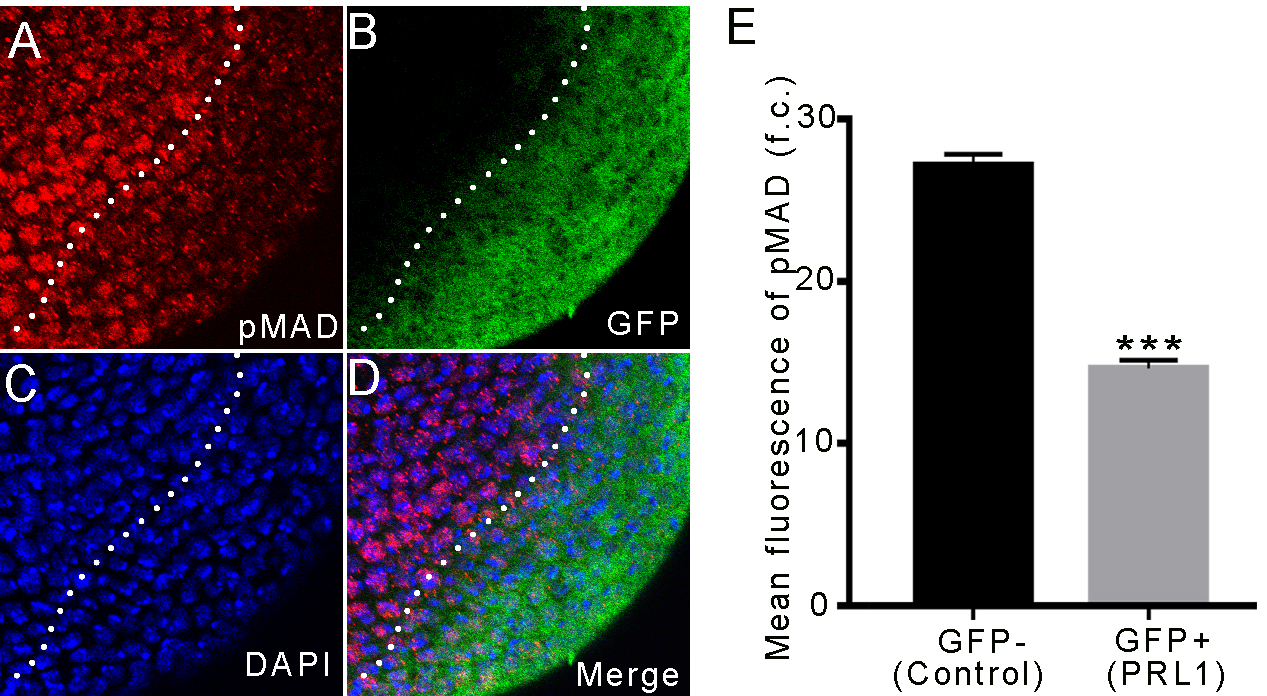 Fig. 4.
Fig. 4. Overexpressing PRL-1 led to decreased expressions of activated
phospho-Mad protein. (A–D) Overexpression of PRL-1 marked by GFP in the brain.
Lower expression of phospho-Mad is observed in overexpressing PRL-1 clone cells
as compared with the wild-type cells around the clone. (A) Phospho-Mad (red). (B)
GFP (green). (C) DAPI (blue). (D) Merge. (E) Mean fluorescence of phospho-Mad; n
= 6. Data are presented as means
As both Dpp/Mad and dAct/Smox pathways both play important roles in fly wing
vein development [26, 27] and share several components [25], we therefore
performed RNAi-mediated knockdown of Smox to confirm the phenotype. We found that
down-regulating the Smox gene led to ectopic vein tissue formation in
the vicinity of L5 and an extra branch at the posterior end of the PCV
(Fig. 3E, green arrows), which showed a
similar phenotype as noted in Sander et al. [26]. When we knocked down
Smox on the background of overexpressing PRL-1, the ectopic vein phenotype was
almost same as that in the down-regulation of Smox alone (Fig. 3F, green arrows).
This may suggest that PRL-1 functions upstream of Smox. To further confirm this
hypothesis, we carried out Real-time PCR experiments to check Smox
levels. Results showed that, except for Smox, the levels of genes such
as Sno oncogene (Snoo), Myoglianin (Myo),
and EcR, that are also involved in Activin signaling
pathway, remained unchanged upon PRL-1 overexpression (Fig. 5A). Additionally,
the transcriptional levels of Smox were shown to be increased in a
dose-dependent manner in the PRL-1-overexpressed wing discs (Fig. 5B). The
overexpression of PRL-1 triggering the up-regulation of Smox probably acts to compensate for decreased Activin signaling. These
results suggest that Drosophila PRL-1 is a regulator of TGF-
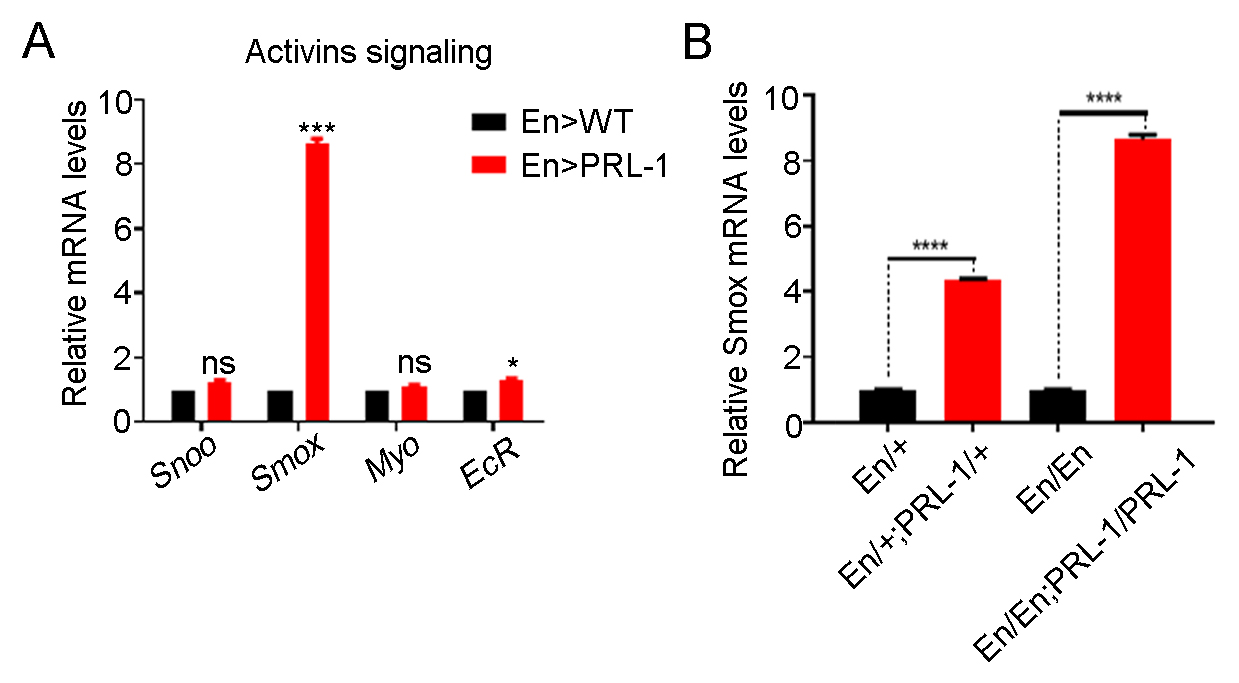 Fig. 5.
Fig. 5.Overexpressing PRL-1 increases the transcriptional
levels of Smox. (A) Real-time PCR analysis of the relative mRNA levels
of genes involved in the Activin signaling pathway showing that only
Smox is obviously increased in overexpressing PRL-1 wing discs; n = 3.
(B) The transcriptional levels of Smox in the discs from different copy
numbers of En-gal4 and UAS-PRL-1 animals indicate that the transcriptional levels
of Smox is a dose dependent effect in the overexpression of PRL-1 wing
disc s; n = 3. Data are presented as means
Drosophila PRL-1 belongs to the Phosphatase of Regenerating Liver (PRL)
family, also known as protein tyrosine phosphatase 4A (PTP4A). In mammals it
consists of three members, PRL-1, 2 and 3. Numerous studies using cultured cells
have demonstrated that the PRLs can induce cell proliferation, migration, and
invasiveness in cancers [29, 31]. Using the Drosophila wing as a model
system, here we report that the homolog of mammalian PRLs, Drosophila PRL-1, functions during wing vein development through its regulation of the
TGF-
Using bioinformatics methods to analyze the conservative properties of PRL phosphatases between human and Drosophila we found that Drosophila PRL-1 shares a 75.7% similarity between all three human PRLs and with many specific structural similarities noted (Fig. 1). This is consistent with Zeng et al. [32] and Lin et al. [33] who found that Drosophila PRL-1 shares high similarities (74%–76%) to all three mammalian PRLs. In our previous study, we found that a holding-up of wings phenotype in the PRL-1 deletion mutant flies could be rescued by overexpressing human PRL-1 or human PRL-2, which implies that human PRL phosphatases may function in a similar neuroprotective capacity [12]. However, in the present study, overexpressing human PRL-1 or human PRL-2 by En-gal4 in the wing failed to result in any similar ectopic wing vein anomalies as seen upon Drosophila PRL-1 overexpression (data not shown). This could be because the expression level is not sufficient, or that some of signal transduction members for human PRLs are absent in Drosophila. Pagarigan et al. [6] reported that overexpressing Drosophila PRL-1 could inhibit growth in a manner dependent on the CAAX motif that is integral to the localization of Drosophila PRL-1 to the apical edge of the lateral membrane.
The Drosophila adult wing has a
stereotypical pattern of longitudinal veins and crossveins which are constructed
mainly of two cell types, vein cells and intervein cells. The specification of
vein territories and the differentiation of wing veins and intervein regions are
regulated by a high number of complex signaling pathways including those of EGFR,
transforming growth factor-
In this scenario, ectopic PRL-1 lowers the expression of the activated phospho-Mad protein, in which a decrease in Dpp signaling and elevation of the transcriptional levels of Smox may act to downregulate the Activin pathway. It has been previously reported that RNAi-mediated Babo knockdown also resulted in decreased Activin signaling and led to a significant increase in the transcription of Smox [40]. These increased transcriptional levels of Smox could lead to a compensation role upon decreased Babo expression. The Activin/Smox signal pathway shares its component Medea with the Dpp/Mad pathway, and Medea has been reported to function as a limiting factor between the Mad and Smox signaling pathways [26]. Overexpressing PRL-1 could potentially cause downregulation of both the Dpp pathway and Activin pathway, therefore, influencing the competitive balance for Medea between Mad and Smox during wing vein pattern formation, as shown in the schematic of Fig. 6.
 Fig. 6.
Fig. 6.Schematic representation of the putative mechanism by which
PRL-1 could regulate the TGF-
Our genetic experiments show that the knockdown of Mad on the background of ectopic PRL-1 leads to the loss of the extra vein, thus reversing the phenotype. This suggests that PRL-1 and Mad may function in parallel pathways to regulate vein formation. However, the detailed molecular mechanism of how PRL-1 affects the phosphorylation of Mad awaits further investigation. PRL-1 and Smox may act in a linear pathway in the regulation of vein formation. The ectopic vein phenotype caused by knocking down Smox was nearly identical to that of the combination of Smox knockdown and overexpression of PRL-1 in the posterior of the wing (Fig. 3E,F). Together, these experiments suggest that dPRL-1 acts, at least in part, upstream of Smox.
Our study identified a novel function of PRL-1 in wing vein formation. We
propose that the main function of PRL-1 in Drosophila wing development
is to affect the phospho-Mad levels and Smox transcriptional levels,
therefore influencing the competitive balance for Medea between Mad and Smox.
This study demonstrates a novel role for PRL-1 in regulating TGF-
YX, HZ and ZL designed the experiments and interpreted the data. HZ, ZL, XY (Xin Yuan) and HW contributed to experiment completion and data analysis. HZ, ZL and YX wrote the initial manuscript. YX and XY (Xiaohang Yang) revised the manuscript. All authors contributed helpful suggestions for this manuscript.
Not applicable.
We thank Chris Wood of the College of Life Sciences, Zhejiang University for checking the English of this manuscript.
This work was supported by National Key R&D Program of China (2018YFC1004900) and the National Basic Research Program of China (2013CB945600, X.Yang.), National Natural Science Foundation of China (31801230).
The authors declare no conflict of interest.
