1 Department of Women’s Health Care, Women and Children Healthcare Hospital of Zhuzhou, 412000 Zhuzhou, Hunan, China
2 Department of Gynecology, Zhuzhou Central Hospital, 412000 Zhuzhou, Hunan, China
3 Department of Laboratory, Zhuzhou Central Hospital, 412000 Zhuzhou, Hunan, China
Abstract
Deoxyribonucleic acid (DNA) damage repair pathways synergistically promote cervical carcinogenesis. The role of tripartite motif-containing 11 (TRIM11) in DNA repair may influence genomic stability in cervical cancer (CC) and modulate treatment response. This study aimed to analyze the expression and prognostic significance of TRIM11 in CC and across multiple cancer types (pan-cancer analysis).
TRIM11 expression patterns in CC were investigated through integrated bioinformatics analyses using two independent cohorts: transcriptomic data from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) dataset GSE67522. Experimental validation of TRIM11 overexpression in clinical CC specimens was performed through molecular techniques, including quantitative PCR and Western blotting. Survival outcomes were assessed using Kaplan-Meier method, revealing significant correlations between TRIM11 expression levels and both progression-free survival (PFS) and overall survival (OS) rates in TCGA CC cases. Functional pathway associations were elucidated through gene set enrichment analysis (GSEA), identifying TRIM11-related oncogenic mechanisms. Furthermore, a comprehensive pan-cancer evaluation employing TCGA multi-omics data systematically characterized the prognostic relevance of TRIM11 across diverse malignancies.
TCGA cohort analysis demonstrated a statistically significant elevation in TRIM11 expression levels in tumor tissues compared to normal controls (p < 0.0001), with consistent validation observed in the GSE67522 cohort (p < 0.0001). Molecular validation experiments confirmed concurrent upregulation of TRIM11 at both the transcriptional (quantitative reverse transcriptase PCR (qRT-PCR), p < 0.05) and proteomic (Western blot, p < 0.05) levels in CC tissues compared to paired adjacent normal samples. Notably, within the context of human papillomavirus (HPV) infection, the GSE67522 dataset highlighted the pivotal role of TRIM11 during malignant transformation, show a significant difference in expression between HPV-positive cancer tissues and matched normal cervical epithelia (p < 0.001). In the TCGA dataset, OS (p = 0.007; HR [high-expression group] = 1.899; 95% confidence interval [CI], 1.189–3.033) and PFS (p = 0.003; HR = 2.035; 95% CI, 1.266–3.273) were significantly longer in patients with CC with lower TRIM11 expression compared to those with higher TRIM11 expression. Subsequently, GSEA in the TCGA dataset showed that TRIM11 is involved in the transforming growth factor beta (TGF-β), calcium, wingless/integrated (WNT), and mitogen-activated protein kinase (MAPK) pathways in CC (p < 0.0001). Pan-cancer analysis showed that TRIM11 expression differed significantly between various tumor tissues and their corresponding normal tissues, and was closely associated with prognosis across several cancer types.
This study demonstrated that TRIM11 is overexpressed in CC, and that its overexpression is associated with poor prognosis. Furthermore, its expression was significantly correlated with prognosis across multiple cancers.
Keywords
- tripartite motif-containing 11
- prognosis
- cervical cancer
- pan-cancer
Cervical cancer (CC) remains a major global health burden, ranking as the fourth
most prevalent malignancy among women. Recent estimates indicate over 600,000 new
diagnoses and 340,000 fatalities in 2024, with disproportionate impacts on
resource-limited regions [1]. Histologically, squamous cell carcinoma dominates
(80% of cases), contrasting with adenocarcinomas (15–20%), which exhibit
divergent molecular pathogenesis [2, 3]. In China alone, 2021 recorded 132,788
incident cases and 49,841 deaths, underscoring its persistent public health challenge [4].
Key risk factors encompass early sexual debut, multiparity,
tobacco use, prolonged oral contraceptive intake, and persistent human
papillomavirus (HPV) infection [5, 6]. Prognosis starkly correlates with staging:
early lesions demonstrate 73–98% 5-year survival, plummeting to
Tripartite motif-containing 11 (TRIM11), an E3 ubiquitin ligase within the TRIM family, orchestrates ubiquitin-mediated protein turnover and immune modulation [10]. Beyond its canonical role in cellular homeostasis, this multifunctional protein intersects with oncogenesis, genetic disorders, and viral pathogenesis [11, 12, 13]. Emerging evidence implicates TRIM11 in tumor progression through post-translational regulation of critical signaling pathways, positioning it as a potential therapeutic target [10]. Dysregulated TRIM11 activates related signaling pathways, thus affecting the phenotype of tumor cells, and is closely associated with tumor development [14]. In lung cancer, TRIM11 expression is associated with tumor size, tumor node metastasis (TNM) stage, lymph node metastasis, and overall survival (OS) [15]. Therefore, we speculate that TRIM11 is a key gene contributing to tumor development in lung cancer. However, the role of TRIM11 in CC remains unclear.
In this study, we analyzed the expression and prognostic value of TRIM11 in CC and various other cancer types.
TRIM11 expression data and associated clinical information were retrieved from two publicly available databases: The Cancer Genome Atlas (TCGA, https://www.cancer.gov/ccg/research/genome-sequencing/tcga) and the GSE67522 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE67522), which provided comprehensive genomic and clinical profiles of cervical cancer patients. TRIM11 protein expressions and mRNA levels were derived from 4 cancer and 4 normal fresh tissues. The workflow is illustrated in Fig. 1.
 Fig. 1.
Fig. 1.
Workflow of the study. TCGA, The Cancer Genome Atlas; TRIM11, Tripartite motif-containing 11; GSEA, Gene set enrichment analysis; qRT-PCR, quantitative reverse transcriptase PCR; CESC, cervical squamous cell carcinoma.
The R package clusterProfiler (https://bioconductor.org/packages/clusterProfiler/) was used to analyze the expression of TRIM11 in CC in The Cancer Genome Atlas-Cervical Squamous Cell Carcinoma (TCGA-CESC) and GSE67522 datasets. The samples in both datasets were divided into high- and low-expression groups based on the median expression level of TRIM11. The survival status of the patients was obtained from the datasets. Subsequently, the prognostic value of TRIM11 in CC was validated in the TCGA-CESC dataset.
Gene set enrichment analysis (GSEA) was conducted using version 3.0 of the
software suite (Broad Institute, Inc., Cambridge, MA, USA, accessible at
http://software.broadinstitute.org/gsea). Samples were stratified into high- and
low-expression cohorts using the median TRIM11 expression level as the threshold
(
Four cervical cancer and four normal fresh tissues were collected and stored at –80 °C until subsequent quantitative reverse transcription PCR (qRT-PCR) and Western blotting analysis. All tissues were collected at Zhuzhou Central Hospital, Zhuzhou, Hunan, China, between June and July 2024. The protocol for tissue harvest was approved by the Ethics Committee of Zhuzhou Central Hospital (No. 20231046), and informed consent was obtained per Declaration of Helsinki guidelines from each eligible participant.
Total RNA was isolated using Trizol reagent (Invitrogen, Cat. #15596026,
Waltham, MA, USA) with homogenization via a KZ-III-FP homogenizer (Servicebio,
Wuhan, Hubei, China). Reverse transcription employed the PrimeScript RT Master
Mix (Takara, Cat. #RR036A, Dalian, Liaoning, China), followed by quantitative
PCR (qPCR) amplification on a QuantStudio 5 System (Thermo Fisher, Waltham, MA,
USA) with
| Gene name | Primer sequence (5′–3′) |
| H- |
GAGCACAGAGCCTCGCCTTT |
| H- |
TCATCATCCATGGTGAGCTGG |
| H-TRIM11-F | GAGAACGTGAACAGGAAGGAG |
| H-TRIM11-R | CCATCGGTGGCACTGTAGAA |
RT-PCR, reverse transcription PCR.
Fresh tissue samples (1–2 mL) were homogenized under cryogenic conditions using a high-throughput tissue homogenizer (KZIII-F, Sevier Biotechnology, Wuhan, Hubei, China) for total protein isolation. Protein concentration was determined via a BCA assay kit (G2026-1000T; Servicebio, Wuhan, Hubei, China). Electrophoresis was performed by loading 40 µg of protein per lane onto 10% Bis-Tris gels (Invitrogen, Waltham, MA, USA) under 120 V for 90 minutes. Separated proteins were electroblotted onto 0.45 µm polyvinylidene fluoride (PVDF) membranes (Millipore, IPVH00010, Burlington, MA, USA) at 25 V for 30 minutes using a semi-dry transfer system (Trans-Blot Turbo, Bio-Rad, Hercules, CA, USA). Membranes were sequentially incubated with a rabbit polyclonal anti-TRIM11 antibody (10851-1-AP, 1:1000, Proteintech, Rosemont, IL, USA) and HRP-Goat anti Rabbit secondary antibody (5220-0336, 1:50,000, Seracare, Milford, MA, USA). Chemiluminescent signals were captured using a JS-1070P imaging system, and band intensity was quantified with IPWIN 6.0 software (Computaleta srl., Milan, Italy).
The standardized pan-cancer datasets in the TCGA, TARGET (https://ocg.cancer.gov/programs/target), and GTEx (https://gtexportal.org/home/) databases (Pan cancer (PANCAN), N = 19,131, G = 60,499) were downloaded from UCSC Xena (https://xenabrowser.net/). The expression data of TRIM11 were extracted from 34 cancer types (Table 2). In addition, TRIM11 expression profiles corresponding to the survival status were obtained from all pan-cancer datasets. Patients in all datasets were divided into high- and low-expression groups based on the median expression level of TRIM11 (cutoff value = 50%). Finally, Kaplan-Meier analysis was performed using the R packages “survminer” and “survival”.
| Cancer types | Abbreviations |
|---|---|
| Adrenocortical carcinoma | TCGA-ACC |
| Bladder urothelial carcinoma | TCGA-BLCA |
| Breast invasive carcinoma | TCGA-BRCA |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma | TCGA-CESC |
| Cholangiocarcinoma | TCGA-CHOL |
| Colon adenocarcinoma | TCGA-COAD |
| Colon adenocarcinoma/rectum adenocarcinoma esophageal carcinoma | TCGA-COADREAD |
| Esophageal carcinoma | TCGA-ESCA |
| Glioblastoma multiforme | TCGA-GBM |
| Glioma | TCGA-GBMLGG |
| Head and neck squamous cell carcinoma | TCGA-HNSC |
| Kidney chromophobe | TCGA-KICH |
| Pan-kidney cohort (KICH+KIRC+KIRP) | TCGA-KIPAN |
| Kidney renal clear cell carcinoma | TCGA-KIRC |
| Kidney renal papillary cell carcinoma | TCGA-KIRP |
| Acute myeloid leukemia | TCGA-LAML |
| Brain lower grade glioma | TCGA-LGG |
| Liver hepatocellular carcinoma | TCGA-LIHC |
| Lung adenocarcinoma | TCGA-LUAD |
| Lung squamous cell carcinoma | TCGA-LUSC |
| Ovarian serous cystadenocarcinoma | TCGA-OV |
| Pancreatic adenocarcinoma | TCGA-PAAD |
| Pheochromocytoma and paraganglioma | TCGA-PCPG |
| Prostate adenocarcinoma | TCGA-PRAD |
| Rectum adenocarcinoma | TCGA-READ |
| Stomach adenocarcinoma | TCGA-STAD |
| Skin cutaneous melanoma | TCGA-SKCM |
| Stomach and esophageal carcinoma | TCGA-STES |
| Testicular germ cell tumors | TCGA-TGCT |
| Thyroid carcinoma | TCGA-THCA |
| Uterine corpus endometrial carcinoma | TCGA-UCEC |
| Uterine carcinosarcoma | TCGA-UCS |
| Acute lymphoblastic leukemia | TARGET-ALL |
| High-risk Wilms tumor | TARGET-WT |
Statistical analyses were conducted using R software version 4.0.3 (R Foundation
for Statistical Computing, 2020, Vienna, Austria). Data visualization was
performed using GraphPad Prism version 9.3.1 (GraphPad Software, Inc., San Diego,
CA, USA), continuous variables are presented as mean
TRIM11 was found to be overexpressed in CC tissues in the TCGA-CESC and GSE67522
datasets (p
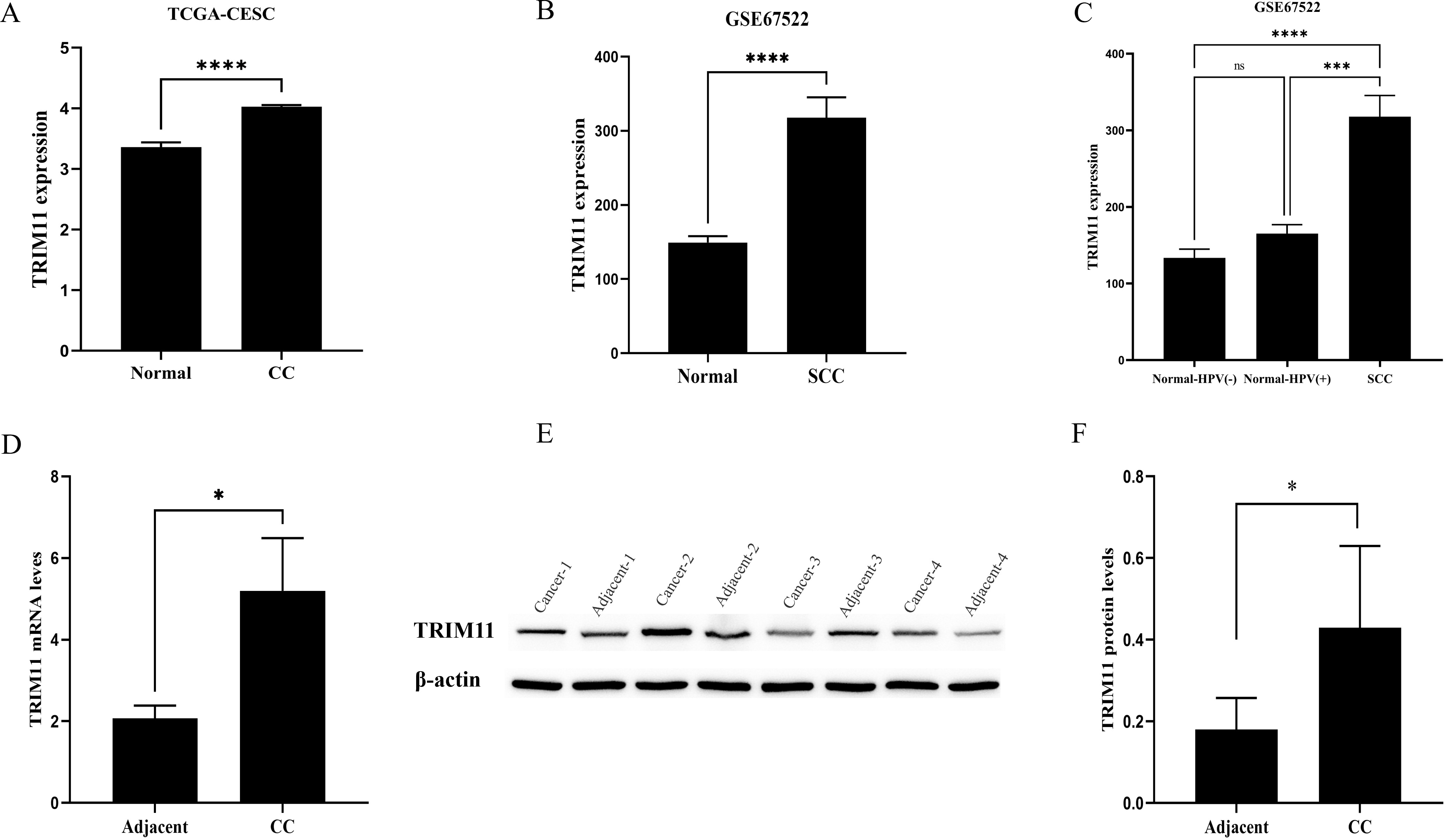 Fig. 2.
Fig. 2.
Expression of TRIM11 in cervical cancer (CC). (A,B) TRIM11 was
overexpressed in CC tissues in the TCGA-CESC and GSE67522 datasets. (C) TRIM11
expression in human papillomavirus (HPV)-positive CC and normal cervical tissues.
(D–F) mRNA and protein expression levels of TRIM11 in CC and adjacent normal
tissues. *, p
In the TCGA-CESC dataset, OS and progression-free survival (PFS) were significantly longer in patients with CC in the low-TRIM11-expression group than in those in the high-TRIM11-expression group (p = 0.007, HR = 1.899, 95% CI [1.189–3.033] and p = 0.003, HR = 2.035, 95% CI [1.266–3.273]; Fig. 3A,B). The area under the receiver operating characteristic (ROC) curve (AUC) at 1, 3, and 5 years was 0.56 (p = 0.048, 95% CI, 0.44–0.68), 0.59 (p = 0.035, 95% CI, 0.50–0.69), and 0.67 (p = 0.012, 95% CI, 0.57–0.78) for OS, respectively, and 0.56 (p = 0.041, 95% CI, 0.47–0.66), 0.61 (p = 0.031, 95% CI, 0.52–0.70), and 0.65 (p = 0.022, 95% CI, 0.54–0.76) for PFS, respectively (Fig. 3C,D).
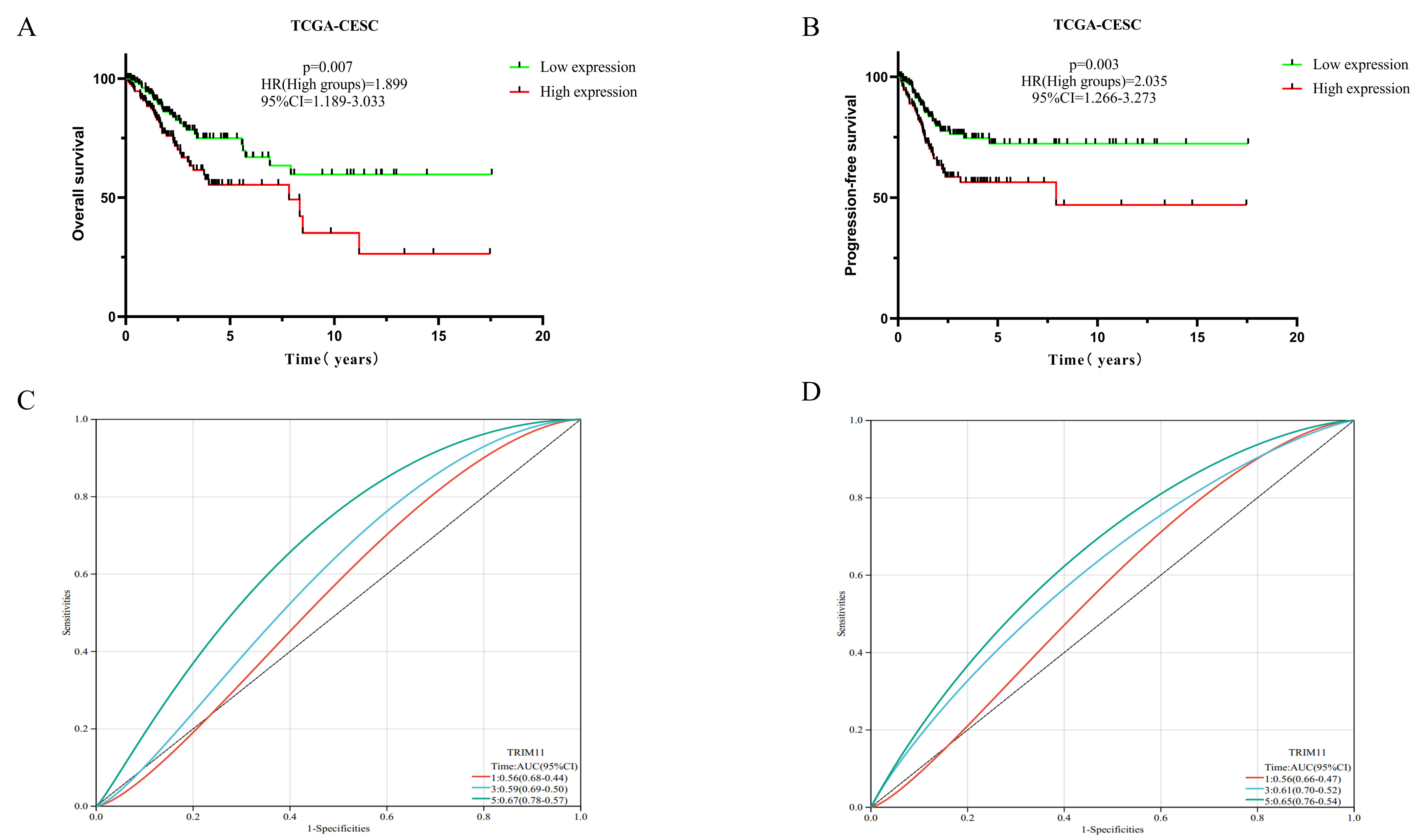 Fig. 3.
Fig. 3.
Effects of TRIM11 on the prognosis of CC. (A,B) The expression of TRIM11 affected OS and PFS in CC. (C,D) ROC curves demonstrating the specificity and sensitivity of TRIM11 in predicting the 1-, 3-, and 5-year survival probabilities in CC. ROC, receiver operating characteristic; AUC, area under the ROC curve; OS, overall survival; PFS, progression-free survival.
GSEA in the TCGA-CESC dataset showed that TRIM11 was significantly associated with various gene sets (Fig. 4), including Vascular-Smooth-Muscle-Contraction (enrichment score (ES) = 0.5834, nominal p-value (NP) = 0.0000), Tgf-Beta-Signaling-Pathway (ES = 0.5727, NP = 0.0000), Melanoma (ES = 0.5588, NP = 0.0000), Prostate-Cancer (ES = 0.5431, NP = 0.0000), Calcium-Signaling-Pathway (ES = 0.5172, NP = 0.0000), Endometrial-Cancer (ES = 0.5158, NP = 0.0000), Melanogenesis (ES = 0.4966, NP = 0.0000), WNT-Signaling-Pathway (ES = 0.4598, NP = 0.0000), Pathways-In-Cancer (ES = 0.4337, NP = 0.0000), and MAPK-Signaling-Pathway (ES = 0.4322, NP = 0.0000).
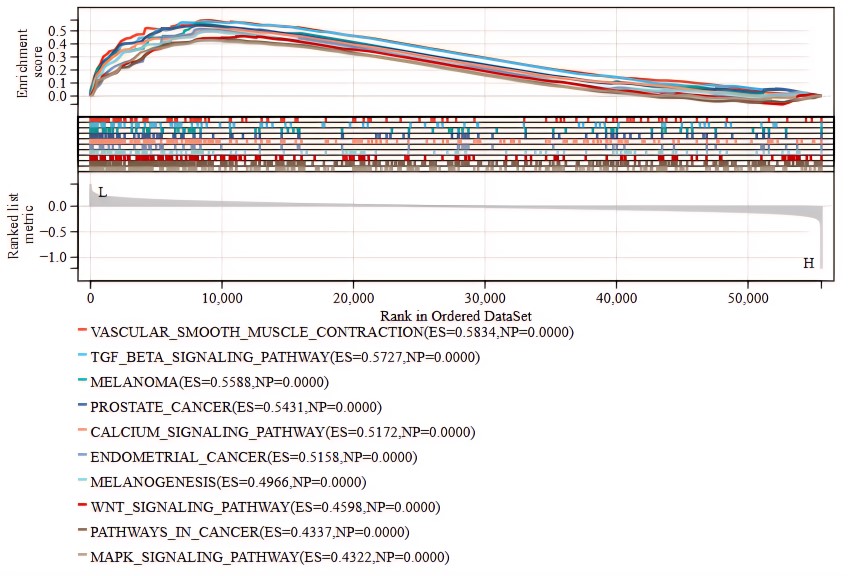 Fig. 4.
Fig. 4.
Gene set enrichment analysis of TRIM11 in CC in the TCGA-CESC dataset. ES, enrichment score; NP, nominal p-value.
No differences were observed in TRIM11 expression among the TCGA-GBM,
TCGA-GBMLGG, TCGA-LGG, TCGA-KIRC, and TCGA-PCPG datasets. However, TRIM11
expression was significantly different in other cancer types. For instance, it
was low in the TCGA-THCA, TCGA-TGCT, and TCGA-KICH datasets (Fig. 5A). Low
expression of TRIM11 was correlated with a poor prognosis in the TCGA-GBMLGG (N =
619, p = 1.1
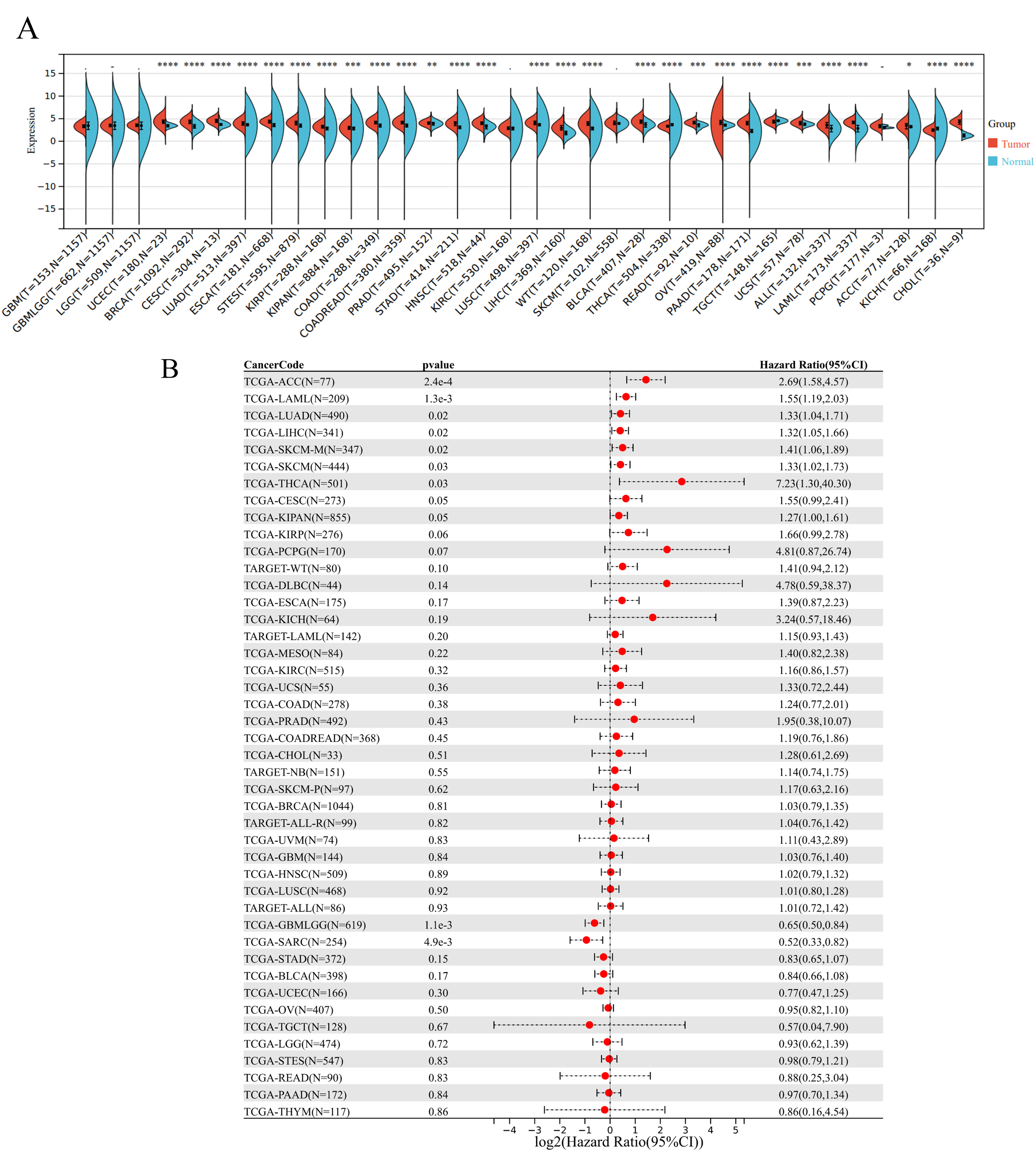 Fig. 5.
Fig. 5.
Pan-cancer analysis of the expression level and prognostic value
of TRIM11. (A) Pan-cancer expression of TRIM11 (*, p
| Cancer code | p-value | Hazard ratio (95% CI) |
|---|---|---|
| TCGA-ACC (N = 77) | 2.4 × 10-4 | 2.69 (1.58, 4.57) |
| TCGA-LAML (N = 209) | 1.3 × 10-3 | 1.55 (1.19, 2.03) |
| TCGA-LUAD (N = 490) | 0.02 | 1.33 (1.04, 1.71) |
| TCGA-LIHC (N = 341) | 0.02 | 1.32 (1.05, 1.66) |
| TCGA-SKCM (N = 444) | 0.03 | 1.33 (1.02, 1.73) |
| TCGA-THCA (N = 501) | 0.03 | 7.23 (1.30, 40.30) |
| TCGA-CESC (N = 273) | 0.007 | 1.89 (1.19, 2.03) |
| TCGA-GBMLGG (N = 619) | 1.1 × 10-3 | 0.65 (0.50, 0.84) |
In recent years, the average age of onset for CC has gradually decreased, with the incidence of CC showing an increasing trend in younger age groups [16]. The development of CC can be effectively controlled through the early examination and prompt treatment of precancerous lesions [17]. Therefore, identifying novel biomarkers closely related to the development of CC is necessary.
As a pivotal member of the TRIM family, TRIM11 has emerged as a promising
diagnostic biomarker across malignancies [18]. Our investigation revealed
pronounced TRIM11 overexpression in cervical cancer (CC) tissues, correlating
with extended PFS and OS in affected patients. Complementary evidence from Zhang
et al. [19] demonstrated elevated TRIM11 expression in SiHa and HeLa CC
cell lines, where genetic silencing substantially attenuated cellular
proliferation, migration, and invasion—underscoring its therapeutic potential.
Mechanistic studies implicate TRIM11 in protein kinase B (AKT) pathway
hyperactivation, thereby driving CC progression. This aligns with our
observations of concordant TRIM11 upregulation at both transcriptomic (mRNA) and
proteomic levels in clinical CC specimens. Notably, while TRIM proteins are
recognized for their antiviral roles [20], TRIM11 exhibits paradoxical
immunomodulatory functions. Interferons (IFNs), central to antiviral defense and
cellular regulation [21], are counterintuitively suppressed by TRIM11 via
inhibition of interferon-beta (IFN-
GSEA showed that TRIM11 was closely related to the TGF-
This investigation corroborates prior findings demonstrating tumor-specific
TRIM11 dysregulation across malignancies, with marked expression disparities
between neoplastic and matched normal tissues. Accumulating evidence positions
TRIM11 as a pan-cancer prognostic modulator, exhibiting tissue-specific oncogenic
behaviors: Lung Cancer: elevated TRIM11 levels correlate with advanced TNM
staging, tumor dimensions, lymph node involvement, and reduced overall survival
[26]. Breast Cancer: TRIM11 upregulation drives unchecked cellular proliferation
through cell cycle deregulation [27]. Thyroid Cancer: in anaplastic thyroid
carcinoma (ATC), TRIM11 silencing attenuates proliferative/migratory capacities
and enhances chemosensitivity [28]. Glioma: high-grade gliomas exhibit
TRIM11-driven malignant progression via enhanced invasion and treatment
resistance, particularly temozolomide (TMZ) in glioblastoma (GBM) [29].
Hepatocellular Carcinoma (HCC): pathological TRIM11 overexpression sustains HCC
aggressiveness, whereas its suppression impedes proliferation and metastatic
potential [24]. These collective findings establish TRIM11 as a multifaceted
therapeutic target, with mechanistic diversity spanning proliferation modulation,
treatment resistance, and metastatic programming. In addition, TRIM11 was found
to inhibit the PI3K/AKT signaling pathway in HCC cells. These findings suggest
that TRIM11 plays an important role in the progression of HCC and serves as a
promising therapeutic target for this disease. In this study, we found that in
TCGA-GBMLGG and TRIM11 exhibited HRs
However, this study has its limitations: (1) small sample size: the study used
limited cervical cancer samples from public datasets and only validated results
in four tissue pairs, which may reduce reliability. (2) Lack of mechanism
validation: while TRIM11 was linked to key pathways (such as TGF-
TRIM11 is overexpressed in cervical cancer and linked to poor survival,
potentially driving progression via pathways like TGF-
Data related to this study are available by requesting the corresponding author via e-mail.
GZ, HC, and YT designed the research study. GZ performed the research. HC and YT provided help and advice on the qRT-PCR Western blotting experiments. HC analyzed the data. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
The study was conducted in accordance with the Declaration of Helsinki. Formal ethical authorization for this investigation was secured from the Institutional Review Board of Zhuzhou Central Hospital (Ethics Approval Code: 20231046). Prior to enrollment, all eligible individuals provided documented informed consent in accordance with institutional protocols.
Not applicable.
This study was financially supported by the Natural Science Foundation of Hunan Province (Grant No.: 2025JJ70023) and Social Investment Project of Zhuzhou City in 2024 (No.: 2024-63).
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.





