1 Department of General Gynecology, Changzhou Maternity and Child Health Care Hospital, Changzhou Medical Center, Nanjing Medical University, 213000 Changzhou, Jiangsu, China
Abstract
The molecular mechanisms of intrauterine adhesions (IUA) are not yet fully understood, and there is a lack of specific diagnostic markers and effective molecularly targeted treatments in clinical practice. This study employed proteomic techniques to analyze differentially expressed proteins (DEPs) and associated signaling pathways in pathological tissues, aiming to identify potential diagnostic markers and therapeutic targets.
This study collected 15 endometrial tissue samples from 10 patients treated at Changzhou Maternity and Child Health Care Hospital from March 2022 to September 2022. The tissue samples were divided into 3 groups: the adhesion group (Adhes group), the peri-adhesion endometrial group (Endome group), and the control group (Control group). Pairwise comparisons of the three groups were performed. Label-free quantitative (LFQ) proteomics was used to identify DEPs, and bioinformatics analyses, such as the Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), and protein domain analysis, were employed to identify the functions and pathways of DEPs. Parallel reaction monitoring (PRM) was used for the quantitative analysis of selected target proteins.
1328, 290, and 1335 DEPs were found in the Adhes vs Control, Endome vs Control, and Adhes vs Endome groups, respectively. Bioinformatics analysis showed that these proteins are primarily involved in key processes such as muscle contraction, cytoskeletal dynamics balance, extracellular matrix (ECM) remodeling, and immune regulation. PRM validation identified out 14 target proteins, among which ADIPOQ, TGFB1, MYLK, and CAMK2G were closely associated with ECM remodeling and cytoskeletal regulation, while lactoferrin (LTF) was involved in immune regulation.
This study identified a series of key proteins associated with IUA and found that they may participate in the disease process through mechanisms such as ECM remodeling, actin cytoskeleton regulation, and immune regulation. The identified target proteins (such as ADIPOQ, TGFB1, MYLK, and CAMK2G) provide potential biomarkers and intervention targets for the diagnosis and treatment of IUA.
Keywords
- intrauterine adhesions
- label-free quantitative proteomics
- parallel reaction monitoring
- differentially expressed proteins
- endometrium
Intrauterine adhesions (IUA), or Asherman syndrome, is characterized by partial or complete endometrial damage leading to adhesions within the uterine cavity or cervical canal. The primary causes are invasive curettage and endometritis [1]. IUA disrupts intrauterine blood supply and angiogenesis, resulting in endometrial fibrosis, atrophy, and the formation of avascular fibrous tissue [2, 3], which reduces the area for embryo implantation and impairs endometrial receptivity [4]. Clinically, IUA presents with decreased menstrual flow, amenorrhea, recurrent miscarriage, secondary infertility, and, in some cases, periodic lower abdominal pain or postpartum complications such as placenta accreta and placental adhesion [5, 6, 7]. The current gold standard for the diagnosis of IUA remains hysteroscopy. The sensitivity and specificity of imaging examinations are limited, and they are prone to overlooking patients with mild adhesions [8]. The treatment of IUA consisting of transcervical resection of adhesion (TCRA) has limitations, which includes a high recurrence rate and a tendency to exacerbate endometrial fibrosis [9]. Moreover, the application of anti-adhesion materials after surgery cannot reverse endometrial fibrosis. Therefore, there is an urgent need to find new ways to more effectively and safely treat IUA. Biomarkers can provide sensitive and highly specific diagnostic tests. More importantly, some biomarkers may serve as therapeutic targets. Regulating key molecules and signaling pathways will help to develop novel therapeutic strategies.
Proteomics can detect the changes in expression of differentially expressed proteins (DEPs) between diseases and normal samples, and obtain the signaling pathways and biological processes of diseases through bioinformatics analysis [10]. Compared with traditional isotope-labeled quantitative proteomics, label-free quantitative (LFQ) proteomics have no limit to the number of samples and do not require expensive stable isotope labels for internal standards. LFQ have a high depth of analysis and dynamic ranges, which can make timely quantification of all protein changes amongst different groups [11, 12, 13]. Parallel Reaction monitoring (PRM) is a proteomic method based on high-resolution mixed mass spectrometry for highly selective quantification of target proteins and target peptides, allowing simultaneous identification of multiple target proteins [14]. There have been few specific proteomics in IUA. In this study, we explored the pathogenesis of IUA at the protein level, hoping to provide a theoretical basis for further study of the pathogenesis of IUA and potential targets for the treatment of IUA.
We selected 5 patients with IUA confirmed by hysteroscopic examination, who were admitted to the Changzhou Maternity and Child Health Care Hospital from March 2022 to September 2022, aged 20–40, who were fertile, and had not taken any estrogen-progestogen hormones within the 6 months prior to sampling and 5 patients who could provide normal endometrial tissue due to uterine fibroids (the presence of intramural or subserous fibroids would not affect the endometrium). The endometrial adhesion tissue of IUA patients was selected as Adhes group, the endometrial tissue 2 cm away from the adhesion tissue was selected as Endome group, and the endometrial tissue of uterine fibroids patients was selected as Control group. We performed pairwise comparisons, divided into three comparison groups: Adhes/Control, Endome/Control, Adhes/Endome group. All specimens were collected 3–7 days after menstruation and confirmed by pathological examination. The study was approved by the Ethics Committee of Changzhou Maternity and Child Health Care Hospital (approval number 2021[88]); Patients were informed and enrolled after obtaining consent and signing written informed consent. Patients with connective tissue diseases, endometrial diseases and endocrine diseases were excluded. All patients underwent routine examinations before surgery, and age, Body Mass Index (BMI), and Hemoglobin (Hb) were recorded. The collected specimens were stored in a –80 ℃ freezer.
Sample were removed and ground into a fine powder. Lysis buffer containing Triton X-100 (Sigma Sangon Biotech, Shanghai, China) and a protease inhibitor cocktail (Merck Millipore, Darmstadt, Germany) was added to the powder, followed by ultrasonication for lysis. The lysate was centrifuged at low temperature, and the supernatant was collected for protein concentration determination using a bicinchoninic acid (BCA) kit (Beyotime, Shanghai, China). A total of 300 µg of protein from each sample was used for enzymatic hydrolysis. Trichloroacetic acid (TCA) (Sigma-Aldrich, St. Louis, MO, USA) was added to each sample to a final concentration of 20%, followed by precipitation and centrifugation to remove the supernatant. The resulting precipitate was washed with pre-cooled acetone (Hangzhou Hanno Chemical, Hangzhou, Zhejiang, China) and air-dried. The dried precipitate was resuspended in 200 mM TEAB (Sigma-Aldrich), sonicated to ensure dispersion, and digested overnight with trypsin (Promega, Madison, WI, USA). The next day, dithiothreitol (DTT) (Sigma-Aldrich) was added and allowed to react for 30 minutes, followed by incubation with iodoacetamide (IAM) (Sigma-Aldrich) for 15 minutes at room temperature in the dark.
The resulting peptides were dissolved in solvent A (0.1% formic acid (Fluka, Buchs, Switzerland), 2% acetonitrile (Thermo Fisher Scientific, Waltham, MA, USA) in water) and separated using an UltiMate 3000 UHPLC system (Thermo Fisher Scientific). The peptides were then ionized using a nanospray ionization (NSI) source and introduced into an Orbitrap Exploris™ 480 mass spectrometer (Thermo Fisher Scientific) for tandem mass spectrometry (MS/MS). The ion source voltage was set to 2.0 kV, and the High-Field Asymmetric Waveform Ion Mobility Spectrometry (FAIMS) compensation voltages (CV) were set to –45 V and –65 V. A high-resolution Orbitrap analyzer was used to detect and analyze the precursor ions of the peptides and their fragment ions. The primary mass spectrometry (MS) scan range was 400–1200 m/z with a resolution of 60,000, while the MS/MS scan range was fixed at 110 m/z with a resolution of 15,000. Data acquisition was performed using a data-dependent acquisition (DDA) mode. The automatic gain control (AGC) was set to 1
The resulting MS/MS data were processed using a Proteome Discoverer (v2.4.1.15) (https://www.thermofisher.cn/order/catalog/product/cn/zh/CSW0064764). The reverse decoy database was added to calculate the false discovery rate (FDR) due to random matching. The FDR for protein, peptide, and propensity score method (PSM) identification was set to 1%. Fisher’s exact test and Pearson’s repeatability test were used to detect and analyze DEPs. A p-value less than 0.05 was considered statistically significant. Proteins with variation in differential expression
The gene ontology (GO) analysis was divided into three categories: biological process, molecular function, and cellular component, to explain the biological role of proteins from different perspectives. The GO annotation is to annotate the identified proteins using the eggnog-mapper software (v2.0) (Heidelberg, Germany) (http://eggnog-mapper.embl.de/) based on the fifth edition of the EggNOG database. The GO ID in each protein annotation result is extracted, and then the proteins are functionally classified according to cellular components, molecular functions, and biological processes. The protein domains are certain components that recur in different protein molecules with similar sequences, structures, and functions and are the units of protein evolution. The Pfam database [15] was used to analyze the enrichment of functional domains of DEPs. Fisher’s exact test method was used to test the significance of domain enrichment of DEPs in the context of identified proteins, and a p-value
The selected target proteins were verified by the PRM analysis. The MS/MS data were retrieved by Maxquant (v1.6.15.0) (https://www.maxquant.org/). The resulting MS data were processed using Skyline (v.3.6) (https://skyline.ms/). Peptide settings: the enzyme was set as Trypsin [KR/P], max missed cleavage set as 2. The peptide length was set as 8–25, Variable modification was set as Carbamidomethyl on Cys and oxidation on Met. Max variable modifications were set as 3. Transition settings: precursor charges were set as 2, 3, ion charges were set as 1, 2, ion types were set as b, y, p. The product ions were set as from ion 3 to last ion, the ion match tolerance was set as 0.02 Da. The above experiments were performed in the Jingjie PTM BioLab Co., Ltd (Hangzhou, Zhejiang, China).
Continuous variables were expressed as the mean
The study included 15 endometrial samples from 5 IUA patients and 5 control patients, and there were no significant differences in terms of age, BMI, and preoperative Hb values among the three groups (p
| Adhes/Endome Group | Control Group | p | |
| Age (years) * | 29.60 | 32.60 | 0.557 |
| BMI (kg/m2) & | 20.83 (3.35) | 23.44 (2.37) | 0.117 |
| Hb (g/L) * | 133.00 | 129.80 | 0.558 |
*Data shown as mean
15 endometrial tissues among 3 groups were performed using LFQ proteomic analysis between the two comparison groups, respectively. Based on 64,325 independent peptides, 7529 proteins were identified after 5 biological duplications. We defined the DEPs as changes with a 1.5-fold up- or down-regulation and a p-value less than 0.05. 789 upregulated DEPs, and 539 downregulated DEPs were found in the Adhes/Control group; 587 upregulated DEPs, and 748 downregulated DEPs were found in the Endome/Adhes group; and 178 upregulated DEPs, and 112 downregulated DEPs were found in the Endome/Control group (Fig. 1A); The samples within the same group were clustered more consistently on the heat map, and there were obvious differences in the expression characteristics between different groups (Fig. 1B). In Fig. 2, it can be seen that the differential expression of proteins in the Adhes/Control group was the most significant.
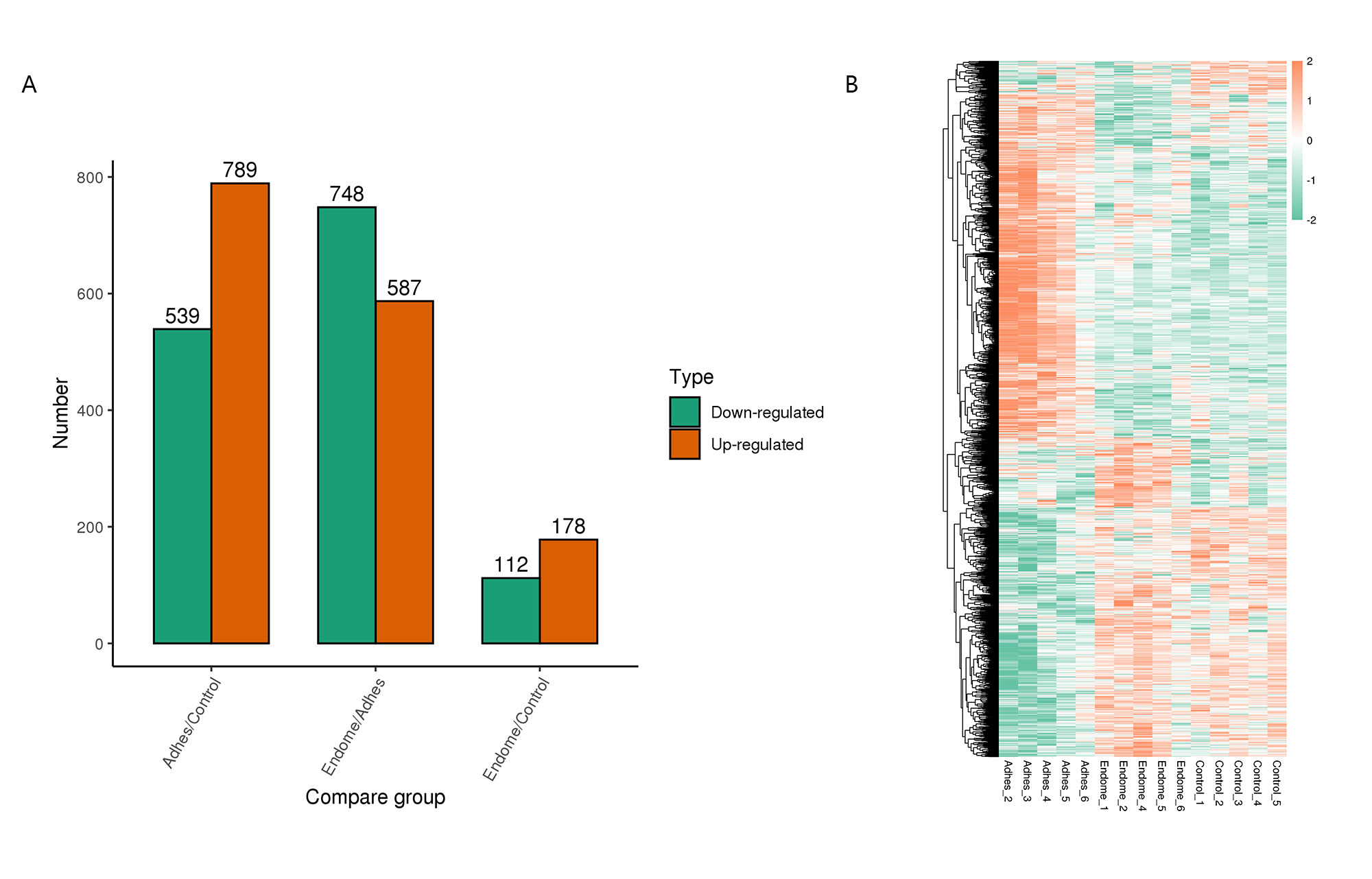 Fig. 1.
Fig. 1. Differential protein analysis of three comparison groups. (A) The green represents down-regulated proteins, while the orange represents the up-regulated proteins. (B) Heat map analysis of differentially expressed proteins (DEPs) in each sample.
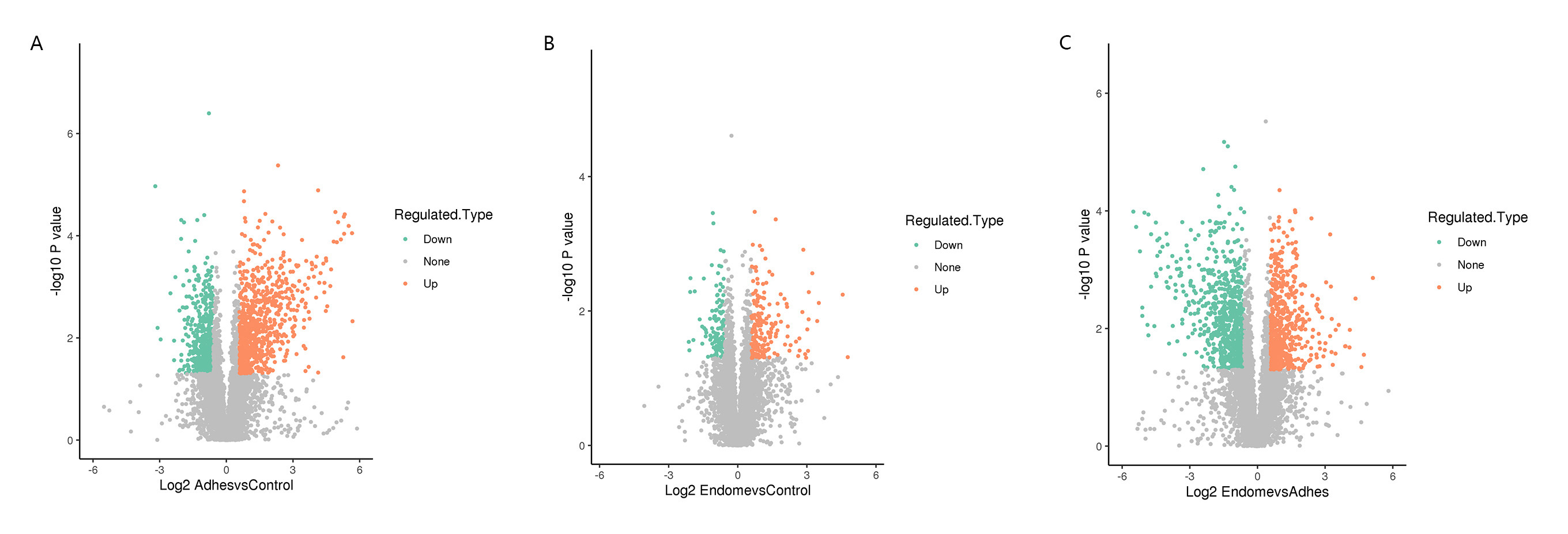 Fig. 2.
Fig. 2. The volcano plot of the DEPs among three comparison groups. Down-regulated proteins were labeled green, and the orange dots represented up-regulated proteins. The gray dots represented proteins with no significant differences. (A) Volcano plot of DEPs in Adhes/Control group. (B) Volcano plot of DEPs in Endome/Control group. (C) Volcano plot of DEPs in Endome/Adhes group.
In order to better understand the biological significance of DEPs, we performed GO enrichment analysis. In the Adhes/Control comparison group, the most significant biological processes were regulation of muscle contraction, extracellular matrix (ECM) organization, and cell junction assembly (Fig. 3A). Similarly, the DEPs mainly enriched in biological processes such as the regulation of transmembrane transport, actin cytoskeleton organization, and regulation of muscle contraction in the Endome/Adhes group (Fig. 3G). In the Endome/Control group, DEPs were mostly enriched in oxygen transport, defense response to fungus, and antimicrobial humoral response on the ontology of biological process (Fig. 3D).
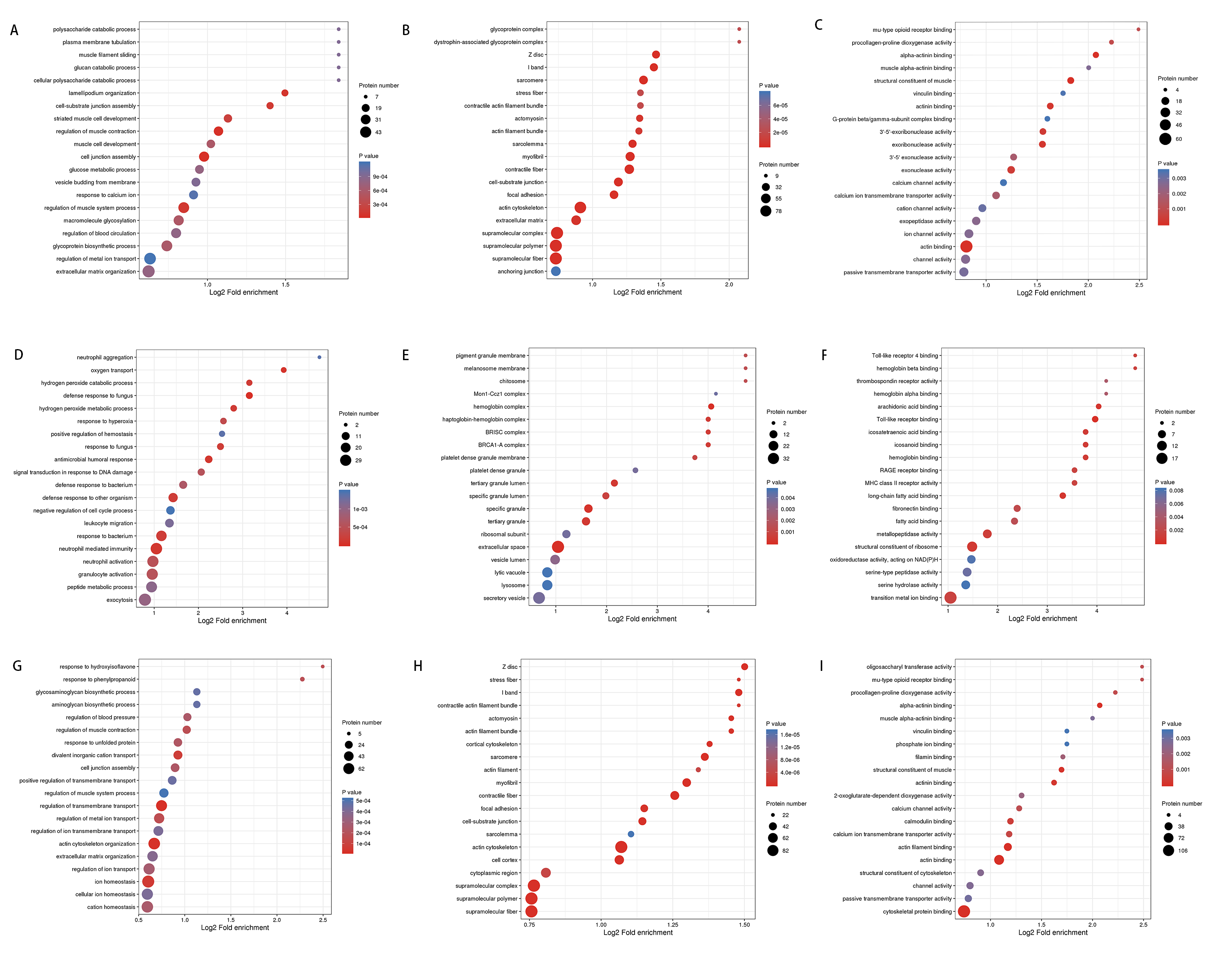 Fig. 3.
Fig. 3. The bubble chart of the gene ontology (GO) enrichment analysis of DEPs in each compare group. The color of the circle represents the p-value, and the size of the circle represents the number of DEPs. (A) Enrichment analysis of biological processes in Adhes/Control group. (B) Enrichment analysis of Cell component in Adhes/Control group. (C) Enrichment analysis of Molecular function in Adhes/Control group. (D) Enrichment analysis of biological processes in Endome/Control group. (E) Enrichment analysis of Cell component in Endome/Control group. (F) Enrichment analysis of Molecular function in Endome/Control group. (G) Enrichment analysis of biological processes in Endome/Adhes group. (H) Enrichment analysis of Cell component in Endome/Adhes group. (I) Enrichment analysis of Molecular function in Endome/Adhes group.
In the Adhes/Control and Endome/Adhes groups, most of the DEPs were concentrated in the ontology of cellular components, such as focal adhesion, cell-substrate junction, and I band (Fig. 3B,H). Among them, focal adhesion was one of the most significant enrichment KEGG pathways of DEPs. For the Endome/Control group, the key cellular components of the DEPs were the hemoglobin complex, tertiary granule lumen, and specific granule (Fig. 3E).
There were several similar enrichments between the Adhes/Control and Endome/Adhes groups in the molecular function, for example, alpha-actinin binding, structural constituent of muscle, and actinin binding (Fig. 3C,I). In the Endome/Control group, the essential molecular functions were Toll-like receptor binding, major histocompatibility complex (MHC) class II receptor activity, and arachidonic acid binding (Fig. 3F).
Bioinformatics analysis demonstrated that the first 3 structural domains of the differential proteins are Lin-11, Isl1, MEC-3 (LIM) domain, calponin homology (CH) domain, the 2OG-Fe (II) oxygenase superfamily in the Adhes/Control and the Endome/Adhes group. The master protein domain was enriched in Globin, the putative peptidoglycan binding domain, and the ferritin-like domain in the Endome/Control group (Fig. 4).
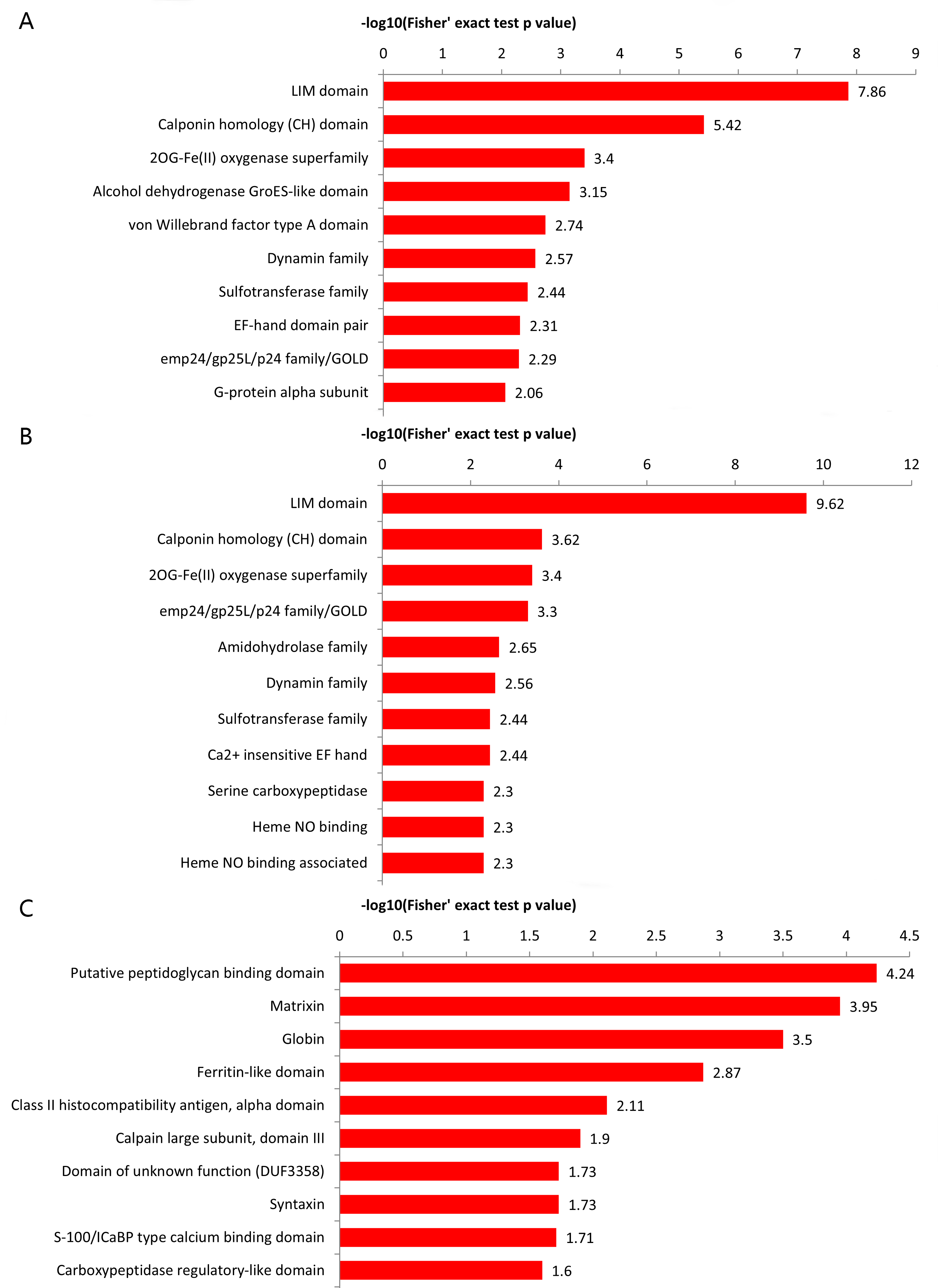 Fig. 4.
Fig. 4. The top 10 protein domain enrichment analysis of DEPs. (A) Protein domain enrichment analysis of Adhes/Control group. (B) Protein domain enrichment analysis of Endome/Adhes group. (C) Protein domain enrichment analysis of Endome/Control group.
We performed cluster analysis of KEGG pathways after enriched analysis to discover the metabolic pathways, biochemical reactions, and signal transduction processes. A cross-sectional comparison of the three groups found that the DEPs had multiple similar KEGG enrichment pathways between the Adhes/Control and the Endome/Adhes groups. In addition to the above focal adhesion, the DEPs was also enriched in regulation of actin cytoskeleton, the cGMP-PKG signaling pathway and the oxytocin signaling pathway. For the Adhes/Control group, the central pathways were the apelin signaling pathway, vascular smooth muscle contraction, and cholinergic synapse. ECM-receptor interaction and protein processing in endoplasmic reticulum were found to be enriched in most proteins of the Endome/Adhes group. The Endome/Control group primarily functioned in the staphylococcus aureus infection, hematopoietic cell lineage and homologous recombination (Fig. 5).
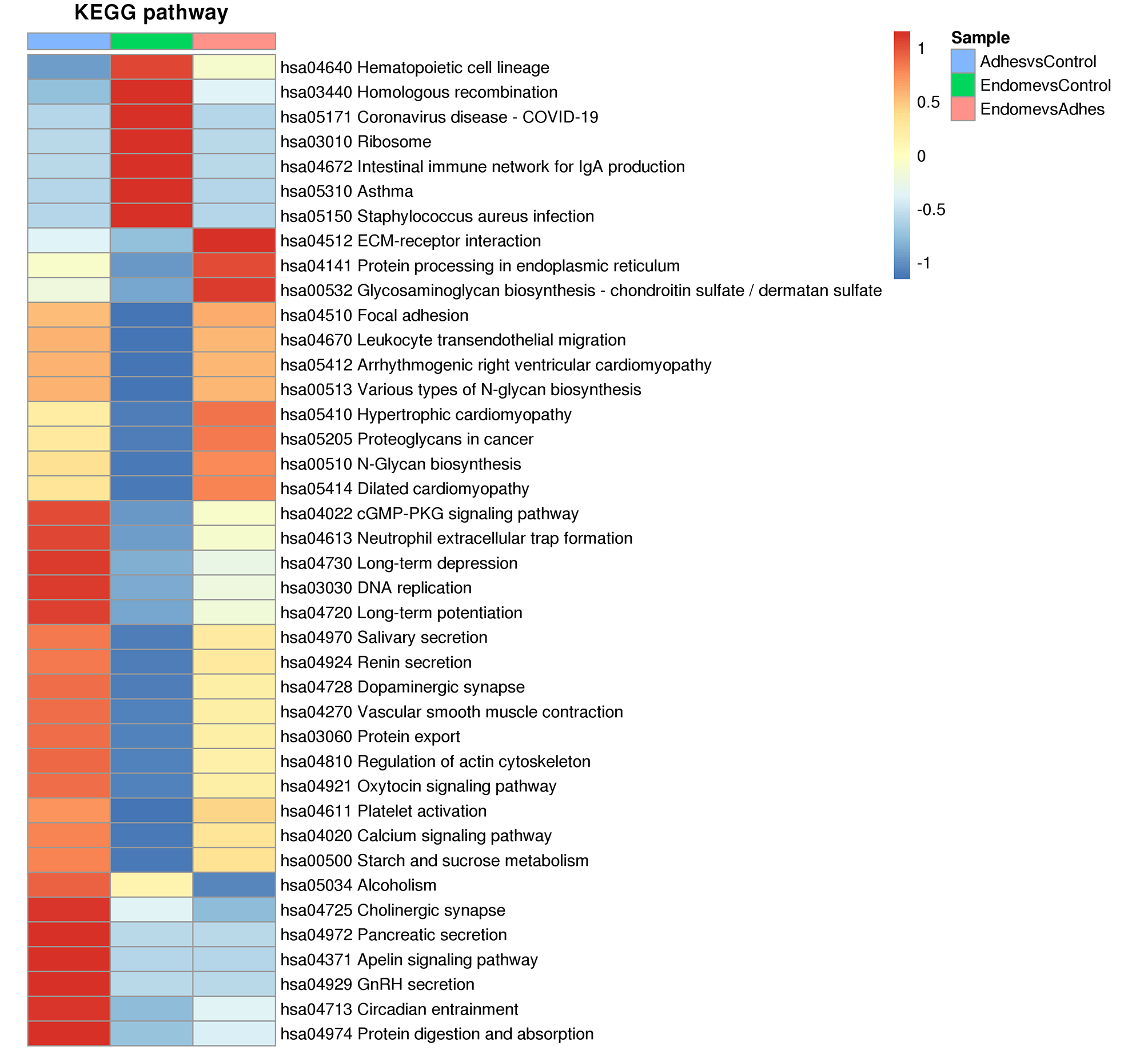 Fig. 5.
Fig. 5. Hierarchical cluster analysis heatmap reveals the DEPs involved in multiple important biological pathways. The color blocks corresponding to the functional description of the enrichment of DEPs in different comparison groups indicate the degree of enrichment. Red represents strong enrichment and blue represents weak enrichment. ECM, extracellular matrix; GnRH, gonadotropin-releasing hormone.
To verify the above proteomic data results, we selected DEPs with lower p-values and higher enrichment multiples, and many specific peptide segments, which are also functional proteins or transcription factors, but do not include cytoskeletal proteins, to pick up to 20 target proteins. Subsequently, 1–2 specific peptide segments were selected to quantify the 20 target proteins using the method of fragment ion peak area. Finally, 14 proteins were successfully detected and quantified (ADIPOQ, ABHD6, CAMK2G, IRAG1, MYLK, SYNPO2, PTPN11, PROS1, SLC4A1, TGFB1, HLA-DRB3, COQ6) (Table 2) through the peak area. The rest of the data is available in Fig. 6. These target proteins were with high fold change and low p-value and most of them have significant enrichment in certain KEGG pathways. The results are in high agreement with those of LFQ proteomics, which appear to serve as reliable confirmations of protein changes.
| Protein accession | Gene name | Protein description | p-value | Ratio | |
| PRM | LFQ | ||||
| Q9Y2Z9 | COQ6 | Ubiquinone biosynthesis monooxygenase COQ6, mitochondrial | 2.63 × 10–2 | 0.51 | 0.34 |
| Q06124 | PTPN11 | Tyrosine-protein phosphatase non-receptor type 11 | 1.08 × 10–2 | 1.12 | 20.59 |
| P01137 | TGFB1 | Transforming growth factor beta-1 | 3.87 × 10–2 | 2.36 | 26.25 |
| P79483 | HLA-DRB3 | HLA class II histocompatibility antigen, DR beta 3 chain | 1.07 × 10–2 | 3.57 | 3.13 |
| P07225 | PROS1 | Vitamin K-dependent protein | 2.77 × 10–5 | 3.70 | 4.91 |
| Q9UMS6 | SYNPO2 | Synaptopodin-2 | 3.81 × 10–2 | 4.65 | 4.59 |
| Q9Y6F6 | IRAG1 | Inositol 1,4,5-triphosphate receptor-associated 1 | 5.22 × 10–3 | 5.96 | 4.22 |
| Q15848 | ADIPOQ | Adiponectin | 3.53 × 10–4 | 8.86 | 6.81 |
| P02730 | SLC4A1 | Band 3 anion transport protein | 5.71 × 10–3 | 9.85 | 3.93 |
| Q15746 | MYLK | Myosin light chain kinase, smooth muscle | 2.26 × 10–4 | 13.06 | 7.95 |
| Q13555 | CAMK2G | Calcium/calmodulin-dependent protein kinase type II subunit gamma | 5.58 × 10–4 | 14.53 | 2.53 |
| Q9BV23 | ABHD6 | Monoacylglycerol lipase ABHD6 | 2.02 × 10–3 | 14.54 | 3.29 |
PRM, parallel reaction monitoring; LFQ, label-free quantitative.
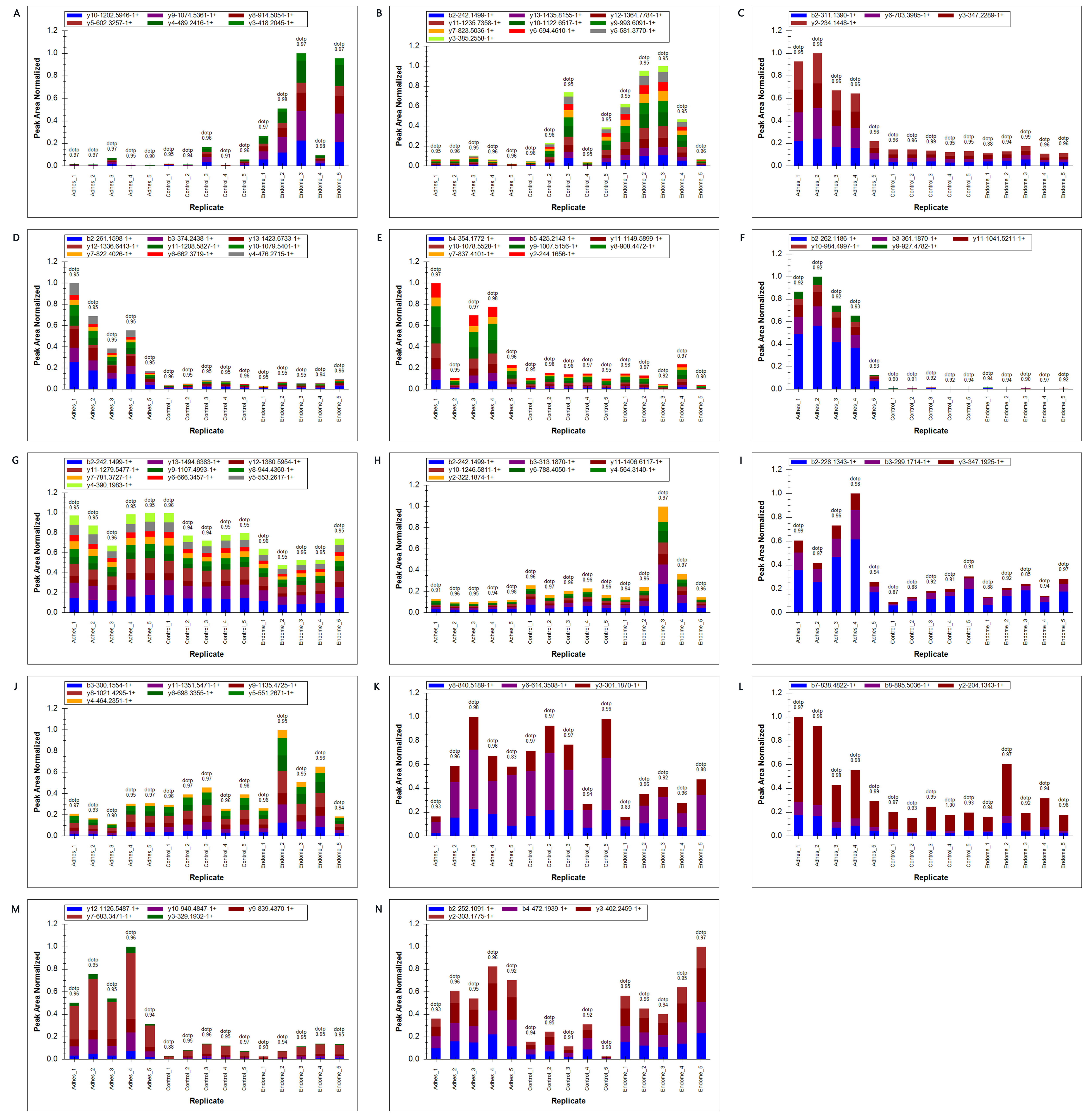 Fig. 6.
Fig. 6. PRM analysis of target protein. (A) LTF. (B) SLC4A. (C) CAMK2G. (D) MYLK. (E) SYNPO2. (F) PGM5. (G) PTPN11. (H) PROS1. (I) IRAG. (J) TGF-
IUA is an endometrial fibrosis disease that significantly affects the reproductive health of women of gestational age. Currently, no cure has been found. There is currently a high rate of recurrence after surgery. Although there have been many studies reporting the biological indicators related to IUA, the specific pathogenesis of IUA has not been clarified. So far there have been few reports on IUA proteomics. In this study, we performed LFQ proteomics and PRM analysis from patients with IUA. 1328, 290, and 1335 DEPs were discovered in the Adhes/Control, Endome/Control, Adhes/Endome groups, respectively. Through comprehensive bioinformatic analysis, similar biological process, molecular function and KEGG pathways were found between the Adhes/Control and the Endome/Adhes groups. Most DEPs were involved in cellular processes and biological regulations, and the role of molecular function regulation and transportation activity. They were found to participate in many signaling pathways, such as regulation of actin cytoskeleton, ECM-receptor interaction, cGMP-PKG signaling pathway and focal adhesion. Zhang et al. [16] conducted transcriptome analysis on IUA tissues and found that actin-mediated cell contraction, focal adhesion and cGMP-PKG signaling pathway were significantly enriched, which is similar to our results. In the Endome/Control group, DEPs were enriched in biological processes such as oxygen transport, defense responses to fungi, and antimicrobial humoral reactions, and are enriched in pathways related to Staphylococcus aureus infection and hematopoietic cell lineage. This suggests that the repair of the endometrium adjacent to adhesions may be accompanied by an enhancement in the immune response. Studies have shown that immune responses play a significant role in endometrial remodeling [17], but the specific role in the repair of the endometrium next to adhesions is still unclear.
Protein domain analysis showed that the main domains of DEPs in the Adhes/Control and Endome/Adhes groups were LIM domain, CH domain, and the 2OG-Fe (II) oxygenase superfamily. The LIM domain is a structural domain widely found in cytoskeletal and signal transduction-related proteins, and is mainly involved in cell adhesion, signal transmission, and the mechanical regulation of the ECM [18, 19]. LIM domain proteins may participate in the formation of IUA by regulating abnormalities in ECM remodeling and cellular connections. The CH domain is commonly found in actin-binding proteins, regulating actin binding and dynamic reorganization, and may influence the formation of adhesion tissues by mediating actin-dependent cell migration and contractile processes [20, 21]. In the Endom/Control group, the key structural domains of DEPs include globin, peptidoglycan-binding domain, and ferritin-like domain, suggesting that immune response, antimicrobial defense, and iron metabolism regulation may play important roles in adhesion endometrial repair. The functional distribution of these domains is highly consistent with the results of the GO and KEGG analyses.
In order to screen for new biomarkers, we conducted PRM verification and obtained 14 target proteins. Some of these target proteins involve multiple key pathways and biological functions in the GO and KEGG analysis. Although the relationship between most target proteins and IUA has not been confirmed, some of them have been proven to be related to other fibrotic diseases. TGF-
PROS1 is a vitamin K-dependent glycoprotein. Urawa M et al. [37] found that it inhibited pulmonary fibrosis in mice and reduced the expression of TGF-
There are some limitations in this study. First, the number of research samples is still insufficient. Second, we only conducted preliminary PRM verification, and the selected target protein has not been independently verified. The detailed function and mechanism of its role in IUA and endometrial fibrosis needs to be further studied. Subsequently, the relevant results of this study will be further verified from the tissue samples, and the biological functions of the differential proteins will be confirmed. The inclusion of more types of research objects provides important support for the subsequent study of molecular biological mechanisms of IUA.
This study reveals the molecular mechanisms related to IUA and the repair of endometrium adjacent to adhesions through proteomics and bioinformatics analysis. The formation of IUA is closely related to the regulation of the actin cytoskeleton and abnormal remodeling of the ECM, while the repair of the endometrium adjacent to adhesions may be achieved by regulating ECM function and the immune response. The target proteins (such as TGF-
IUA, intrauterine adhesions; LFQ, label-free quantitative; PRM, parallel reaction monitoring; DEPs, differentially expressed proteins; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; EMT, epithelial-mesenchymal transition.
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
YL designed the research study. FYL and CXZ performed the research. JXY and YWR provided help and advice on analyzing the data. FYL wrote the original draft. YL reviewed and edited the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Written informed consent from every patient was obtained before the surgery. The Nanjing Medical University and the Changzhou Maternity and Child Health Care Hospital Committee approved all the experiments. Approval No.: Ethics Committee of Changzhou Maternity and Child Health Care Hospital 2021[88]. All procedures involving human participants in this study were conducted in strict adherence to the ethical guidelines established by the 1964 Helsinki Declaration and its subsequent amendments.
We would like to thank all the staff members who took part in this study.
This research received no external funding.
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.






