1 Gynecological Tumor Radiotherapy Second Ward, Cancer Hospital Affiliated of Xinjiang Medical University, State Key Laboratory of Pathogenesis, Prevention and Treatment of High Incidence Diseases in Central Asia, 830000 Urumqi, Xinjiang, China
2 Department of Gynecology, Second Affiliated Hospital of Xinjiang Medical University, State Key Laboratory of Pathogenesis, Prevention and Treatment of High Incidence Diseases in Central Asia, 830000 Urumqi, Xinjiang, China
Abstract
Background: Cervical cancer, a malignancy of gynecological origin, typically necessitates a therapeutic approach combining surgery and chemoradiotherapy as the primary intervention. However, the 5-year survival rate remains suboptimal, prompting researchers to explore novel strategies for the early diagnosis and treatment of cervical cancer. This study delves into the investigation of human papillomavirus (HPV)-mediated DNA methylation modifications within the promoter region of the long non-coding RNA MAGI2-AS3 during cervical cancer development. This paper is an experimental study, laboratory based, using cell lines. Methods: A lentivirus overexpression vector encoding HPV16 E6/E7 proteins was established for transfecting cervical epithelial cells. The methylation status of DNA in the MAGI2-AS3 promoter region was assessed using MassARRAY, and the MAGI2-AS3 gene expression was measured through quantitative real time polymerase chain reaction (qRT-PCR). Subsequently, the correlation between methylation levels and gene expression in cervical cancer was analyzed. Results: (1) Relative to the HPV-negative cervical cancer cell line C33A, MAGI2-AS3 expression significantly decreased in the HPV-positive cervical cancer cell line Siha. (2) The methylation rate of 16 CpG sites in the HPV-positive cervical cancer cell line Siha was notably higher compared to the HPV-negative cervical cancer cell line C33A. (3) To further substantiate the regulatory impact on DNA methylation and expression within the MAGI2-AS3 promoter region, we silenced the expression of HPV16 E6 in the HPV-positive cervical cancer cell line Siha using HPV16 E6 siRNA. The ensuing qRT-PCR analysis revealed a significant up-regulation of MAGI2-AS3 expression in the HPV16 E6 siRNA group when contrasted with the negative control group. MassARRAY analysis was employed to gauge the DNA methylation levels in the promoter region of the MAGI2-AS3 gene in HPV-positive cervical cancer cells (Siha) following HPV16 E6 silencing. The results demonstrated a significantly lower methylation rate at the CpG_29 site in the HPV16 E6 siRNA group compared to the HPV16 E6 siRNA group. Conclusions: The study establishes a close association between HPV infection and elevated methylation levels coupled with diminished expression of MAGI2-AS3. The influence of HPV infection on the malignant transformation of cervical epithelial cells is potentially mediated through the regulatory modulation of MAGI2-AS3 expression via DNA methylation.
Keywords
- lncRNA
- MAGI2-AS3
- cervical cancer
- methylation
Cervical cancer stands as one of the prevailing gynecological malignancies, marked by a heightened mortality rate [1]. Human papillomavirus (HPV) infection emerges as the primary instigator of cervical cancer, with the pivotal role played by the E6 and E7 genes of HPV16 in instigating the malignant transformation of host cells, thereby precipitating cervical epithelial lesions [2]. Recent investigations reveal that HPV E6/E7 mRNA detection, alongside HPV-DNA detection, exhibits commensurate sensitivity, commendable stability, and heightened specificity in screening cervical lesions [3].
DNA methylation, a prominent epigenetic mechanism, has been identified as a major contributor to various diseases, including cancer, through aberrant methylation of host genes. Hyper-methylation of the CpG sites within tumor suppressor genes represents a recurrent anomaly in tumour cells, leading to the silencing of these genes and ensuing tumorigenesis. HPV may precipitate cellular malignant transformation by directly or indirectly modulating gene DNA methylation levels [4].
Long non-coding RNAs (lncRNAs), particularly the recently discovered lncRNA MAGI2-AS3 of WW and PDZ domain 2, have garnered attention in recent years. A mounting body of evidence underscores the pivotal role of lncRNAs in the progression of cervical cancer, as they contribute to enhanced cell proliferation, migration, and invasion, thereby influencing the onset and advancement of cervical cancer [5]. lncRNA MAGI2-AS3, recently identified as a tumour suppressor in various cancer types, is subjected to scrutiny in this study [6]. Accordingly, our investigation delves into elucidating the role of lncRNA MAGI2-AS3 in cervical cancer and unravelling the specific mechanism through which HPV infection modulates the malignant transformation of cervical epithelial cells by regulating MAGI2-AS3 expression levels through DNA methylation. This inquiry not only provides novel molecular targets but also establishes a theoretical foundation for the diagnosis, treatment, and prevention of cervical cancer.
This paper is an experimental study, laboratory based, using cell lines.
The CO
Fetal bovine serum (Excell Bio, Shanghai, China), Minimum Essential Medium (MEM), and trypsin (GIBCO, Waltham, MA, USA), as well as siRNA negative control, HPV16 E6 siRNA-73, HPV16 E6 siRNA-161, HPV16 E6 siRNA-303, and lipofectamine RNAiMAX (Thermo Fisher, Waltham, MA, USA) were utilised as reagents and consumables.
Human cervical squamous carcinoma cells (SiHa) were procured from Prosai Bio (Wuhan, Hubei, China) and
cultivated using MEM medium supplemented with 10% fetal bovine serum (FBS) and
1% Penicillin/Streptomycin. The cells were maintained at 37 ℃, 5% CO
Human cervical cancer cells (C33A) were obtained from Prosai Bio and
cultured using MEM medium supplemented with 10% FBS and 1% P/S. The cells were
maintained under conditions of 37 ℃, 5% CO
The cell lines employed in this study underwent rigorous testing to confirm the absence of mycoplasma contamination.
The cell line utilised in this study was verified through STR analysis.
The culture medium in the flask was discarded, and 3 mL of aseptic phosphate buffered saline (PBS) buffer was added for repeated washing. After discarding the PBS buffer, 1 mL of trypsin was introduced to the flask, and the trypsin solution was evenly spread over the cell layer. Following a gentle shake of the cell culture bottle, digestion occurred in the incubator. After 1 minute, 1 mL of medium containing 10% FBS was added to halt trypsin digestion. The unexfoliated cells were thoroughly rinsed with a suction head. The cell suspension was then transferred to a 15 mL centrifuge tube, centrifuged at 1000 rpm for 5 minutes, the liquid was carefully removed, and the cells were resuscitated with 2 mL of complete culture medium. Cell density was adjusted, and the cells were inoculated into the culture bottle and cultured in the incubator.
SiHa and C33A cells were collected, adjusted to a cell density of 1
| Primer name | Sequence, (5′ to 3′) | Product size |
| MAGI2-AS3-F | TTTCTTCAGCCTCTGTGCGA | 107 bp |
| MAGI2-AS3-R | CAGGTCCCGCTATTTCTGCT | |
| hsa GAPDH_F | GGAGCGAGATCCCTCCAAAAT | 197 bp |
| hsa GAPDH_R | GGCTGTTGTCATACTTCTCATGG |
(1) Cell Plank: SiHa cells, exhibiting a robust growth state with a confluence
rate of 90%, were subjected to trypsin digestion. A single-cell suspension was
prepared with an antibiotic-free medium at a concentration of 5
(2) siRNA/Lipofectamine RNAiMAX Complex Configuration: Three 1.5 mL EP tubes were designated as A, B, and C tubes. In A tube, 25 µL of serum-free and antibiotic-free medium, along with 0.25 µL of 20 µM Block-iTAlexa Fluor Red reagent (final concentration 0.01 µM), were added and gently mixed. The B tube received 2.5 µL of 20 µM Block-iTAlexa Fluor Red reagent (final concentration 0.1 µM) and was gently mixed. For the C tube, 50 µL of serum-free and antibiotic-free medium, along with 1.5 µL of Lipofectamine RNAiMAX reagent, were added and gently mixed. Subsequently, 25 µL from the C tube was transferred to both A and B tubes, mixed gently, and left to stand for 10 minutes at room temperature.
(3) Transfection: The combined siRNA/Lipofectamine RNAiMAX complex was added to
the wells of the 24-well culture plate containing cells at a rate of 50 µL
per well. The cell culture plate was gently shaken, and the cells were incubated
in a 37 ℃, saturated humidity, and 5% CO
(4) Determination of Optimal Transfection Conditions: To establish the optimum transfection conditions for siRNA, control siRNA, labelled with varying concentrations of red fluorescence, was used to transfect HPV-positive cervical cancer cells (SiHa). After 48 hours, the red fluorescence within the cells was observed using a fluorescence microscope. Experimental outcomes revealed that siRNA achieved the best transfection efficiency at a concentration of 0.1 µM and a transfection time of 48 hours (refer to Fig. 1). Consequently, the transfection concentration of 0.1 µM siRNA and a transfection time of 48 hours were chosen as the optimal conditions for subsequent experiments.
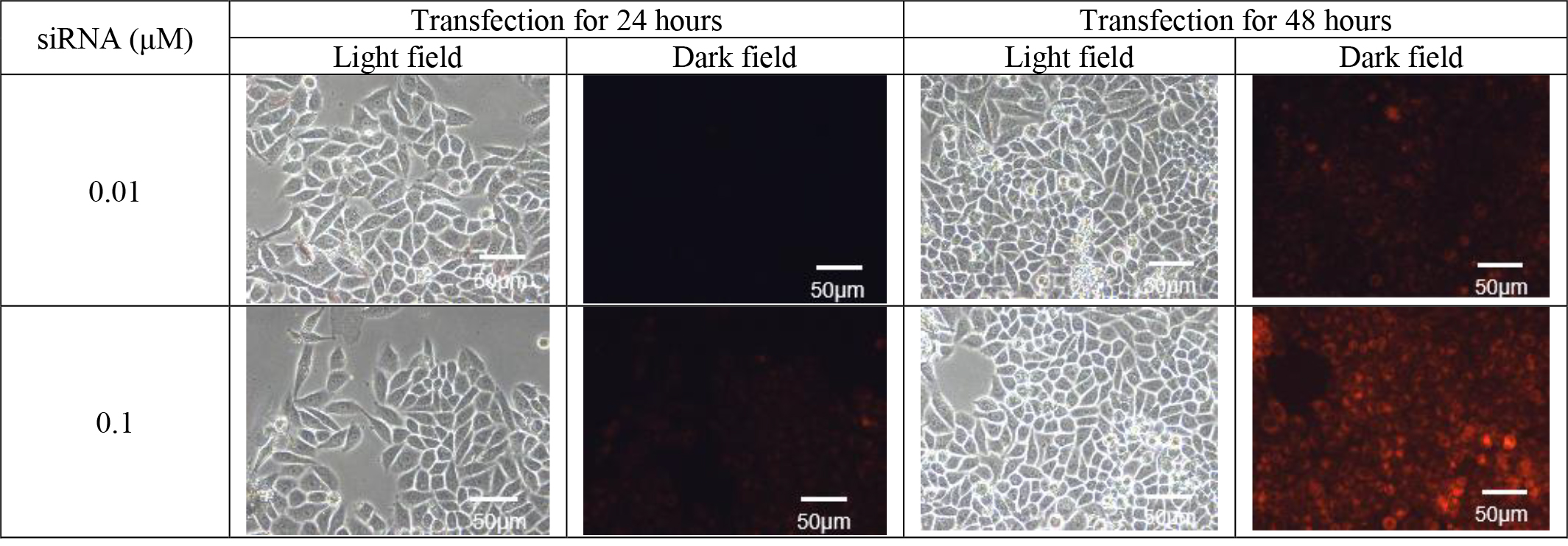 Fig. 1.
Fig. 1.
Transfection fluorescence picture (100
(5) Determination of Transfection Conditions: The optimal transfection parameters were established, with a siRNA concentration of 0.1 µM and a transfection duration of 48 hours selected as the most efficacious conditions for subsequent experiments.
(1) Blank Group: SiHa cells were collected after standard culture for 48 hours.
(2) SiRNA-NC Group: Cells were collected 48 hours after SiHa transfection with 0.1 µM siRNA negative control.
(3) RPS27-siRNA Group: Cells were collected 48 hours after SiHa transfection with 0.1 µM HPV16 E6 siRNA-73.
SiHa cells were collected, and the cell density was adjusted to 1
SiHa cells were collected, and the cell density was adjusted to 1
All data were presented as mean
The experimental findings revealed that the expression level of MAGI2-AS3 in
SiHa cells was (0.187
| Experimental grouping | MAGI2-AS3 |
| C33A cells | 1.030 |
| Siha cells | 0.187 |
| T value | 3.613 |
| p value | 0.022 |
Note: The t-value denotes the T-value of the T-test statistic, while the p-value signifies the level of significance. A significant p-value indicates a substantial difference between the two groups.
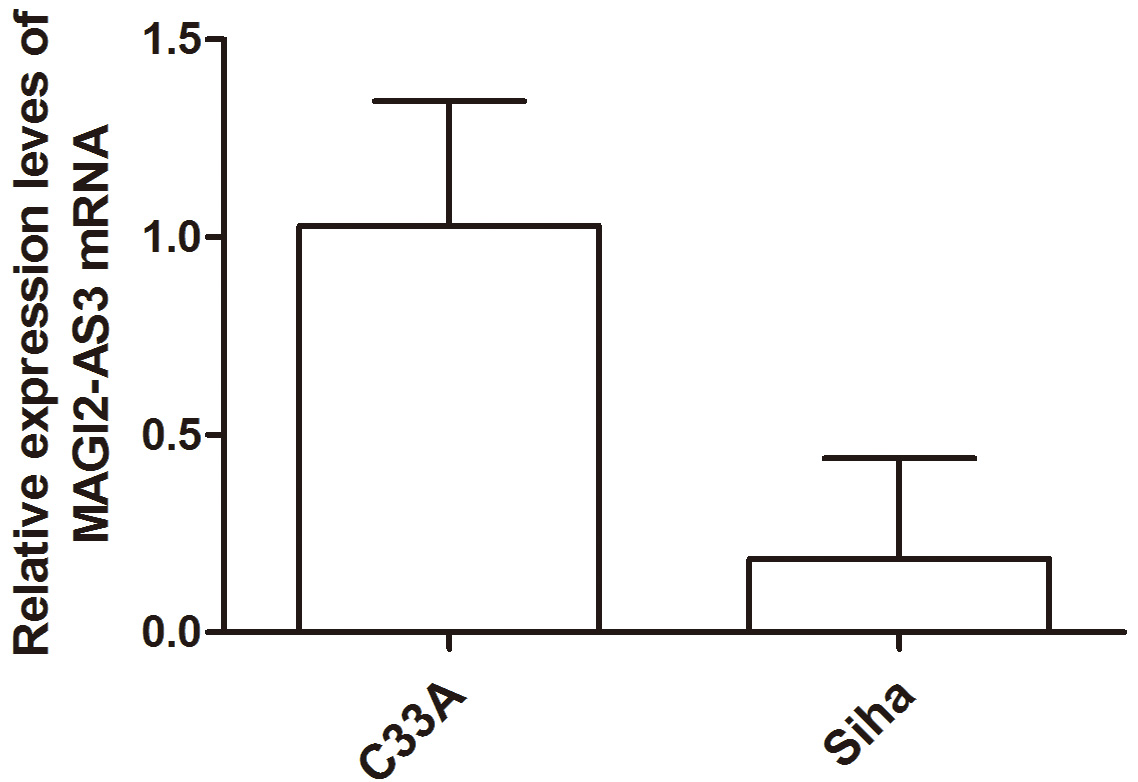 Fig. 2.
Fig. 2.Histogram Analysis of MAGI2-AS3 Expression Level in Each Cell Group.
Experimental findings unveiled the detection of a total of 16 CpG loci within the selected region. The methylation rate at these sites in HPV-positive cervical cancer cells (SiHa) exhibited a significant increase compared to that in HPV-negative cervical cancer cells (C33A) (refer to Table 3).
| CpG Island | Group | CpG island methylation rate (%) | T value | p value |
| CpG_2 | Siha | 65.2 |
12.473 | 0.000 |
| C33A | 13.2 | |||
| CpG_4 | Siha | 57 |
2.873 | 0.021 |
| C33A | 24.4 | |||
| CpG_7 | Siha | 43.25 |
7.493 | 0.000 |
| C33A | 3.75 | |||
| CpG_8.9 | Siha | 78.25 |
6.281 | 0.007 |
| C33A | 4 | |||
| CpG_10 | Siha | 82.6 |
20.906 | 0.000 |
| C33A | 4.6 | |||
| CpG_11 | Siha | 56.8 |
5.154 | 0.001 |
| C33A | 2.4 | |||
| CpG_14.15 | Siha | 34.6 |
14.174 | 0.000 |
| C33A | 4.2 | |||
| CpG_16.17.18 | Siha | 32.6 |
3.055 | 0.016 |
| C33A | 6.8 | |||
| CpG_19.20 | Siha | 18.6 |
6.490 | 0.000 |
| C33A | 3.8 | |||
| CpG_21 | Siha | 37.8 |
9.656 | 0.000 |
| C33A | 4.8 | |||
| CpG_22 | Siha | 24.8 |
2.718 | 0.026 |
| C33A | 6 | |||
| CpG_23 | Siha | 54.2 |
5.557 | 0.001 |
| C33A | 24 | |||
| CpG_24.25 | Siha | 49.4 |
3.095 | 0.015 |
| C33A | 34 | |||
| CpG_27.28 | Siha | 39.8 |
11.041 | 0.000 |
| C33A | 12 | |||
| CpG_29 | Siha | 62.2 |
4.996 | 0.001 |
| C33A | 37 | |||
| CpG_30 | Siha | 79.8 |
8.628 | 0.000 |
| C33A | 38.6 |
Note: The t-value serves as a statistical measure, while the p-value indicates significance. The significance level was set at 0.05.
The outcomes revealed that the silencing effect of HPV16 E6 siRNA-73 surpassed
the other two siRNAs, and this difference was statistically significant
(p
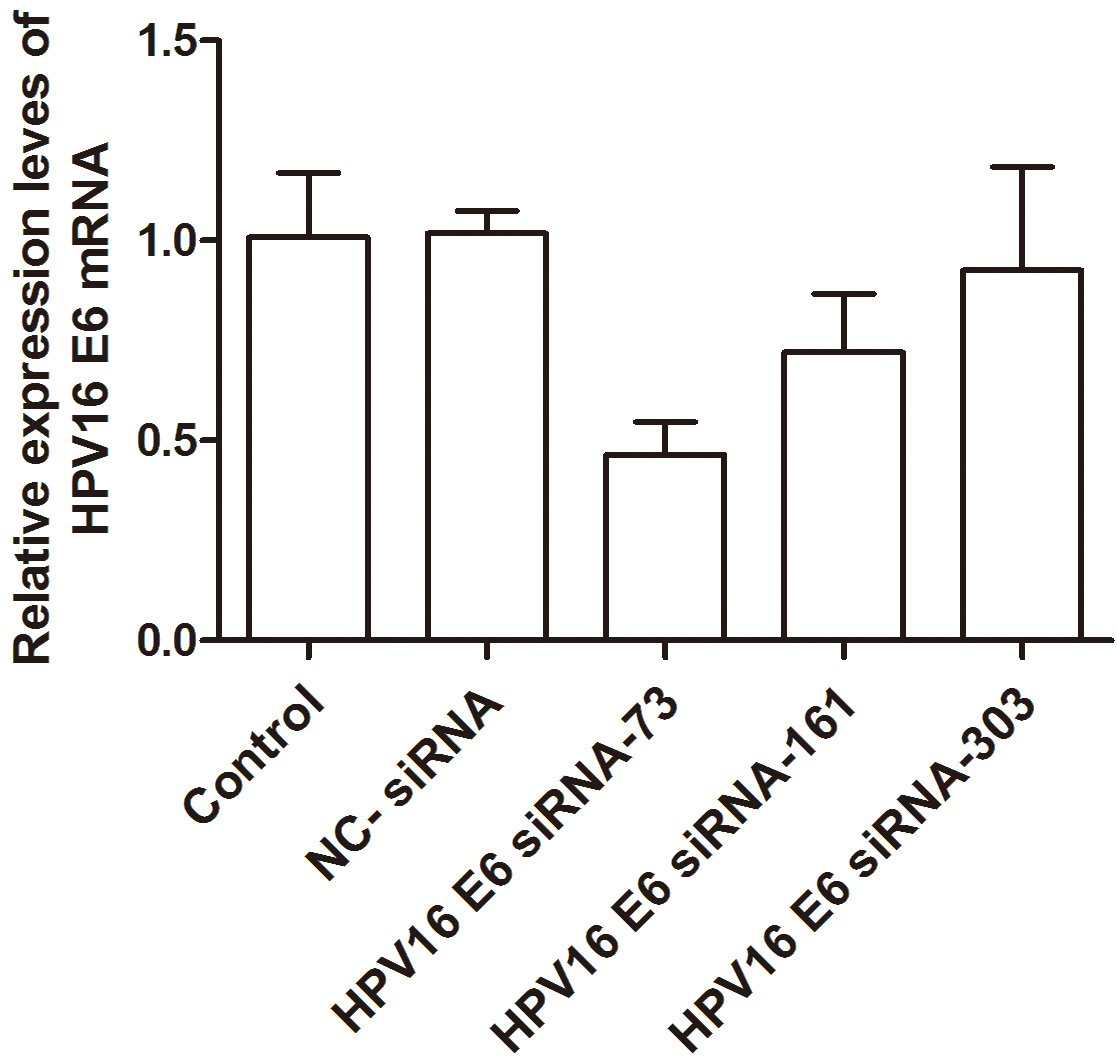 Fig. 3.
Fig. 3.Histogram analysis of HPV16 E6 expression level in each group of cells. HPV, human papillomavirus; NC, negative control.
| Experimental group | HPV16 E6 |
| Blank group | 1.009 |
| Negative control siRNA | 1.018 |
| HPV16 E6 siRNA-73 | 0.465 |
| HPV16 E6 siRNA-161 | 0.721 |
| HPV16 E6 siRNA-303 | 0.925 |
Note:
According to the experimental results, the target HPV16 E6 siRNA-73 was selected for follow-up experiments.
Table 5 shows the fluorescence quantitative primer information. The findings indicated that the expression level of MAGI2-AS3 in the blank group
was (1.002
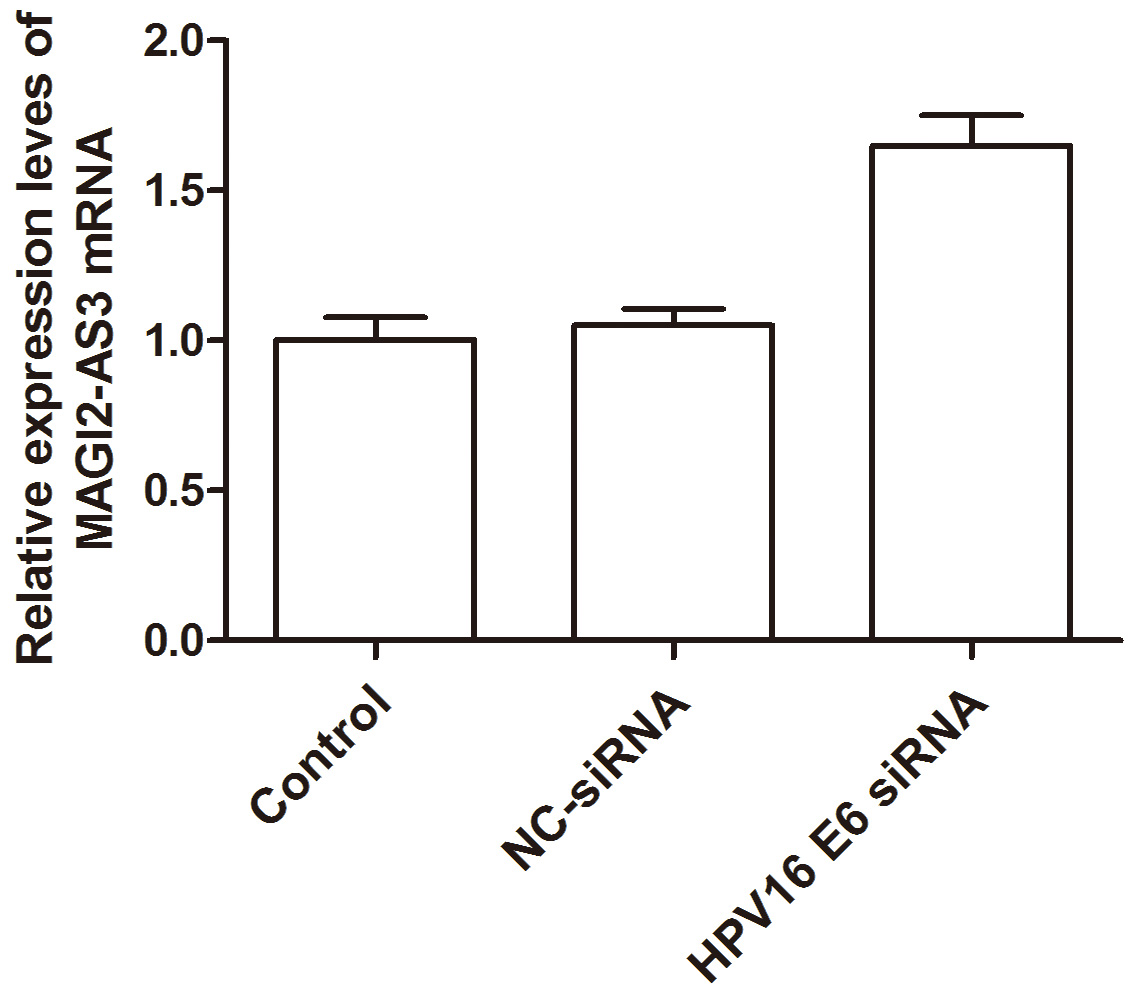 Fig. 4.
Fig. 4.Histogram analysis of MAGI2-AS3 expression level in each cell group.
| Primer name | Sequence (5′ to 3′) | Product size |
| MAGI2-AS3-F2 | TTTCTTCAGCCTCTGTGCGA | 107 bp |
| MAGI2-AS3-R2 | CAGGTCCCGCTATTTCTGCT | |
| hsa GAPDH_F2 | GGAGCGAGATCCCTCCAAAAT | 197 bp |
| hsa GAPDH_R2 | GGCTGTTGTCATACTTCTCATGG |
| Experimental group | MAGI2-AS3 |
| Blank group | 1.002 |
| Negative control siRNA | 1.051 |
| HPV16 E6 siRNA | 1.650 |
Note:
In comparison with the negative control siRNA group, a marked reduction in the
methylation rate of CpG 29 site was observed in the HPV16 E6 siRNA group
(p
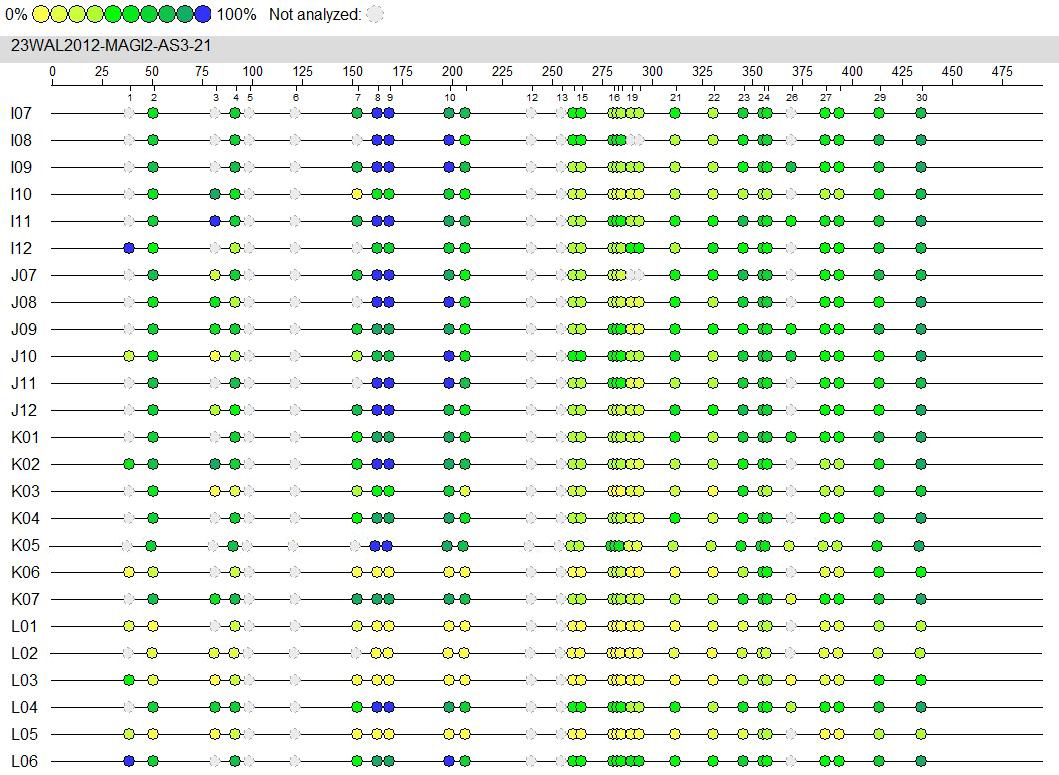 Fig. 5.
Fig. 5.The CpG site is the location of the MAGI2-AS3 promoter region.
| CpG Island | Group | CpG island methylation rate (%) | F value | p value |
| CpG_2 | Blank group | 64 |
0.076 | 0.927 |
| siRNA-NC group | 61.4 | |||
| siRNA-HPV16 E6 group | 63.2 | |||
| CpG_4 | Blank group | 69.6 |
1.497 | 0.263 |
| siRNA-NC group | 57 | |||
| siRNA-HPV16 E6 group | 52.6 | |||
| CpG_7 | Blank group | 62.8 |
0.652 | 0.547 |
| siRNA-NC group | 43.333 | |||
| siRNA-HPV16 E6 group | 62.667 | |||
| CpG_8.9 | Blank group | 91.6 |
1.994 | 0.179 |
| siRNA-NC group | 80 | |||
| siRNA-HPV16 E6 group | 91 | |||
| CpG_10 | Blank group | 89.2 |
1.763 | 0.213 |
| siRNA-NC group | 81.6 | |||
| siRNA-HPV16 E6 group | 90.2 | |||
| CpG_11 | Blank group | 68.4 |
10.634 | 0.002 |
| siRNA-NC group | 48.6 | |||
| siRNA-HPV16 E6 group | 62.4 | |||
| CpG_14.15 | Blank group | 37.8 |
0.489 | 0.625 |
| siRNA-NC group | 35 | |||
| siRNA-HPV16 E6 group | 38.4 | |||
| CpG_16.17.18 | Blank group | 42.8 |
1.151 | 0.349 |
| siRNA-NC group | 35 | |||
| siRNA-HPV16 E6 group | 34.6 | |||
| CpG_19.20 | Blank group | 40.6 |
2.549 | 0.127 |
| siRNA-NC group | 26.333 | |||
| siRNA-HPV16 E6 group | 24.6 | |||
| CpG_21 | Blank group | 44.8 |
1.587 | 0.245 |
| SiRNA-NC group | 38.2 | |||
| siRNA-HPV16 E6 group | 45.2 | |||
| CpG_22 | Blank group | 39.6 |
0.079 | 0.925 |
| siRNA -NC group | 40.4 | |||
| siRNA-HPV16 E6 group | 38.2 | |||
| CpG_23 | Blank group | 61.6 |
1.935 | 0.187 |
| siRNA-NC group | 52 | |||
| siRNA-HPV16 E6 group | 62.4 | |||
| CpG_24.25 | Blank group | 52 |
1.799 | 0.207 |
| siRNA-NC group | 51.8 | |||
| siRNA-HPV16 E6 group | 60.6 | |||
| CpG_27.28 | Blank group | 48.2 |
0.308 | 0.740 |
| siRNA-NC group | 46 | |||
| siRNA-HPV16 E6 group | 45.2 | |||
| CpG_29 | Blank group | 61 |
4.953 | 0.027 |
| siRNA-NC group | 69 | |||
| siRNA-HPV16 E6 group | 54.2 | |||
| CpG_30 | Blank group | 82.2 |
1.500 | 0.262 |
| siRNA-NC group | 79.4 | |||
| siRNA-HPV16 E6 group | 84.4 |
Note: F value is a statistic, p value is significant. The significant level was 0.05.
Cervical cancer stands as one of the prevalent malignant tumours in gynecology, with HPV infection serving as a pivotal risk factor in its onset and progression. In recent years, cytological screening has facilitated the early identification of cervical cancer and precancerous lesions, leading to enhanced treatment outcomes and prognoses. However, gene methylation screening exhibits superior sensitivity and specificity in this context [7]. DNA methylation involves the binding of cytosine to methyl groups within the CpG dinucleotides of the genome, inducing alterations in gene expression levels without modifying the DNA sequence. CpG islands, situated in the human genome, typically remain unmethylated in normal cells. Notably, hypermethylation of the CpG regions within tumour suppressor genes is a prevalent occurrence in tumour cells, resulting in the silencing of these genes and the initiation of tumorigenesis [8]. Recent investigations suggest that HPV DNA methylation, along with host DNA methylation, constitutes a crucial carcinogenic mechanism of HPV [9]. Methylation serves as a dependable marker for diagnosing, monitoring disease progression, and predicting the prognosis of cervical cancer. Furthermore, the modulation of methylation status can inform the development of effective treatment strategies, imparting profound guidance in the treatment of cervical cancer [10]. lncRNA is an RNA molecule exceeding 200 bp in length [11]. A substantial body of evidence indicates that lncRNA is intricately involved in various biological processes, encompassing tumorigenesis, differentiation, and metastasis [12]. Within hepatocellular carcinoma cells, lncRNA-H19 orchestrates the activation of the CDC42/PAK1 pathway by targeted miR-15b modulation, thereby fostering cell proliferation, migration, and invasion [13]. Growing evidence underscores the pivotal role of lncRNA in the progression of cervical cancer [14]. These molecules enhance cell proliferation, migration, and invasion, significantly contributing to the initiation and advancement of cervical cancer [5]. In the context of cervical cancer, lncRNA-CTS promotes epithelial-mesenchymal transition and cancer cell metastasis through the regulation of the miR-505/ZEB2 axis [15]. The lncRNA ZNF667-AS1 pathway mitigates the reduction of miRNA-93-3p-dependent PEG3 in cervical cancer, thereby restraining tumour cell invasion [16]. In recent years, the newly discovered lncRNA MAGI2-AS3 has emerged as closely associated with tumour occurrence and progression, regulating malignant biological behaviours such as tumour proliferation, apoptosis, invasion, and metastasis [17]. MAGI2-AS3, located within the chromosomal region 79452957-79471208 of human genome number 7, has been implicated in various cancers [17].
In murine models, hepatoma cells rely on lncRNA MAGI2-AS3 to inhibit cell viability and metastasis through the miR-374b/SMG1 signalling pathway [6]. In non-small cell lung cancer, lncRNA MAGI2-AS3 inhibits cell proliferation and invasion through the miRNA-23a-3p/PTEN axis [18]. Furthermore, MAGI2-AS3 has been found to promote the progression of gastric and colorectal cancer [19]. A study has identified an association between lncRNA MAGI2-AS3 and poor prognosis in bladder cancer [20]. Interestingly, MAGI2-AS3 exhibits a tumour-inhibitory role in high-grade serous ovarian and breast cancers [21]. However, the functional mechanisms of MAGI2-AS3 in cervical cancer remain unclear. This study employed qRT-PCR to assess the expression levels of the MAGI2-AS3 gene in HPV-negative cervical cancer cells (C33A) and HPV-positive cervical cancer cells (SiHa). The findings revealed a significantly lower expression of MAGI2-AS3 in HPV-positive cervical cancer cells (SiHa) compared to HPV-negative cervical cancer cells (C33A), suggesting a correlation between reduced MAGI2-AS3 expression and the onset and progression of cervical cancer. High methylation level is easy to cause HPV virus infection, which is easy to lead to cervical cancer. Concurrently, MassARRAY was utilised to evaluate the DNA methylation levels in the promoter region of the target gene MAGI2-AS3 in HPV-negative cervical cancer cells (C33A) and HPV-positive cervical cancer cells (SiHa). Results showed a notable increase in the methylation rate of these sites in HPV-positive cervical cancer cells (SiHa) compared to HPV-negative cervical cancer cells (C33A), highlighting a potential link between heightened methylation rates and the development of cervical cancer. The role of MAGI2-AS3 as a tumour suppressor and a biomarker for clinical diagnosis in cervical cancer cells warrants further investigation.
In conclusion, our hypothesis posits a close association between HPV infection and elevated methylation levels coupled with diminished expression of MAGI2-AS3. The regulatory influence of HPV infection on MAGI2-AS3 expression levels is postulated to occur through DNA methylation mechanisms, thereby influencing the malignant transformation of cervical epithelial cells.
In adherence to the journal’s stipulations, we commit to providing our data to facilitate the reproducibility of this study upon approval from our institution.
JZ: conceptualization, data curation, formal analysis, investigation, methodology, writing — original draft, writing - review & editing. QX: participate in the completion of writing — original draft, interpretation of data. QZ: conception, funding acquisition, resources. All authors contributed to editorial changes in the manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work. All authors read and approved the final manuscript.
All subjects gave their informed consent for inclusion before they participated in the study. This study was approved by the Ethics Committee of the Second Affiliated Hospital of Xinjiang Medical University (Ethics Approval Number: 2022H024).
Not applicable.
This study was funded by Natural Science Foundation of Xinjiang Uygur Autonomous Region (2023D01C122) and Department of Gynecology, Second Affiliated Hospital of Xinjiang Medical University, State Key Laboratory of Pathogenesis, Prevention and Treatment of High Incidence Diseases in Central Asia (SKL-HIDCA-2022-GJ4), Open topic of the Key Laboratory of Neurological Diseases in Xinjiang (XJDX1711-2260) and the ”Tianshan Talents” medical and health high-level personnel training plan (TSYC202301B128).
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.





