1 Department of Obstetrics and Gynecology, Nanfang Hospital, Southern Medical University, 510515 Guangzhou, Guangdong, China
2 Department of Obstetrics and Gynecology, Dalian Municipal Women and Children’s Medical Group, 116035 Dalian, Liaoning, China
Abstract
Background: We investigated the composition and dynamic changes of vaginal microbiota (VM) in pregnant women who underwent in vitro fertilization (IVF), as well as VM in relation to preterm birth. Methods: Sixteen women who conceived after IVF and 6 women who conceived naturally were recruited to the study. Vaginal samples from all participants were collected in the first, second, and third trimesters of pregnancy (T1, T2 and T3, respectively). The V3–V4 region of 16S rRNA was sequenced to assess the VM. Results: In all participants, the alpha-diversity indices Chao1 and observed species of VM were significantly higher in T1 compared to T2 and T3. Non-metric multi-dimensional scaling (NMDS) analysis of beta-diversity revealed the VM structure during T1 was significantly different between IVF and control groups, but then gradually converged during T3. A greater abundance of potential pathogenic bacteria and lower abundance of commensal bacteria was observed in the IVF group compared to control group during T1. Moreover, a higher abundance of Lactobacillus_iners, Escherichia_coli and Alloscardovia_omnicolens was found in preterm birth women who underwent IVF. Conclusions: The VM diversity decreased with increasing gestation in women who underwent IVF and in healthy controls. IVF-induced dysbiosis of the VM occurs mainly during T1 of gestation and may be related to preterm birth.
Keywords
- in vitro fertilization (IVF)
- VM
- preterm birth
- pregnancy outcomes
- longitudinal study
Infertility is one of the conditions of the reproductive system that generates disability due to impairment of function [1]. This condition is estimated to affect 186 million individuals and 48 million couples worldwide [2]. In vitro fertilization (IVF) is an important technology that can assist women and couples to achieve pregnancy. Currently, over 5 million children have been born through IVF technology [3], and its use is constantly increasing. In light of the increasing use of IVF, the potential for IVF-induced health problems in these infants has been evaluated in recent studies. The main findings were that IVF was associated with increased risks of malformation and functional disorders, and with worse peripartum outcome [4]. The specific causes remains unknown, but may be related to parental factors or to the IVF technology utilized.
In recent years, the human microbiota has received considerable research interest because of its central role in numerous aspects of health and disease. These include protection from pathogens, nutrient metabolism, promotion of immune system development, and its influence on various mental and neurological functions [5, 6, 7]. The vaginal microbiota (VM) composition is believed to be associated with the reproductive health of women. Emerging evidence indicates that the maternal microbiota composition prior to delivery influences the development of infant immunity [8]. Maternal vaginal microbes can translocate to the placenta and amniotic fluid, thereby influencing preterm birth [9]. Current evidence suggests that preterm birth is more common in IVF pregnancies than in spontaneous pregnancies. The VM could therefore be a causative factor in the link between IVF pregnancy and risk of preterm birth [10].
The female vagina is colonized by an array of commensal microorganisms, among which Lactobacillus is the most dominant species in healthy women of reproductive age. The VM is particularly dynamic, with fluctuations occurring in response to sexual development and sexual intercourse, pregnancy, hormone levels, as well as personal hygiene. Pregnancy is a special physiologic period for women and is accompanied by hormonal and immune changes that can modulate the structure and function of microbiota, thereby making it different to that of non-pregnant women. During pregnancy, the VM is less diverse and more stable, with predominantly Lactobacillus species [11]. Women who undergo IVF may experience routine clinical interventions, including medication to stimulate ovulation and embryo implantation. All of these factors can affect the structural adjustment of female vaginal microbes. Amplicon sequencing was recently used to analyze the VM of females undergoing IVF. The results suggest that VM may be useful in predicting the outcome of IVF, since women with low percentages of Lactobacillus were less likely to have successful embryo implantation [12]. The VM composition during the first trimester (T1) of gestation could also explain the association between IVF pregnancy and preterm birth risk [10]. Longitudinal analysis of the VM in 45 preterm births and 90 term births conducted by the integrative Human Microbiome Project found that preterm women had significantly lower levels of Lactobacillus crispatus, but higher levels of BVAB1, Sneathia amnii, TM7-HI, and a group of prevotella species [13]. So far, however, very few longitudinal studies have been carried out on the VM of women who undergo IVF. In the present work, we performed the first longitudinal cohort study to analyze the dynamic changes in VM during all three trimesters (T1-T3) of gestation in women undergoing IVF.
Enrolled in this study were 16 pregnant women who underwent a first cycle of
fresh embryo transfer due to male infertility, and 6 pregnant women with
spontaneous pregnancies. All participants attended the Dalian Women’s and
Children’s Medical Center from 2017 to 2021. This research was approved by the
institutional ethics committee (# 20160021) and written consent was given by all
participants for provision of samples and clinical information. Inclusion
criteria for the study were: (1) without symptoms of vaginitis, single live
births, and primigravid; (2) without sex for two weeks, without antibiotics or
probiotics for one month, no medication history for vulva and vaginal disorders
for one month, no systemic use of hormone drugs or immune-suppressants; (3) no
history of pregnancy complications such as vaginitis, gestational hypertension,
gestational diabetes mellitus, or pregnancy with abnormal thyroid function; (4)
no history of cigarette use, drinking or illicit drugs, eating a balanced diet;
(5) ethnicity is Dalian local Chinese Han population. The exclusion criteria were
as follows: (1) age
The standard long IVF regimen was used. A gonadotropin-releasing hormone (GnRH) agonist regimen was the main protocol used in this study, and all patients underwent embryo transfer in fresh cycles. Controlled ovarian hyper-stimulation, oocyte retrieval, and embryo transfer were carried out. Participants were treated with downregulation from the mid-luteal phase of the previous cycle. When the pituitary reached desensitization, recombinant follicle stimulating hormone (FSH) was begun at 150–225 IU/day. Human chorionic gonadotropin (hCG) was given (4000–10,000 IU) once two or more follicles had reached a size of 18 mm. Oocytes were extracted 34–36 h after the hCG trigger, and this was followed by intracytoplasmic sperm injection (ICSI). Participants were injected progesterone (lot NO. 1220507, Shanghai General Pharma, Shanghai, China) at 40 mg/day 48 h after oocytes fertilization. The progesterone supplementation was continued until 10 weeks of gestation after pregnancy was achieved.
Vaginal swab samples were obtained in T1 (10–13 + 6 weeks), T2 (20–27 + 6 weeks), and T3 (28–33 + 6 weeks). These were obtained from the posterior fornix and lateral vaginal wall under direct visualization, then stored at –80 °C until DNA extraction.
The QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) was used to extract genomic DNA as per the manufacturer’s instructions. Agarose gel electrophoresis (1%) was used to evaluate DNA concentration and purity. The V3–V4 region of the 16S rRNA gene underwent high-throughput sequencing. This region was amplified using a specific primer with barcode 314F-806R (V3–V4 primers: 314F 5’-CCTAYGGGRBGCASCAG-3’ 806R 5’-GGACTACNNGGGTATCTAAT-3’). Phusion® High-Fidelity PCR Master Mix (New England Biolabs) was used for PCR amplification. The PCR products were purified using the Qiagen Gel Extraction Kit (Qiagen, Hilden, Germany). TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA, USA) was used to generate sequencing libraries and their quality was analyzed with the Qubit@ 2.0 Fluorometer (Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA) and Agilent Bioanalyser 2100 system. Sequencing of the library was performed with the Illumina HiSeq2500 platform, which generated 250-bp paired-end reads.
Sequencing reads were merged by FLASH to obtain raw tags (v1.2.7,
http://ccb.jhu.edu/software/FLASH/). Filtering to obtain high-quality clean tags
was performed by QIIME (v1.7.0, http://qiime.org/). Tags were compared to the
Gold database, and effective tags were identified using the UCHIME algorithm.
Sequences were analyzed using Uparse software (Uparse v7.0.1001,
http://www.drive5.com/uparse/) and assigned to the same operational taxonomic
units (OTUs) when similarities were
Alpha-diversity was evaluated using the observed species, Chao1, ACE, Shannon, and Simpson indices. These were calculated with QIIME (Version 1.7.0) and displayed using R software (Version 2.15.3, https://www.r-project.org). Beta-diversity was calculated with the same software packages used to determine alpha-diversity. Statistical analysis of differences in species between IVF and control groups was done using the Student’s t-test, while mapping between the two groups was done using R software (Version 2.15.3).
Sixteen women who conceived using IVF because of male infertility were enrolled, together with 6 women who conceived naturally. Table 1 shows the participant details. No significant differences between IVF and control groups were observed for age, body mass index (BMI), and birth weight. The levels of pregnancy hormones, including estrogen and progesterone, were significantly higher in women who conceived through IVF than in women who conceived naturally.
| IVF group (n = 16) | Control group (n = 6) | p | ||
|---|---|---|---|---|
| Age (years) | 31.42 |
31.50 |
0.18 | |
| BMI (kg/m |
26.57 |
27.50 |
0.07 | |
| Duration of COH (days) | 19.89 |
- | - | |
| Number of oocytes obtained (n) | 8.89 |
- | - | |
| Determine gestational weeks | 4.39 |
5.12 |
0.77 | |
| Pregnancy day hormone level | ||||
| Estrogen (pg/mL) | 693.68 |
419.17 |
0.01 | |
| Progesterone (ng/mL) | 23.17 |
0.00 | ||
| Cesarean section (n) | 9 | 1 | ||
| Vaginal delivery (n) | 7 | 5 | ||
| Birth weight (g) | 3055.26 |
2900 |
0.60 | |
| Preterm delivery |
3 | 0 | - | |
BMI, body mass index; COH, controlled ovarian hyperstimulation; IVF, in
vitro fertilization. p
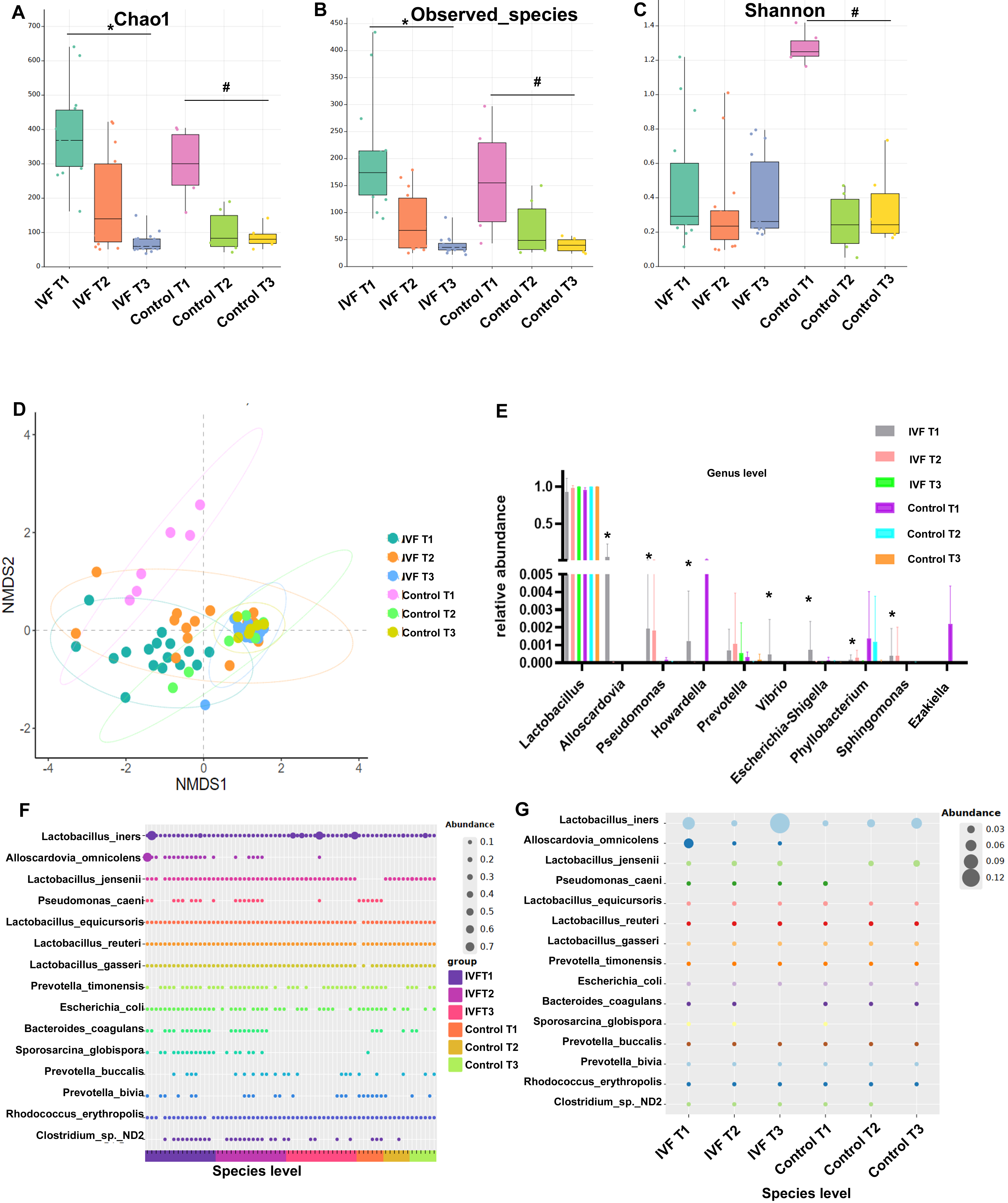
Analysis of the alpha-diversity of VM revealed that the indexes for Chao1 and for observed species were both significantly higher during T1 compared to T2 and T3. The Shannon index was significantly lower in the controls during T2 compared to T1, but no significant difference was seen in the IVF group (Fig. 1A–C). Non-metric multi-dimensional scaling (NMDS) analysis of beta-diversity was performed to assess the similarity in VM between all samples. A significant difference in the VM structure was found between the IVF and control groups during T1. However, the two groups gradually converged during T2, and by T3 were distributed in the same area (Fig. 1D).
 Fig. 1.
Fig. 1.VM composition of all participants. (A) The alpha-diversity
index Chao1. (B) The observed species index. (C) The Shannon index. (D)
Non-Metric Multi-Dimensional Scaling (NMDS) analysis of VM using Bray-Curtis
distance metric. (E) Analysis of relative abundance at the species level in all
participants. (F) Each dot represents the sample from one participant. (G)
Relative abundance at the species level was analyzed by group, with each dot
representing a group. The dot size indicates relative abundance. * p
To further explore the dynamic characteristics of VM, the relative taxon abundance was assessed at phylum, genus, and species levels. Dominant phyla identified in all samples with this analysis were Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes. No significant differences in the abundance of VM were observed in the same group between different trimesters, or between groups (data not shown). Lactobacillus was the dominant genera in all samples. The distribution of the top 10 genera in the vagina was further analyzed. In women who underwent IVF, the proportions of Alloscardovia, Pseudomonas, Vibrio and Escherichia-Shigella in the total VM were found to be significantly greater during T1 than T2 or T3. During T1, the proportions of Howardella, Phyllobacterium, Sphingomonas and Ezakiella in IVF women were significantly less than the controls (Fig. 1E). Evaluation of sequence reads at species level revealed that Alloscardovia_omnicolens was present only in the IVF group, and that its abundance decreased in T3. In both the IVF and control groups, Pseudomonas_caeni, Bacteroides_coagulans, and Sporosarcina_globispora were detected in T1, but in in T3 they were not detectable in most participants. During T1 and T2, Clostridium sp. ND2 was more abundant in the IVF group than the controls. Clostridium sp. ND2 decreased during T3 in both groups and was not detectable in control participants (Fig. 1F,G).
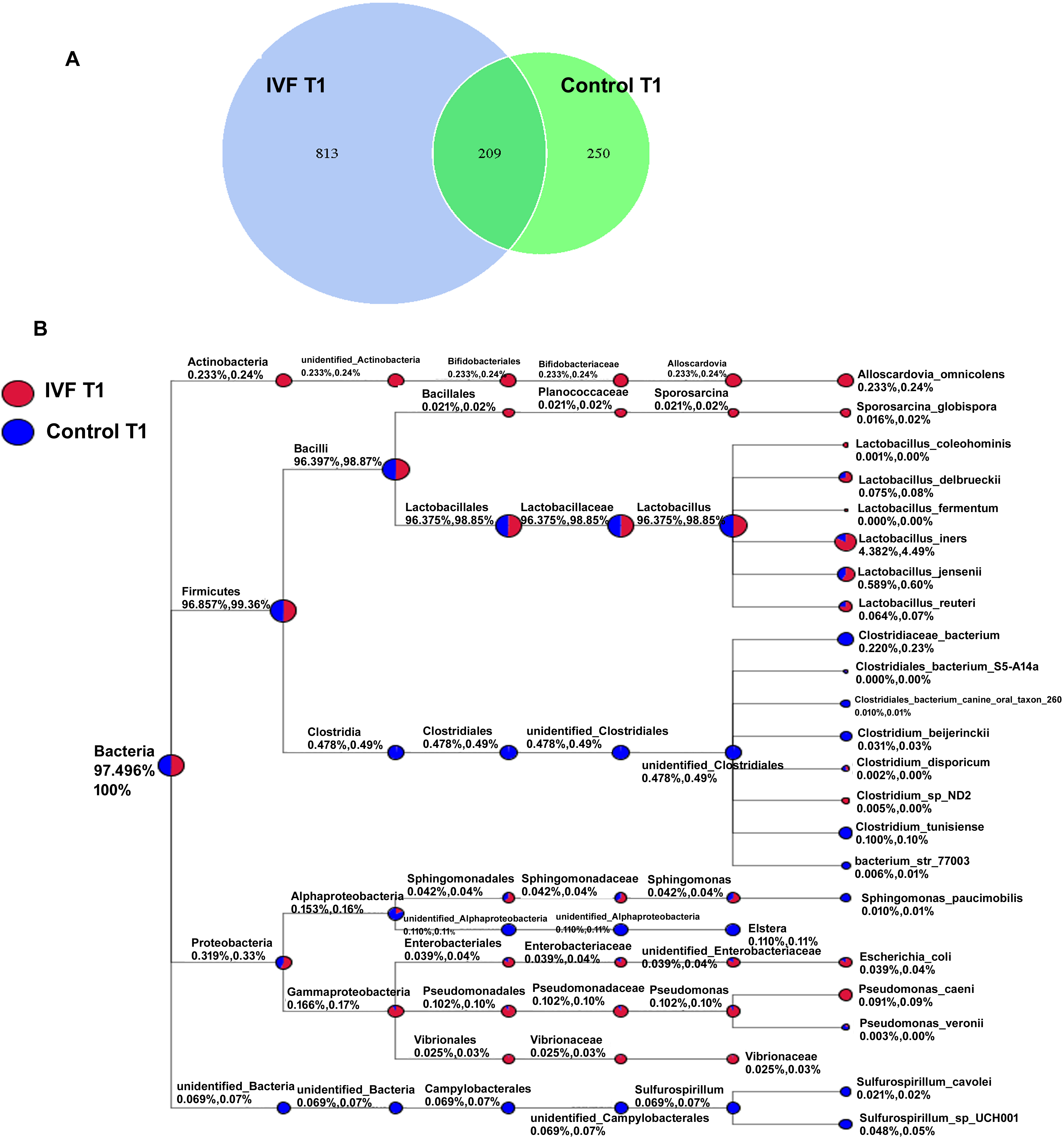
The overall changes in VM during the different trimesters of gestation were quite similar in the IVF and control groups, with the largest differences being apparent mainly in T1. We therefore analyzed for differences in VM between the two groups during T1 in more detail. The OTU of IVF at T1 was 1022 and for the controls it was 459, with the two groups sharing 209 OTU (Fig. 2A). The taxonomic classification tree was used to compare microbial taxonomy between IVF and control groups. IVF women showed a higher abundance of Alloscardovia, Sporosarcina, unidentified_Enterobacteriaceae, Pseudomonas and Vibrio at T1 compared to the control group, but a lower abundance of unidentified_clostridiales, Elstera and Sulfurospirillum (Fig. 2B).
 Fig. 2.
Fig. 2.Vaginal bacteria in the IVF and control groups during T1 gestation. (A) Venn diagram showing unique and shared genera in the IVF and control groups. (B) Taxonomic classification tree showing the microbial taxonomy of the IVF and control groups.
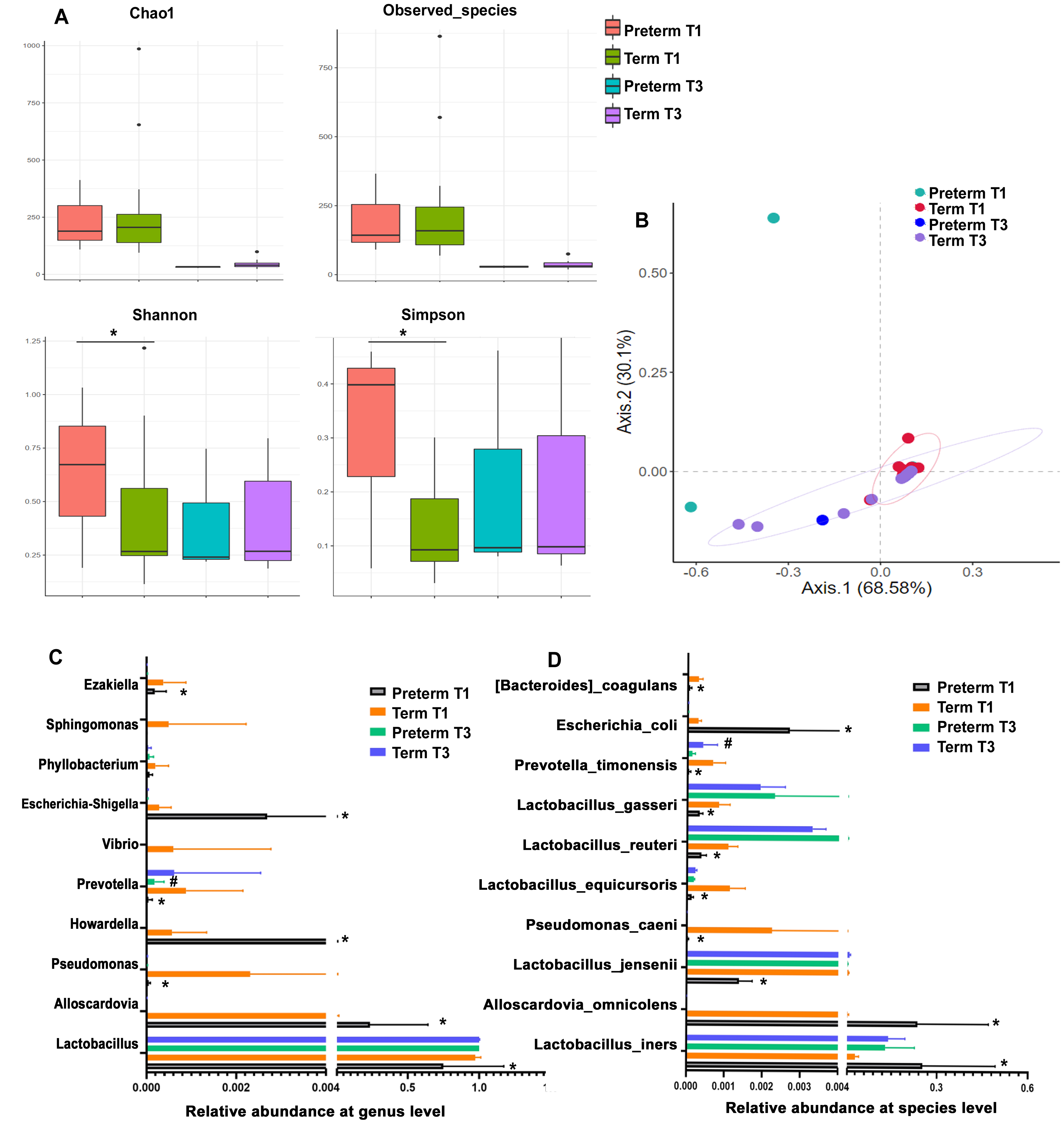
Three spontaneous preterm births occurred in the IVF group. Earlier studies have reported an association between VM and preterm birth. We therefore compared the VM between preterm and term mothers during T1 and T3 of gestation. Preterm and term delivery women showed no significant difference in either the Chao1 or ACE alpha-diversity indexes for VM. However, the Shannon index and Simpson index in preterm women at T1 were significantly higher than those of term delivery women. PCoA analysis of beta-diversity revealed the VM of two of the three preterm women at T1 was significantly different to that of term delivery women (Fig. 3A,B). Taxonomy-based analysis at genus level showed the abundance of Escherichia_Shigella, Howardell and Alloscardovia was higher in VM of preterm women compared to term delivery women, whereas the abundance of Sphingomonas, Prevotella, Pseudomonas, Vibrio and Lactobacillus was lower. At species level, the abundance of Escherichia_coli, Alloscardovia_omnicolens, Lactobacillus_iners was higher in VM of preterm women at T1 than term delivery women, whereas abundance of Bacteroides_coagulans, Lactobacillus_jensenii, Lactobacillus_gasseri, and Lactobacillus_equicursoris were all lower (Fig. 3C,D).
 Fig. 3.
Fig. 3.Comparison of vaginal bacteria between preterm and term delivery
women in the IVF group during T1 and T3 of gestation. (A) Alpha-diversity
indices Chao1, ACE, Shannon and Simpson for VM. (B) Principal Coordinates
Analysis of the VM in preterm and term delivery women at T1 and T3. (C) Relative
abundance of VM at genus level. (D) Relative abundance of VM at species level. *
p
The VM is a complex, dynamic community that changes constantly in response to hormonal fluctuations. Its composition is known to play important roles in vaginal and reproductive health, and has been associated with the risks of miscarriage, preterm birth, and sexually transmitted disease [14]. Currently, IVF is an effective method for conception in women who fail to conceive naturally. Dynamic changes in the VM of women who become pregnant through IVF are still poorly understood. Using high-throughput sequencing, we found that VM is dominated by Lactobacillus species during pregnancy. The alpha-diversity in women who undergo IVF decreased with increasing gestation time. The variation in microbiota diversity was the same in the IVF and control groups. The greatest difference between the two groups was observed during T1, and this gradually decreased during gestation. Women in IVF group received hormonal treatment to stimulate ovulation and followed a progesterone supplementation treatment until 10 weeks of gestation. The first trimester vagina samples were collected from 10 weeks to 13 + 6 weeks. During this period, although the clinical practice and hormone supplementation were no longer performed, the effect of IVF intervention on vaginal microbiota was still existed, which may be the reason for the difference in microbiota between the two groups. The vaginal microbiota difference between the two groups gradually decreased with the increase of gestation days. At T3 of gestation, beta-diversity analysis revealed that the two groups clustered in the same area, thus suggesting a high degree of similarity. These results indicate that the influence of IVF on the VM occurred mainly during T1 of gestation and then gradually decreased as gestation progressed.
The differences in VM between IVF and control groups were greatest in T1. We
therefore analyzed the microbiota in greater detail during this period. The IVF
group showed a higher abundance of Alloscardovia, Sporosarcina,
unidentified_Enterobacteriaceae, Pseudomonas and Vibrio was compared to
controls, and a lower abundance of unidentified_clostridiales, Elstera and
Sulfurospirillum. Alloscardovia is a recently reported microorganism with unknown
pathogenic implications. Alloscardovia omnicolens is present in premature rupture
of membranes (PPROM) and positively related to PPROM [15]. Pseudomonas belonging
to the phylum
Previous studies have showed that pretem birth is more common in IVF pregnancies as compared to naturally coinceived pregnancies [21, 22]. Furthermore, VM dysbiosis was associated with preterm birth and pregnancy loss [23, 24]. In the current study, three spontaneous preterm births occurred in the IVF group. Comparison of structure differences between women who underwent preterm birth or term delivery found that the greatest VM differences occurred during T1. This included higher species complexity in the vaginal ecosystem of preterm birth women, a greater abundance of pathogenic bacteria (Escherichia_Shigella and Alloscardovia), and lower abundance of commensal bacteria (Prevotella and Lactobacillus). We also found higher abundance of Lactobacillus_iners and lower abundance of Lactobacillus_jensenii, Lactobacillus_gasseri and Lactobacillus_equicursoris in VM of preterm women compared to term delivery women. The presence of a Lactobacillus_iners-dominated VM during gestation has been linked to abortion, spontaneous preterm delivery with intact membranes, and premature membrane rupture [25]. Lactobacillus_iners has therefore been suggested as a marker of vaginal cleanliness and leukocyte esterase [26]. In the present study we observed an increase in Lactobacillus_iners-dominated VM and pathogenic microorganisms, which may be a cause of spontaneous premature delivery. Preterm parturition is a syndrome induced by various factors. The prediction and prevention of preterm birth is a global challenge. Our data suggested that specific microbial taxa may be useful in defining the risk of preterm birth. Future studies may use probiotics or prebiotic to manipulate vaginal microbiota and prevent preterm birth.
The main limitation of this study is the small sample size. The small number of patients conceived through IVF due to male infertility together with the exclusion of patients with pregnancy complications further reduced the sample size. Thus, it should be noted that our conclusion should be seen in the context of this limitation. To our knowledge, this is the first longitudinal retrospective study to assess variability of vaginal microbiota in woman who undergoing IVF. For future studies, it will be desirable to increase the cohort size and to consider baseline samples prior to IVF or pregnancy in order to validate our findings.
In summary, the dynamic alterations in VM of IVF women during pregnancy were found to be similar to those of normal pregnant women. Furthermore, the diversity of VM was found to decrease during the gestation period. Lactobacillus was the predominant vaginal bacteria observed in all pregnant women in the present study. The major differences in VM between IVF and control groups were observed to occur during T1 of gestation, where IVF women had more abundant potential pathogenic bacteria and less abundant commensal bacteria. More abundant Lactobacillus_iners, Escherichia_coli and Alloscardovia_omnicolens was also linked to preterm birth in women undergoing IVF. Further investigations into the causal relationships between VM and pregnancy outcomes are required, in addition to studies of the underlying mechanism.
We uploaded the raw 16S rRNA gene sequencing data to the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA728871, accession no. PRJNA728871).
ZW contributed to conception and design of the study. YT performed the experiments and analyzed the data. QS and XS contributed to acquisition of data and revise the manuscript. YT and ZW wrote the manuscript. All authors read and agreed to the final version and are accountable for what is published.
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Dalian Women’s and Children’s Medical Center Group (approval number 20160021). All participants gave written informed consent prior to entering the study.
The authors extend their sincere gratitude to all medical workers for their enthusiastic help to assist in samples collection in our hospital.
This work was supported by National Key R&D Program of China (No. 2022YFC2704504), and Dalian Science and Technology Innovation Fund (No. 2019J13SN83).
The authors declare no conflict of interest.
References
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.