-
- Academic Editor
-
-
-
†These authors contributed equally.
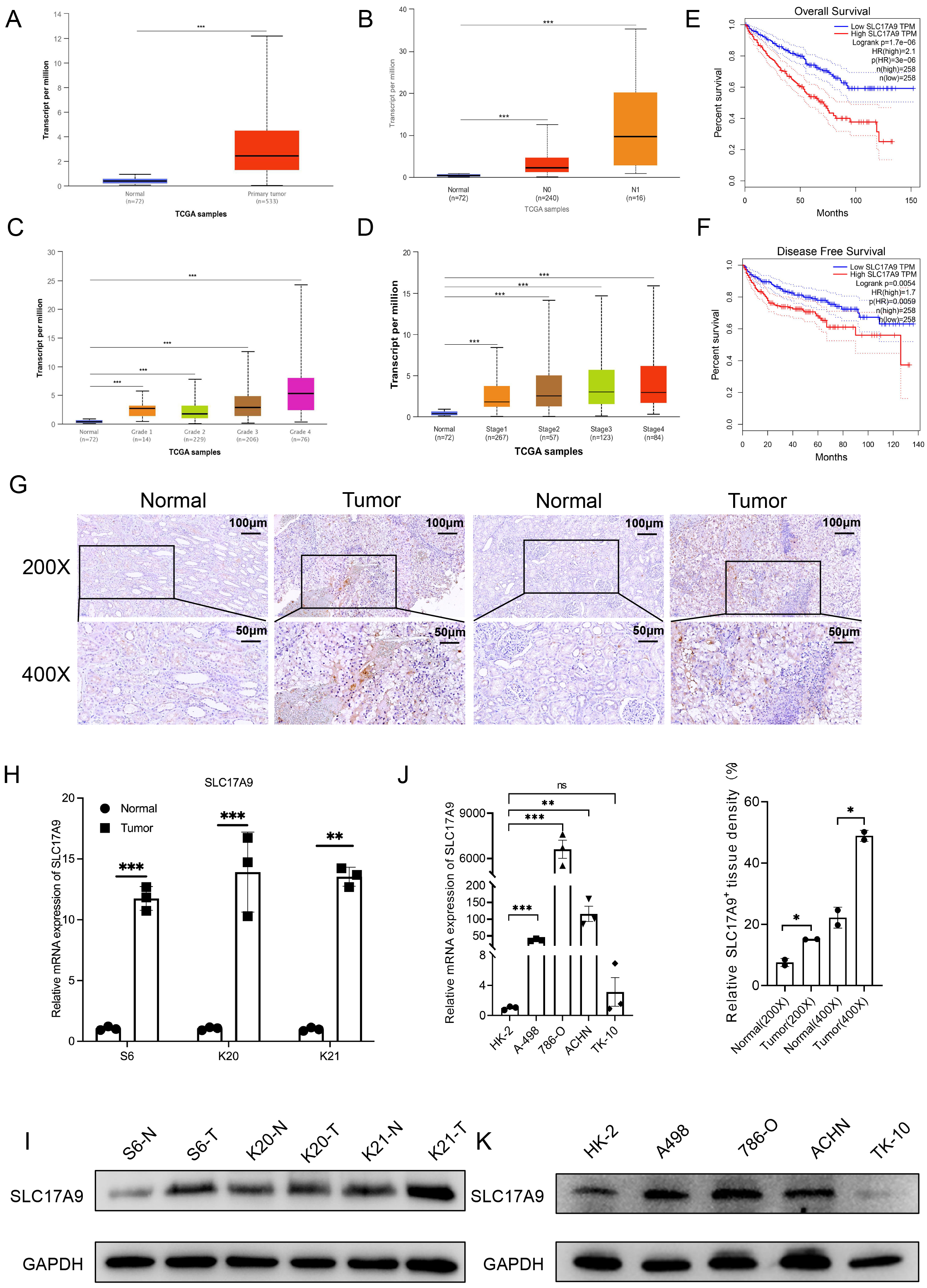
The vesicular nucleotide transporter Solute Carrier Family 17 Member 9 (SLC17A9) has recently been recognized as a significant modulator of oncogenic pathways, with its elevated expression levels being closely linked to the aggressiveness of clear cell renal cell carcinoma (ccRCC). A comprehensive understanding of the role of SLC17A9 and its associated protein markers presents substantial potential for the advancement of targeted therapeutic interventions.
Our study commenced with a comprehensive bioinformatics analysis to identify differentially expressed genes potentially associated with ccRCC. Leveraging The Cancer Genome Atlas (TCGA) database, we predicted the clinical relevance of these cancer-associated genes and validated their expression profiles through multiple experimental methodologies. Functional assays were conducted to assess the impact of these genes on renal cancer cell lines. Additionally, we generated cell lines overexpressing oncogenes and identified downstream targets through RNA sequencing, followed by mechanistic exploration of their interactions. Finally, bioinformatics tools were subsequently employed to assess the diagnostic and prognostic significance of these genes in patients with ccRCC.
The bioinformatics analysis revealed SLC17A9 as a highly expressed oncogene in ccRCC, serving as a robust prognostic marker. Experimental validation demonstrated that SLC17A9 promotes ccRCC cell growth, proliferation, and migration. Lentivirus-based experiments revealed Potassium Voltage-Gated Channel Subfamily H Member 1 (KCNH1) as a downstream target regulated by SLC17A9 (p < 0.05). Database analysis further confirmed KCNH1’s oncogenic role in ccRCC, with significant implications for patient survival. Notably, SLC17A9 and KCNH1 collaboratively drive the initiation and progression of renal cancer. Elevated expression of SLC17A9 and KCNH1 correlates with poorer prognosis (p < 0.001), whereas lower expression levels are associated with favorable outcomes in ccRCC patients. These findings highlight SLC17A9 and KCNH1 as critical biomarkers and potential therapeutic targets in ccRCC.
SLC17A9 and KCNH1 serve as critical prognostic biomarkers in ccRCC, with SLC17A9 driving tumor progression through KCNH1 regulation. Their upregulated expression predicts poor clinical outcomes, while reduced levels correlate with improved survival, highlighting their dual role as therapeutic targets.