- Academic Editor
-
-
-
†These authors contributed equally.
Primary liver cancer (PLC) exhibits a high incidence and mortality rate. Early diagnosis and effective treatment are crucial for improving patient survival rates. This study aims to identify biomarkers of hepatitis B-related liver cancer and establish a new method for molecular subtype classification based on differential metabolite-related regulatory gene expression profiles.
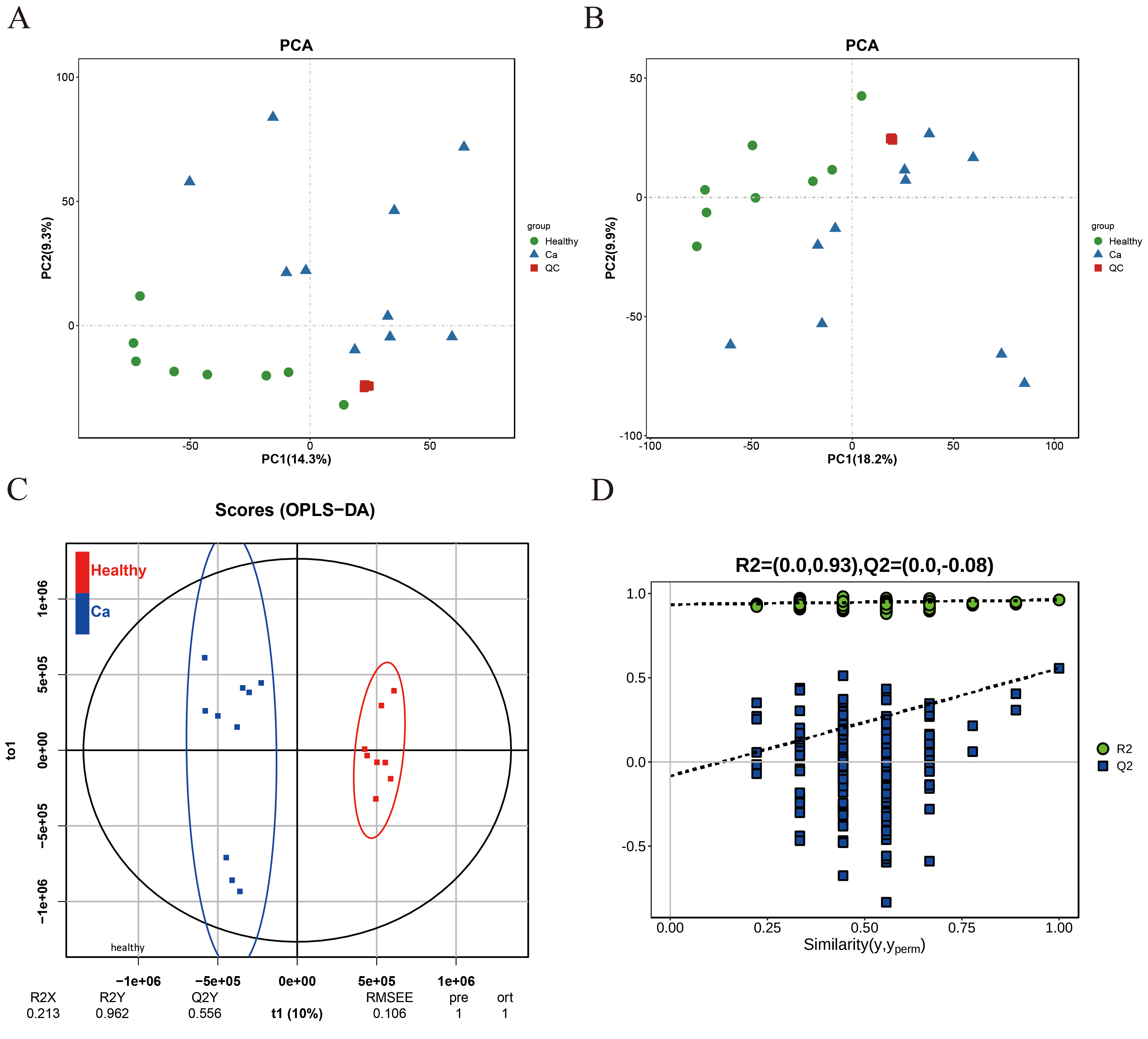
This study collected sterile midstream urine samples from patients with hepatitis B-related liver cancer who had not received standardized systematic antiviral therapy or anticancer therapy, as well as from healthy controls. Potential biomarkers were identified through liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based metabolomics, followed by Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis performed on the differential metabolites. Gene expression data of 371 hepatocellular carcinoma (HCC) samples in The Cancer Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) database were clustered using gene annotations for differential metabolites derived from the Human Metabolome Database (HMDB). The Kaplan-Meier (KM) survival curve was employed to assess the prognosis of different HCC molecular subtypes. Expression differences of subtype-specific genes and their enrichment in Hallmark, KEGG and Gene Ontology (GO) pathways were analyzed. The Tumor Immune Dysfunction and Exclusion (TIDE) scoring tool was used to evaluate the subtypes’ response to immunotherapy. Sensitivity to sorafenib was also compared across the different subtypes.
A total of 53 differential metabolites were identified (p < 0.01), which were significantly enriched in seven metabolic pathways (p < 0.05). Three potential biomarkers were discovered: Suberic acid, 2′-O-methylcytidine, and 3′-Sialyllactose. Regulatory genes associated with these differential metabolites clustered HCC samples from the TCGA-LIHC database into two molecular subtypes (C1 and C2). KM survival analysis indicated that patients in the C2 subtype exhibited higher overall survival compared to those in C1. Differential genes between the two subtypes were significantly enriched in Hallmark, KEGG and GO pathways. The TIDE scoring tool revealed a higher likelihood of immune escape in C1 subtype patients. Molecular targeted drug analysis suggested that sorafenib may be more effective in patients with the C1 subtype.
Suberic acid, 2′-O-methylcytidine, and 3′-Sialyllactose hold promise as metabolic biomarkers for hepatitis B-related liver cancer. Understanding the diversity of the human liver cancer gene expression profile from a metabolomic perspective has potential applications for developing novel clinical treatment strategies.