-
- Academic Editor
-
-
-
†These authors contributed equally.
Sepsis is a life-threatening condition characterized by a dysregulated host response to infection, often leading to multiorgan dysfunction. Despite their clinical importance, early diagnostic biomarkers that reflect organ-specific damage remain inadequately characterized.
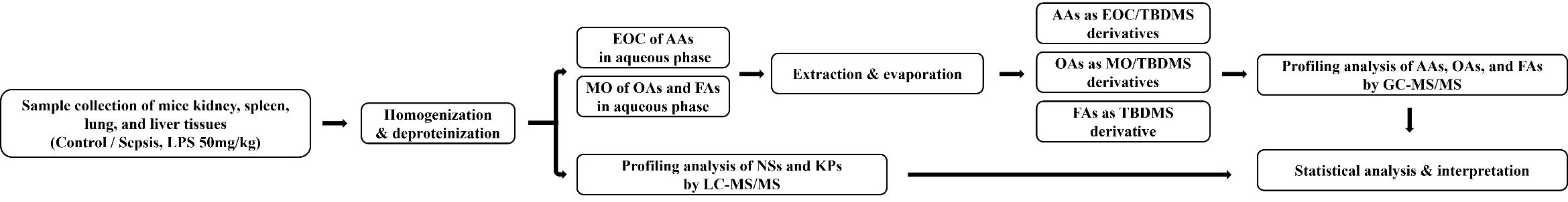
Targeted metabolomic profiling of amino acids, organic acids, fatty acids, nucleosides, and kynurenine pathway metabolites was performed on lung, kidney, spleen, and liver tissues obtained from a lipopolysaccharide-induced mouse model of sepsis, using liquid chromatography-tandem mass spectrometry and gas chromatography-tandem mass spectrometry. Univariate and multivariate statistical analyses (principal component analysis and partial least squares discriminant analysis) were performed to identify potential biomarkers, followed by pathway analysis to elucidate their biological relevance.
Twenty-nine metabolites were significantly altered across the four tissues, exhibiting organ-specific metabolic signatures. Tyrosine, epinephrine, 5-hydroxytryptophan, and kynurenic acid in the kidney; serine, 4-hydroxyproline, normetanephrine, xanthosine, uridine, adenosine, succinic acid, cis-aconitic acid, linoleic acid, and eicosadienoic acid in the spleen; alanine, α-aminobutyric acid, ornithine, uridine, adenosine, 5′-deoxy-5′-methylthioadenosine, succinic acid, and cis-aconitic acid in the lung; and α-aminobutyric acid, pipecolic acid, uridine, inosine, adenosine, glycolic acid, and oxaloacetic acid in the liver were identified as potential biomarkers reflecting organ-specific dysfunction in sepsis.
This study highlights the distinct organ-specific metabolic alterations in sepsis and identifies candidate biomarkers that may reflect early organ dysfunction. These findings provide a foundation for the development of precise diagnostic and medical strategies for sepsis.