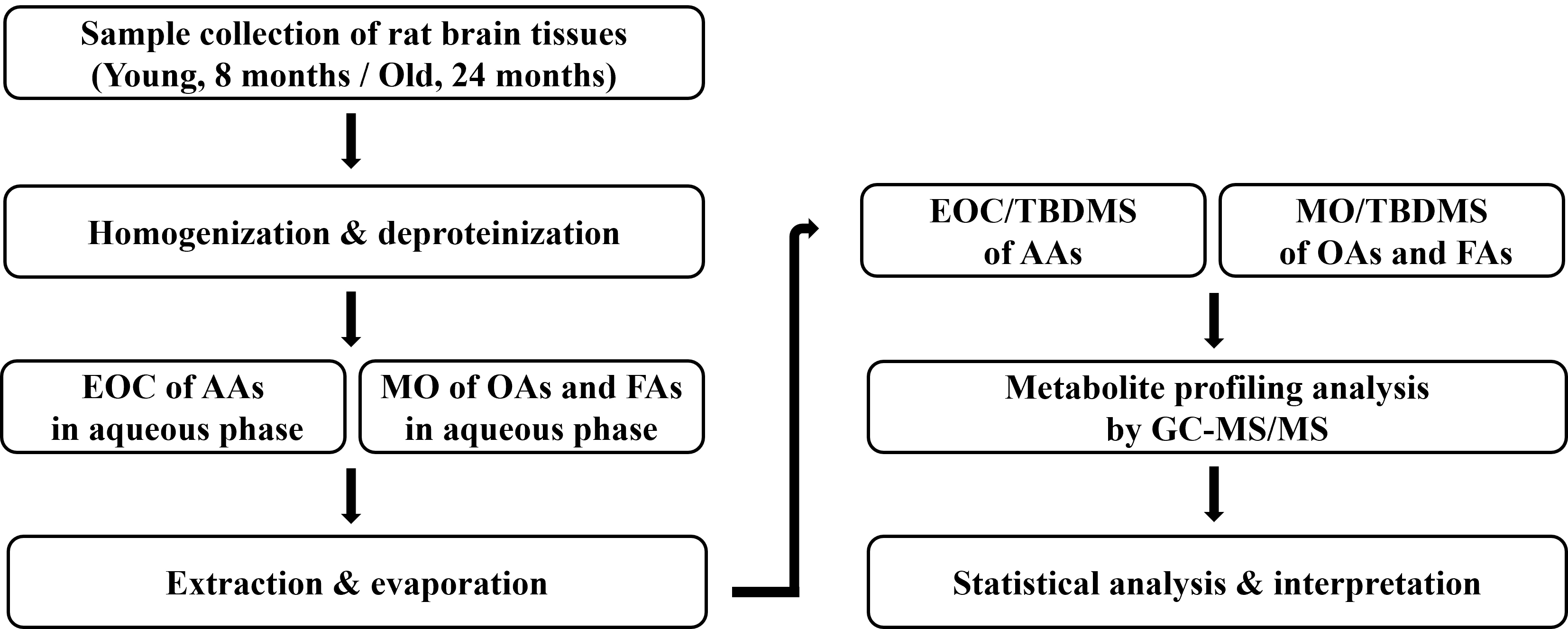
Background: Aging is a progressive process characterized by weakness in
brain function. Although metabolomics studies on the brain related with aging
have been conducted, it is not yet fully understood. A systematic metabolomics
study was performed to search for biomarkers and monitor altered metabolism in
various brain tissues of the cortex, cerebellum, hypothalamus, and hippocampus of
young (8 months old) and old rats (22 months old). Methods: Simultaneous
profiling analysis of amino acids (AAs), organic acids (OAs), and fatty acids
(FAs) in the brain tissues of young and old rats were performed by gas
chromatography-tandem mass spectrometry. Results: Under optimal
conditions, AA, OA, and FA profiling methods showed good linearity (r
0.995) with limit of detection of 30 and 73.2 ng and limit of
quantification of 90.1 and 219.5 ng, respectively. Repeatability varied
from 0.4 to 10.4 and 0.8 to 14.8% relative standard deviation and accuracy
varied from –11.3 to 10.3 and –12.8 to 14.1% relative error, respectively. In
the profiling analysis, total 32, 43, 45, and 30 metabolites were determined in
cortex, cerebellum, hypothalamus, and hippocampus, respectively. In statistical
analysis, eight AAs (alanine, valine, leucine, isoleucine, threonine, serine,
proline, and phenylalanine) in the cortex and four metabolites (alanine,
phenylalanine, 3-hydoxypropionic acid, and eicosadienoic acid) in the cerebellum
were significantly evaluated (Q-value 0.05, variable importance in projection
scores 1.0). In all brain tissues, the score plots of orthogonal partial
least square discriminant analysis were clearly separated between the young and
old groups. Conclusions: Metabolomics results indicate that mechanistic
targets of rapamycin complex 1, branched chain-amino acid, and energy metabolism
are related to inflammation and mitochondrial dysfunction in the brain during
aging. Thus, these results may explain the characteristic metabolism of brain
aging.