- Academic Editor
Liver cancer, particularly hepatocellular carcinoma (HCC), represents a global health challenge. The tumor microenvironment (TME) plays a pivotal role in the progression and therapeutic resistance of HCC. Interventional therapies have emerged as pivotal modalities in the treatment of liver cancer, especially in cases that are unsuitable for surgical resection. The evolution of these techniques has been markedly enhanced by the integration of artificial intelligence (AI), which has the potential to increase precision, improve outcomes, and personalize patient care. This review covers modern interventional therapies for liver cancer, highlighting recent advances in minimally invasive procedures. It describes the intricate liver TME and emphasizes the importance of characterizing its diversity and identifying therapeutic targets. Additionally, we discuss how AI can decipher TME complexities, predict responses, categorize patients, and personalize treatments. By elucidating connections between the TME, therapeutic interventions, and AI, this review aims to improve the management and care of patients with liver cancer.
Liver cancer remains one of the most challenging malignancies to treat, posing a significant global health burden. Advanced therapeutic strategies are urgently needed to improve patient outcomes and survival rates. Interventional therapies, such as trans-arterial chemoembolization (TACE) and radiofrequency ablation (RFA), have emerged as pivotal treatments in the management of liver cancer, particularly for cases in which traditional surgical options are not viable [1, 2].
The integration of artificial intelligence (AI) in oncology heralds a transformative era in cancer management, offering novel insights into disease complexity and treatment personalization [3, 4, 5, 6]. The tumor microenvironment (TME) in liver cancer plays a critical role in disease progression and therapeutic response, shaping the interactions between malignant and non-malignant cells within the hepatic landscape [7, 8, 9].
This review discusses the evolving landscape of interventional therapy for liver cancer, with a focus on the integration of AI to navigate the complexities presented by the liver TME. The current state of interventional therapies, the intricate nature of the liver TME, and the application of AI in relation to the TME and interventional therapy will be described in detail.
Liver cancer, primarily hepatocellular carcinoma (HCC), is a leading cause of cancer-related mortality worldwide. Its incidence is closely linked to the prevalence of chronic liver diseases, such as hepatitis and cirrhosis [10]. The pathophysiology of liver cancer involves a complex interplay of genetic, epigenetic, and environmental factors, contributing to the heterogeneity of this disease [11]. Interventional therapies for liver cancer have been developed to target tumors in a minimally invasive manner, often providing a therapeutic option for patients who are not candidates for resection or transplantation. TACE delivers chemotherapeutic agents directly to the tumor, combined with embolic materials to restrict the tumor’s blood supply [12, 13]. RFA and microwave ablation (MWA) use thermal energy to induce cellular death within the tumor [14, 15]. Trans-arterial radioembolization (TARE) involves the delivery of radiation-emitting beads to the tumor vasculature, offering a targeted radiological approach [16].
Current interventional treatments for HCC comprise several locoregional therapies, each with unique mechanisms. TACE combines the targeted delivery of chemotherapy with the occlusion of arterial blood supply, thereby inducing ischemic necrosis in tumors by starving them of nutrients and oxygen, while ensuring a high concentration of cytotoxic agents [17]. Liver cancer ablation therapy encompasses thermal ablation techniques, including RFA and MWA. This procedure entails percutaneous access to intrahepatic lesions, employing either high or low temperatures to induce thermal or cryogenic coagulation and effectively destroying the tumor cells [17, 18]. Moreover, TARE delivers Yttrium-90 microspheres directly into the hepatic artery, providing localized radiation that damages tumor DNA while preserving most of the surrounding healthy tissue [19]. Together, these approaches form an integrated framework for the effective treatment of HCC (Fig. 1).
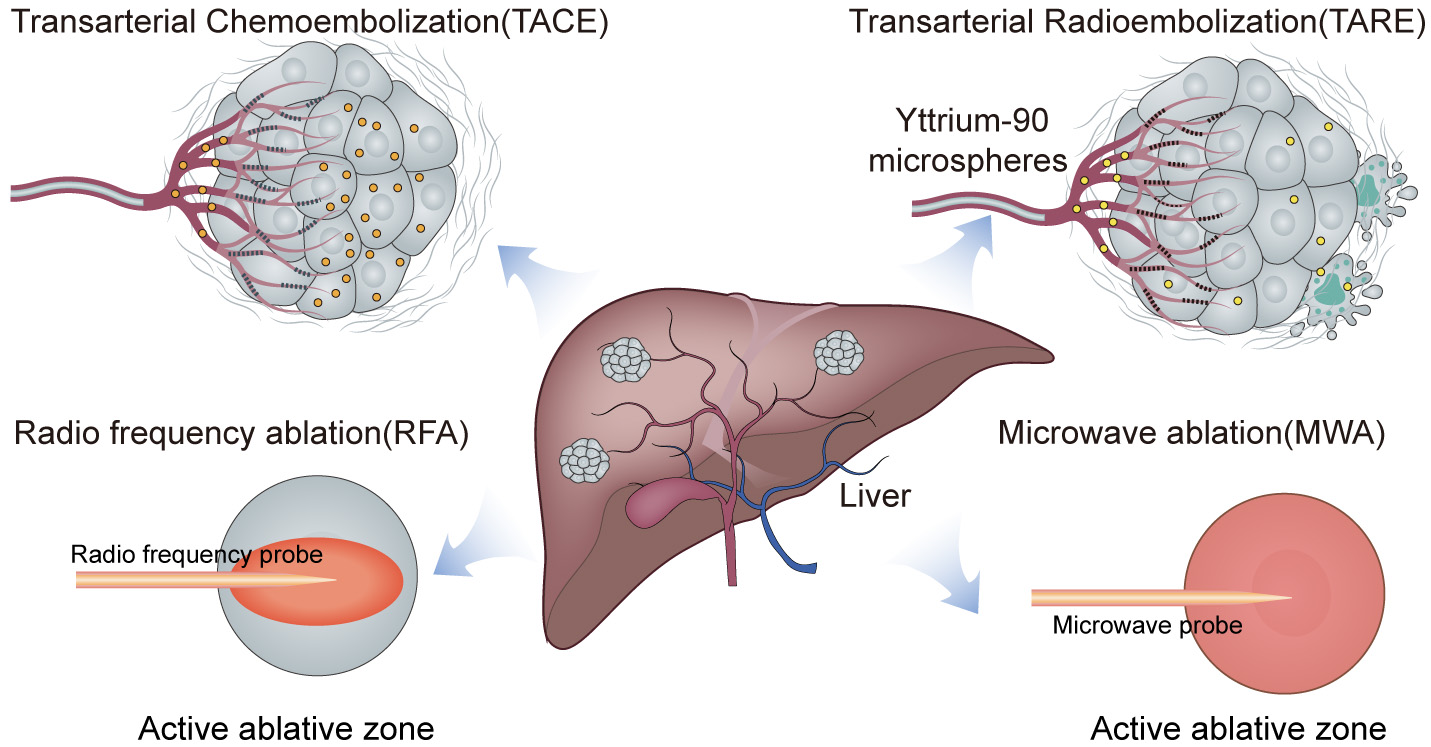 Fig. 1.
Fig. 1.
Schematic overview of interventional therapies in liver cancer.
Recent developments in interventional oncology have focused on enhancing the
efficacy and safety of established therapies, as well as the introduction of
innovative techniques such as irreversible electroporation (IRE) and
nanoparticle-assisted therapies [2, 20]. IRE is a non-thermal ablation technique
that disrupts cellular membranes using short, high-voltage pulses which lead to
apoptosis [21]. The COLDFIRE-2 trial demonstrated that IRE is an effective and
relatively safe salvage treatment for unresectable colorectal liver metastases
(CRLMs)
Interventional therapies have demonstrated significant improvements in local tumor control and overall survival rates for select patient populations with HCC [25]. However, challenges persist in optimizing treatment selection, managing post-procedural complications, and addressing tumor recurrence. The heterogeneity of tumors, variability in patient response, and the evolving landscape of combination therapies necessitate continued research and the refinement of interventional strategies to improve clinical outcomes. The outcomes of interventional therapies can vary significantly due to factors such as tumor size and location, and the presence of underlying liver disease. Challenges in achieving complete tumor eradication and managing post-treatment recurrences are considerable obstacles in clinical practice [26].
The TME is an intricate ecosystem comprised of various cellular and acellular components that surround and interact with malignant cells. In the most prevalent form of liver cancer, HCC, the TME plays a crucial role in tumorigenesis, progression, and the response to treatment [27]. Cellular and acellular components coalesce in this highly specialized niche to create a unique microhabitat that can influence tumor behavior, including proliferation, invasion, and resistance to therapies. Understanding the contribution of the HCC TME is essential for the development of more effective therapeutic strategies.
The cellular constituents of the HCC TME include immune cells, endothelial cells, cancer-associated fibroblasts (CAFs), and other stromal components. These cells engage in dynamic crosstalk with the cancer cells, mediated by a plethora of signaling molecules such as cytokines, growth factors, and extracellular matrix (ECM) components [28]. The TME can foster a pro-tumorigenic environment, thus facilitating cancer cell proliferation, survival, and invasion.
Cancer Cells: Liver cancer cells, primarily HCC, exhibit a multitude of genetic aberrations and phenotypic diversity. These cells often harbor mutations in key oncogenes and tumor suppressor genes, such as TP53 and CTNNB1, which drive the malignant phenotype and modulate interactions with the surrounding stromal cells [29].
Immune Cells: The immune cell milieu within the liver TME is a complex assembly.
Tumor-infiltrating lymphocytes (TILs), including CD8+ cytotoxic T cells, CD4+
helper T cells, and regulatory T cells (Tregs) play pivotal roles in tumor
surveillance and immunosuppression. Tumor-associated macrophages (TAMs) and
myeloid-derived suppressor cells (MDSCs) often exhibit an immunosuppressive
phenotype that can hinder anti-tumor immune responses [30]. TAMs can polarize
into M1 (anti-tumorigenic) or M2 (pro-tumorigenic) phenotypes depending on local
cytokine cues, with M2 often prevailing in liver cancer [31]. These immune cells
can also contribute to the formation of an immunosuppressive milieu by secreting
cytokines such as transforming growth factor-beta (TGF-
Fibroblasts: CAFs in liver cancer are known to originate from hepatic stellate
cells (HSCs) or portal fibroblasts, among others. Upon activation by factors such
as TGF-
Endothelial Cells: Liver cancer is characterized by a hypervascular phenotype, with endothelial cells being critical for neovascularization. Vascular endothelial growth factor (VEGF) is a key driver of angiogenesis in the liver TME, and its expression correlates with tumor progression and poor prognosis [35].
ECM: The ECM in liver cancer is not a passive scaffold but rather an active participant in tumorigenesis. It can form a physical barrier that impedes the delivery of chemotherapeutic agents to cancer cells. Moreover, TME cells can secrete soluble factors that activate survival pathways within cancer cells, conferring resistance to apoptosis [36] and playing a pivotal role in the development of resistance to therapies. Alterations in ECM components, such as increased crosslinking of collagen fibers, can lead to greater stiffness of the liver tissue, which has been associated with enhanced cellular proliferation and migration. The ECM also serves as a reservoir for growth factors that are released upon matrix degradation, further influencing tumor cell behavior [37].
Signaling Molecules: Cytokines such as IL-6 and TNF-
The interplay between these cellular and non-cellular components establishes a TME that is permissive for growth and metastasis of liver tumor cells. In particular, the immune landscape is a critical determinant of the tumor’s ability to evade host defenses. Moreover, the ECM and signaling milieu contribute to the mechanical and biochemical signaling that supports cancer cell survival and dissemination.
Therefore, targeting the TME is a promising avenue for enhancing the efficacy of existing treatments and overcoming therapeutic resistance. Novel strategies that modulate the immunosuppressive TME, or disrupt the stromal support of cancer cells, are currently being investigated and have the potential to improve outcomes for HCC patients [39]. A deeper understanding of the complex interactions within the TME is essential for the development of innovative therapies that can effectively target both the cancer cells and their supportive environment. For example, disruption of immunosuppressive networks within the TME can increase the efficacy of immunotherapeutic approaches, such as immune checkpoint inhibitors [40]. Similarly, targeting the ECM and its associated signaling pathways may disrupt the protective niche for cancer cells, thereby inhibiting tumor growth and metastasis [41]. Advanced technologies, such as spatial multi-omics, now enable analysis of the TME at unprecedented resolution, revealing the spatial organization and functional states of individual components and their interactions within the TME [42]. Such insights are invaluable for the development of next-generation therapies that can selectively target the most relevant aspects of the TME in liver cancer. By utilizing a multi-omic approach, the complex signaling networks and cellular interactions that underpin the liver TME can start to be unraveled. This integrative analysis may provide a more holistic view of the diverse mechanisms in tumorigenesis, paving the way for more personalized and effective treatment modalities for liver cancer patients.
Interventional therapies for cancer, such as chemotherapy, radiotherapy, and targeted therapies, have profound effects on the TME, which in turn affects therapeutic efficacy and tumor resistance (Fig. 2) [43]. A better understanding of these interactions is crucial for optimizing treatment strategies.
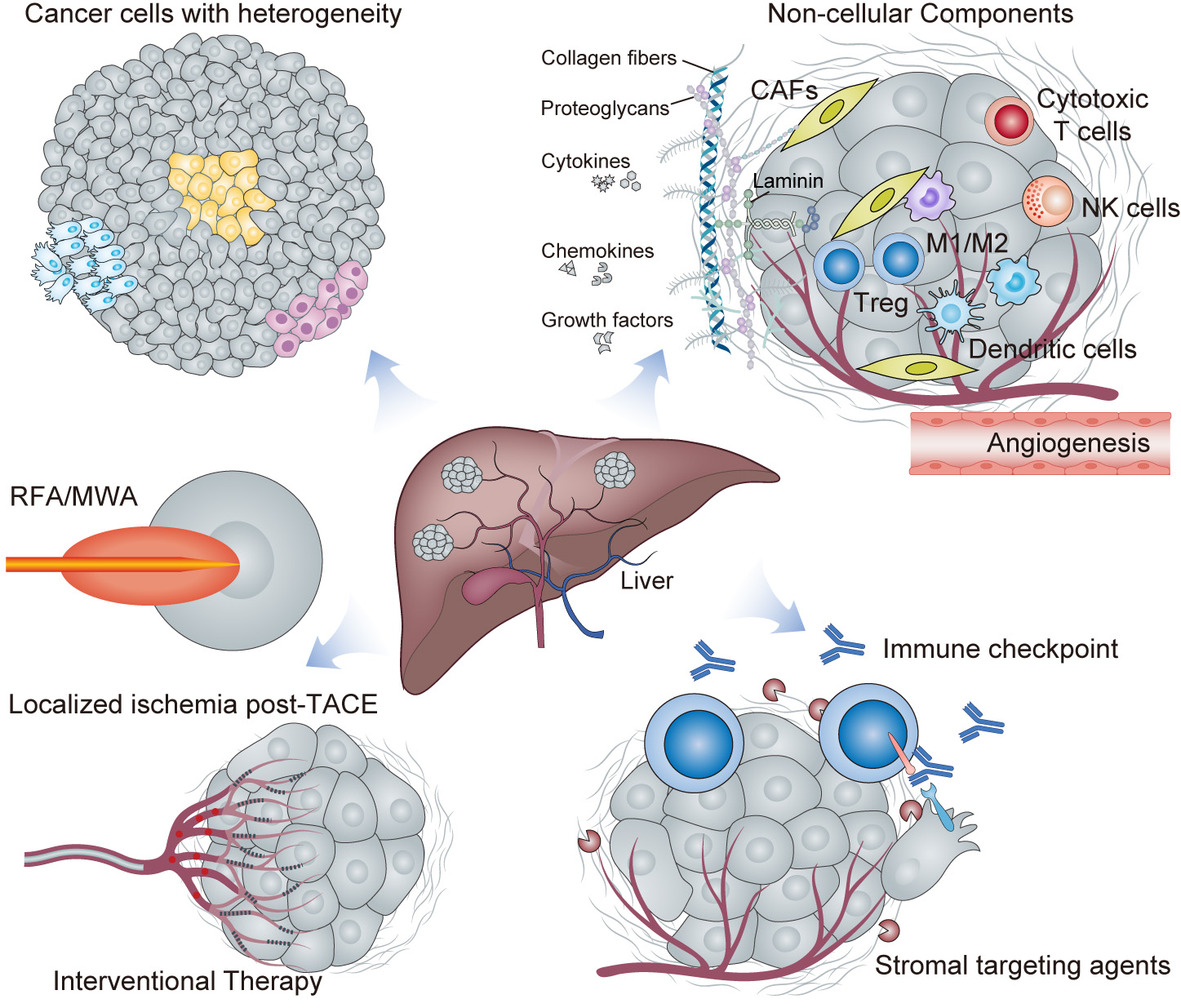 Fig. 2.
Fig. 2.
Complex interplay in the liver cancer tumor microenvironment (TME). This figure provides a comprehensive portrayal of the hepatocellular carcinoma (HCC) TME within a liver lobule, highlighting the cellular heterogeneity, immune cell diversity with both anti- and pro-tumor roles, the transition of fibroblasts to cancer-associated fibroblasts (CAFs), and tumor angiogenesis. The non-cellular matrix contains extracellular matrix (ECM) components and signaling molecule gradients. The effects of interventional therapies such as TACE and RFA/MWA on the TME are depicted schematically, showing altered signaling and cell fate. Finally, therapeutic modulation strategies such as immune checkpoint blockade and stromal disruption are represented to show TME “normalization” post-treatment. This figure encapsulates the dynamic and complex interactions occurring within the HCC TME, and which are essential for understanding and developing targeted therapeutic interventions.
Although interventional therapies are designed to target cancer cells, they also inevitably affect components of the TME, leading to both beneficial and detrimental outcomes [44, 45].
(a) Immune Modulation: Chemotherapeutic agents can cause immunogenic cell death, leading to the release of tumor antigens and activation of dendritic cells. This in turn can stimulate an adaptive immune response against the tumor [46]. Radiotherapy has also been reported to increase tumor immunogenicity by inducing the expression of MHC class I molecules and promoting T cell infiltration [47].
(b) Stromal Disruption: Many chemotherapeutic drugs, such as those targeting microtubules, can also affect CAFs and alter the secretion of ECM components, thus disrupting the physical barriers that impede drug penetration [18]. Similarly, angiogenesis inhibitors can normalize the aberrant tumor vasculature, potentially improving the delivery of chemotherapeutic agents [48].
(c) Metabolic Alterations: TME is often characterized by hypoxia and nutrient deprivation, leading to metabolic adaptations in cancer cells. Some targeted therapies, such as those inhibiting the HIF pathway, are aimed at disrupting these adaptations [49]. However, the interventions can also lead to compensatory metabolic changes within the TME that may promote treatment resistance.
TME can significantly impact the response to interventional therapies and the development of resistance.
(a) Immunosuppressive Networks: Immune components of the TME, particularly Tregs
and MDSCs, can create an immunosuppressive milieu that limits the efficacy of
immunotherapies such as checkpoint inhibitors [50]. Additionally, the secretion
of immunosuppressive cytokines like TGF-
(b) ECM Barrier and Drug Penetration: In many tumors, the dense ECM acts as a physical barrier that impedes the diffusion of therapeutic agents, thus contributing to treatment resistance. Collagen cross-linking and hyaluronan accumulation are known to increase interstitial fluid pressure, further reducing drug delivery [52].
(c) Hypoxic Adaptation: Hypoxia is a common feature of the TME and can lead to
resistance to radiotherapy and chemotherapy. Hypoxic conditions stabilize
HIF-1
(d) Vascular Abnormalities: Abnormal tumor vasculature can limit the delivery of interventional agents to the tumor site. The use of anti-angiogenic therapies, while initially beneficial, may lead to a more invasive phenotype or the selection of more aggressive cancer cell clones due to increased intra-tumoral hypoxia [54].
In conclusion, the interplay between interventional therapies and TME is an important factor in the development of cancer treatment. Research techniques that advance our knowledge of the TME, such as spatial multi-omics, should lead to a better understanding of these interactions and help to design better and more personalized interventions [55, 56]. Although data from spatial multi-omics research is still limited, it has the potential to provide clear evidence of TME spatial heterogeneity, and consequently the implications for different treatment modalities. A greater awareness of the potential value from combining multi-omic data with clinical outcomes should lead to a reconsideration of future cancer therapies, including the discovery of new biomarkers and therapeutic targets in the TME.
The advent of AI has heralded a transformative era in the field of oncology, with profound implications for cancer diagnosis, treatment, and research. AI encompasses machine learning (ML), deep learning (DL), and other computational approaches. It is poised to redefine the paradigm of cancer care by enhancing human expertise and enabling the extraction and recognition of meaningful patterns from complex biomedical data [57].
The utility of AI in oncology is multi-faceted (Fig. 3), ranging from improved precision of diagnostic imaging to the personalization of therapeutic regimens, as well as the identification of novel targets for drug development.
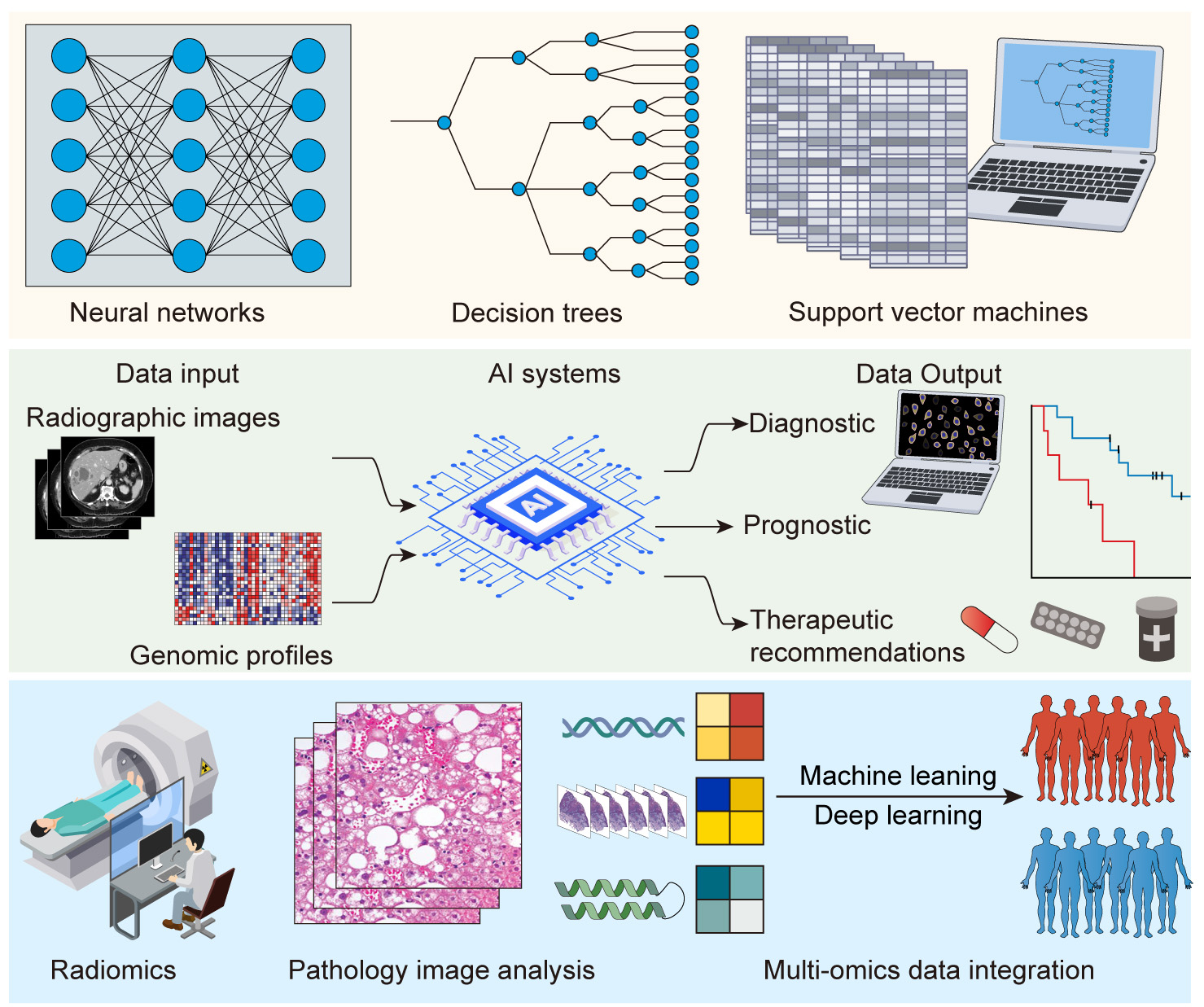 Fig. 3.
Fig. 3.
Artificial intelligence (AI) and oncology interface. Upper panel: schematic representation of AI algorithms (e.g., neural networks, decision trees, support vector machines (SVM)). Middle panel: flowchart illustrating the process of data input (e.g., radiographic images, genomic profiles) into AI systems and output (e.g., diagnostic, prognostic, and therapeutic recommendations). Lower panel: examples of AI applications, such as radiomics, pathology image analysis, and multi-omics data integration.
In the realm of diagnostics, ML approaches can be broadly categorized into supervised and unsupervised learning paradigms [58]. Supervised models include Random Forests (RF), Gradient Boosting Machines (GBM) and Support Vector Machines (SVM). These are able to predict survival, classify tumors, and assess treatment response. Unsupervised models such as k-means and hierarchical clustering can be used to classify TME subtypes. Principal Component Analysis (PCA) is used to reduce variability in genomic and proteomic datasets. However, challenges exist due to class imbalances in clinical datasets, along with the need for strong cross-validation to avoid overfitting. Recently, the DL model has emerged as the predominant approach for diagnostic imaging. Convolutional neural networks (CNNs) that use hierarchical learning to detect subtle tumor features such as microvascular invasion or necrosis have demonstrated exceptional proficiency in interpreting radiographic images, sometimes exceeding the performance of experienced radiologists [59] and reporting higher accuracy for the detection of HCC [60]. EmerGNN is a flow-based graph neural network (GNN) approach that builds on conventional methodologies for predicting drug–drug interactions in emerging drugs by effectively leveraging biomedical networks [61]. Additionally, Vision transformers (ViTs) provided better classification results over histopathology slides by utilizing global attention to capture long-range dependences [62]. For the longitudinal imaging of liver tumors, recurrent neural network (RNN) models such as long short-term memories (LSTM) can manage temporal change [63]. As for reinforcement learning (RL), the RL-assisted design of reactive oxygen species-responsive polymeric nanocarriers was shown to enhance the anti-tumor efficacy of radiotherapy in HCC treatment [64].
Radiomics is the process of extracting many quantitative features from medical images (e.g., Computed Tomography (CT), MRI, PET) for data analysis and diagnosis support. By analyzing these features, clinicians are able to identify characteristic tumor patterns that could potentially be used to predict disease prognosis and treatment response, and to assess other clinically relevant outcomes. Radiomics transforms imaging data into quantifiable information, thus aiding personalized medical decision-making. For example, researchers have developed useful 3D-ResUNet models that enhance volumetric CT/MRI analysis [65]. Attention models assist with locating features from the heterogeneity of tumors. However, challenges persist due to datasets with limited annotation, and to the costs associated with computation of high resolution 3D models. Radiomics workflows typically involve a domain for disentangling, extracting, and quantifying certain features (e.g., texture, shape, wavelet) from imaging data using software tools, and then leveraging these quantified imaging features through predictive analytics with ML/DL models. Dynamic contrast-enhanced (DCE) MRI data can be used with temporal CNNs to longitudinally evaluate perfusion changes pre- and post-TACE treatment [66]. Standard pre-processing steps include normalization, artifact remediation, and sampling strategies for patch sampling of images to reduce the processing burden of large amounts of imaging data. However, the lack of standardization between institutions for radiomics feature extraction has led to issues with reproducibility.
The utility of AI also extends to the prognostic domain, where it can assimilate and analyze diverse datasets, including genomic, transcriptomic, proteomic, and clinical data. This integration of multi-omics and clinical information facilitates the development of predictive models that stratify patients according to their risk and likely response to treatment, thus guiding the selection of tailored therapeutic strategies [67]. Multi-omics approaches may also use GNNs to construct models for the multi-omic interactions (genes, proteins, and immune cells) occurring in the TME [68]. Multi-model transformers help to integrate imaging, genomics, and clinical data into clinical stratification. General challenges with data-fusion include the handling of batch effects across varying omics platforms, and having sufficient real scale and harmonized datasets to train robust models. AI-based interventional tools allow real-time processing, thereby enabling precision therapy. Although these AI methods demonstrate strong technical capabilities in the research setting, the next stage in their development is testing with real-world case examples.
In the therapeutic domain, AI applications are redefining drug discovery and development. AI platforms can rapidly screen vast chemical libraries to identify potential anti-cancer compounds, predict their efficacy, and optimize drug design, thus significantly accelerating the pipeline from laboratory bench to patient bedside [69].
Despite these promising advances, the integration of AI into clinical oncology is not without challenges. Ethical considerations such as patient privacy and data security, and the potential for algorithmic biases, necessitate careful scrutiny [70]. Furthermore, bridging the gap between the capabilities of AI and its practical implementation requires robust validation studies, regulatory oversight, and the education of healthcare professionals to work synergistically with AI tools [71]. As the potential of AI is harnessed, the associated ethical and logistical challenges must be navigated with prudence and foresight to ensure this technological revolution benefits patients across the spectrum of cancer care.
In order to integrate AI into interventional therapy for liver cancer, several data quality and heterogeneity challenges must first be addressed. Imaging protocols, biopsy sampling, and electronic health records can all introduce biases due to variable data collection protocols across institutions, thereby complicating the training of AI models. Additional problems are the lack of annotated datasets and class imbalance issues. With supervised learning, there is often a scarcity of correctly labeled data regarding health conditions, post-treatments, and HCC subtypes. Interpretability is another challenge, since “black box” DL algorithms such as CNNs can provide high accuracy in specific AI tasks, but may reduce clinician trust in AI models. Although regulatory bodies require both AI transparency and explainability, current interpretability methods may oversimplify the complicated biological interactions found in the TME, while requiring more computational resources. Moreover, the computational resource requirements can be challenging. In particular, the high-performance graphic processing units (GPUs) and cloud-based systems required by the 3D-ResUNet model to perform tumor segmentation are not feasible in many clinical settings. Data privacy issues are another consideration in multi-institutional collaborations. Regulations such as the General Data Protection Regulation (GDPR) and the Health Insurance Portability and Accountability Act (HIPAA) prevent the sharing of patient data because it would improve AI model generalizability. Population-specific biases embedded in trained AI models should also be considered. Moreover, TME systems evolve during treatment. This necessitates the use of adaptive AI models that learn continuously, which is not a feature of most current static models. Addressing these challenges will require standardization of data collection, the use of hybrid-interpretable AI, and optimization of hardware so that AI tools are sufficiently reliable for deployment in clinical practice in a transparent and efficient manner.
AI has undergone rapid advances in cancer research, offering new perspectives for precision treatment. However, AI applications in interventional therapy for liver cancer still face challenges, such as data integration, model optimization, and clinical validation. The following section discusses how AI could improve interventional therapy in liver cancer (Fig. 4).
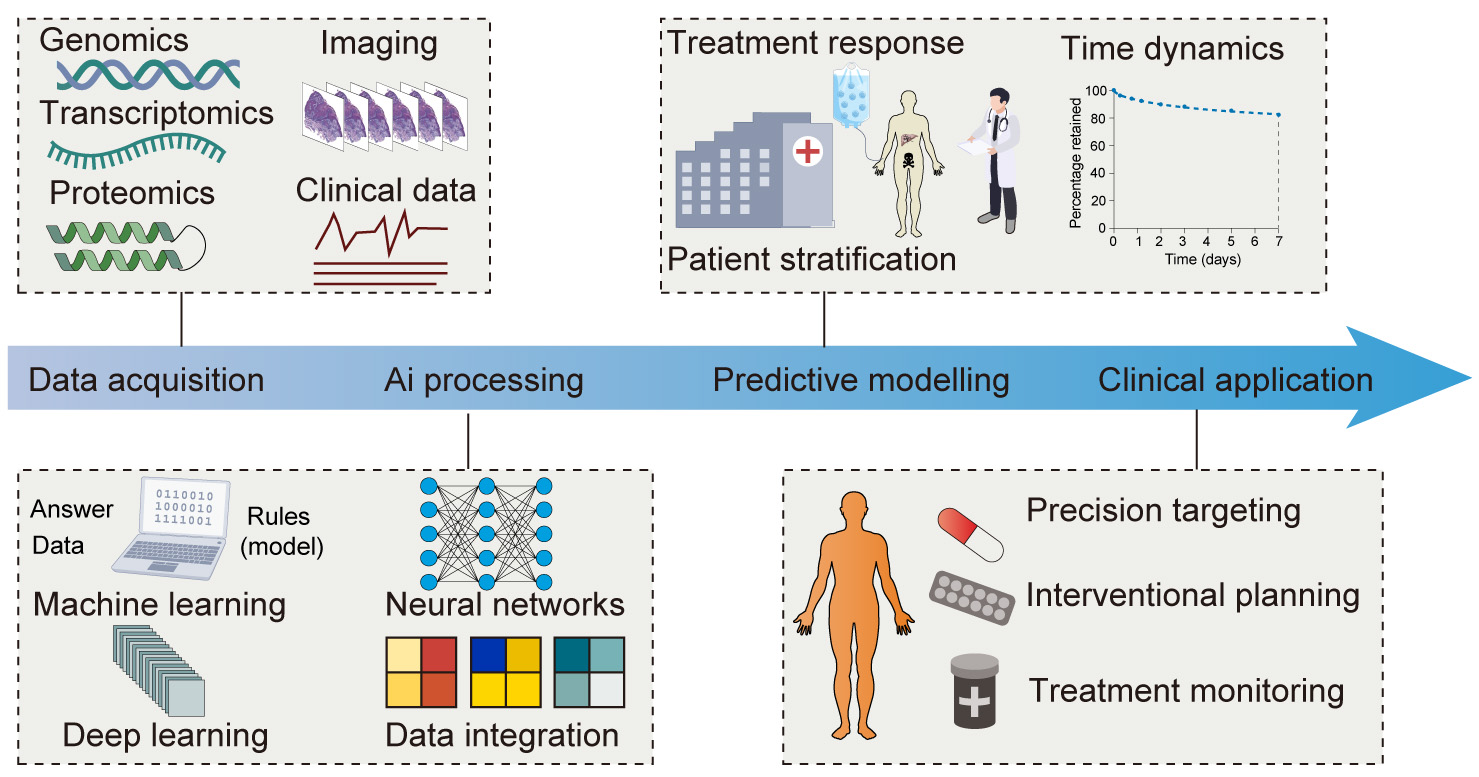 Fig. 4.
Fig. 4.
Harnessing AI for interventional oncology and TME-informed therapeutic enhancement. Flowchart showing an overview of the application of AI to improve interventional oncology through a TME-informed approach. The process begins with data acquisition, where a diverse array of inputs such as genomics, transcriptomics, proteomics, imaging, and clinical data are collected. This is followed by AI processing, which employs sophisticated techniques like machine learning (ML), deep learning (DL), and neural networks to achieve comprehensive data integration and analysis. The subsequent predictive modelling phase utilizes the processed data to predict treatment responses, understand the TME dynamics, and stratify patients more accurately. The final clinical application stage translates AI-derived insights into practice to improve precision targeting, interventional planning, and continuous treatment monitoring. This integrative framework highlights the transformative role of AI in personalizing cancer treatment and optimizing therapeutic interventions.
AI has transformed the diagnosis of HCC through unique image analysis. The diagnostic accuracy of AI algorithms is now on par with or even exceeds that of assessing radiologists across all major imaging modalities (Table 1, Ref. [72, 73, 74, 75, 76, 77, 78]).
| Modality/Model | Key feature | Performance | Clinical impact | Reference |
| CT with CNN | HCC detection | AUC: 0.89–0.92 | Improved early detection | [72] |
| 3D-ResUNet (CT) | Tumor segmentation | Dice: 0.88 ( |
Surgical planning | [73] |
| Radiomics analysis | Prediction of microvascular invasion | AUC: 0.974 | Pre-treatment risk assessment | [74] |
| Multiparametric MRI + DL | Lesion classification | AUC: 0.99 | Differential diagnosis | [75] |
| CEUS-based AI | Classification of focal liver lesions | Accuracy: 79–92% | Non-invasive characterization | [76] |
| AI segmentation model | Tumor/liver parenchyma delineation | Dice: 83.05% (tumor), 92.72% (liver) | Procedural planning | [77] |
| Clinical-radiomics model | INR, albumin + 10 radiomic features | AUC: 0.878 (vs. 0.785 clinical) | 30% reduction in overtreatment | [77] |
| Real-time radiomics | Intra-procedural tumor targeting | 35% improved coverage | 22% lower recurrence | [78] |
Note: CT, Computed Tomography; CNN, convolutional neural network; AUC, Area Under the Curve; DL, deep learning; CEUS, Contrast-enhanced ultrasound; INR, international normalized ratio.
CNNs analyze medical images through multiple layers. Early layers detect basic features like edges, while deeper layers identify complex tumor patterns. For example, studies have reported AUCs ranging from 0.89 to 0.92 with CNN models that assist in detecting HCC with CT-based diagnoses [72]. 3D-ResUNet enables volumetric segmentation through skip connections. These connections maintain spatial details while processing the full 3D context. This model yielded a Dice score of 0.88, with threshold tumors greater than 20 mm [73]. Radiomics complements DL by mathematically quantifying thousands of subvisual texture features from medical images. Radiomics-based analysis also improved HCC detection by predicting microvascular invasion with an AUC of 0.974 [74]. Multiparametric MRI images combined with DL technology achieved an extremely high AUC of 0.99 for lesion classification [75]. The addition of methods to assess peritumoral radiomics with a C-index of 0.73 to 0.77 improves recurrence prediction. Advanced ultrasound methods facilitated by AI have also been empowered. For example, contrast-enhanced ultrasound (CEUS)-based AI predicted multi-classification of focal liver lesions with an accuracy ranging from 79–92% [76], while highlighted real-time CNN visual predictive (analyses) for intra-procedural tumor targeting were associated with a 35% improvement in coverage. In short, the increasing incorporation of AI into all forms of medical imaging indicates ongoing transformation and improvement in the accuracy and precision of HCC diagnosis across imaging platforms.
AI-based radiomics is also changing the prediction of treatment response to TACE for HCC patients. Radiomics models use CT or MRI images to analyze features like texture, shape, and enhancement patterns. When combined with ML or DL, these models can predict which patients will benefit most from TACE. Several authors have demonstrated strong predictive performance for these AI models. For example, a segmentation model based on AI showed good performance, with average Dice scores of 83.05% for tumors and 92.72% for liver parenchyma. A combined clinical-radiomics model (using international normalized ratio (INR), albumin, and 10 radiomic features) performed better than clinical or radiomics models alone. It had an AUC of 0.878 for predicting post-TACE liver failure, compared to 0.785 (clinical model) and 0.815 (radiomics model) [77]. The clinical use of AI models has also demonstrated positive outcomes. For example, a 30% reduction in overtreatment was achieved by identifying non-responders early, and the use of real-time radiomics during TACE procedures led to a 35% improved coverage of tumors and 22% decrease in tumor recurrence [78]. These results indicate that durable and meaningful changes can be achieved by incorporating AI-based radiomics in clinical TACE treatment strategies for HCC patients, including prediction of tailored treatment response, and optimized treatment planning.
Collectively, these studies underscore the transformative impact of AI when integrated with interventional therapy. By harnessing the power of AI to interpret vast and complex data from HCC, clinicians can make informed decisions that lead to more precise and effective treatment. As AI algorithms continue to be refined and integrated into the clinical setting, we anticipate a new era of personalized and adaptive oncology care that is driven by deeper insights into the biology of the TME.
The TME is a complex mosaic of cellular and acellular components that play pivotal roles in tumorigenesis, metastasis, and response to therapy. Deciphering the interplay between different TME constituents within this intricate milieu and identifying potential therapeutic targets are critical for improving cancer treatment. The application of AI has emerged as an invaluable approach in untangling the complexities of the TME and identifying novel targets for interventional therapies (Table 2, Ref. [76, 77, 78, 79, 80]).
| Modality/Model | Key features | Performance | Clinical impact | Reference |
| DL model (Zhang et al.) | PD-L1, CD8A gene signatures | AUC: 0.92 | Predicts atezolizumab/bevacizumab response | [76] |
| Pathological-biomarker-finder | Novel tissue biomarkers | Accuracy: 88% | Identifies immune-active TME | [77] |
| Multi-omics integration | “Cold” vs. “hot” tumor classification | Correlates with PD-1 response | Immunotherapy stratification | [78] |
| ATLS-8 model | CD8+ TIL quantification | 5-yr OS: 65.03% vs. 34.05% (HR = 2.40) | Prognostic stratification | [79] |
| IRGPI (LASSO regression) | Immune-related gene prognostic index | C-index: 0.75 | Predicts immune cell infiltration | [80] |
Note: TIL, tumor-infiltrating lymphocyte; IRGPI, immune-related gene prognostic index.
AI, and especially DL and ML, has reshaped the prediction of immunotherapy
response in HCC, enabling the analysis of complex features of the TME and
biomarker signatures. For example, Zhang et al. [76] created a DL model
that analyzed histopathology images of HCC to identify immune-associated gene
signatures (PD-L1, CD8A) that correlated with response to
atezolizumab/bevacizumab combination therapy, achieving an AUC of 0.92. Liang
et al. [77] produced an interpretable DL framework called
Pathological-biomarker-finder that identified novel tissue biomarker signatures
associated with immune activity in the TME with 88% accuracy. Another pivotal
development is the quantification of CD8 TILs using AI. The ATLS-8 method
stratifies patients into prognostic groups. Those with high ATLS-8 had a 5-year
overall survival (OS) of 65.03%, while those with low ATLS-8 had a 5-year OS of
34% (HR = 2.40, 95% CI: 1.37–4.19, p = 0.015) [79]. AI has also
improved radiomics-based models, with clinical-radiomics models performing better
than solely clinical-based models (AUC: 0.89 vs. 0.60, p
AI has the capacity to integrate and interpret vast amounts of data derived from the TME, including but not limited to genomic, transcriptomic, proteomic, and imaging data. Such integrative analysis can reveal hidden patterns and biomarkers that are indicative of tumor behavior and therapeutic response. Multi-omics integration has also been instrumental in transition, as demonstrated by Xie et al. [78] who employed AI to integrate genomics and radiomics data. These authors successfully distinguished immune-excluded (‘cold’) tumors from immune-inflamed (‘hot’) tumors, which are classifications associated with the response to PD-1 inhibitors. Dai et al. [80] demonstrated the accuracy of AI integration into prognostic formatting by employing LASSO regression to create an immune-related gene prognostic index (IRGPI). This index predicted TME immune cell infiltration and the efficacy of immunotherapy with a C-index of 0.75. By applying interferon-gamma and antigen presentation gene expression signatures, DL models can stratify HCC patients into either immunotherapy responders or non-responders (e.g., nivolumab clinical trials) [81].
Beyond predicting the response to immune-based therapy, AI has also played a role in the prediction of treatment effectiveness. Combining radiomic features with clinical data in CT- and MRI-based DL models were found to improve the prediction of response to therapy. Nevertheless, significant challenges remain, including data heterogeneity, the ‘black-box’ nature of CNNs, and prospective validation. It will also be critical to standardize datasets and develop explainable AI (XAI) models to promote their clinical application [82]. Overall, AI has demonstrated multi-faceted roles in elucidating the complexity of the TME. It offers a lens through which the subtle and intricate interactions occurring within the TME can be observed and interpreted. By leveraging AI to reveal the underlying biological processes and to identify novel therapeutic targets, the precision and effectiveness of interventional therapies for liver cancer can be improved.
The notion of manipulating the TME to enhance the efficacy of cancer therapies is an area of active investigation. AI-driven approaches are increasingly being employed to model the TME and predict how manipulations might influence treatment outcomes. Such approaches include the use of computational simulations to predict the impact of altering TME factors concurrent with interventional strategies.
AI algorithms have the potential to simulate a variety of TME manipulations, ranging from the targeting of specific cell populations to the modulation of signaling pathways. For example, Mpekris et al. [83] utilized AI to model the vascular structure of the TME and its response to anti-angiogenic therapy in combination with chemotherapy. This computational approach helped to predict how changes in the vascular landscape of the TME could improve drug delivery and efficacy. Another AI-driven method employs agent-based modeling to simulate interactions within the TME, including the response of immune cells to immunotherapies. For example, an AI model was used to simulate the effects of immune checkpoint inhibitors on the spatial and temporal dynamics of T cells within the TME [84]. This predictive model provided insights into how reinvigoration of the immune system could be optimized when used in combination with other interventions.
AI has also been instrumental in predicting how genetic and epigenetic modification of the TME can influence therapy response. AI models were used to identify epigenetic alterations in the TME that could serve as biomarkers for response to epigenetic therapy [85]. By integrating multi-omics data, AI can guide the development of therapeutic strategies that modulate TME to improve patient outcomes.
In conclusion, AI-driven approaches are proving invaluable for modeling TME manipulation and providing a predictive framework for concurrent interventions. By simulating the dynamic and complex interactions within the TME, AI can help to design more effective combination therapies, ultimately paving the way for personalized treatment plans that are responsive to the unique characteristics of each patient’s TME.
AI is likely to revolutionize the personalization of interventional treatments in oncology over the coming years. It is poised to play a central role in tailoring therapies to individual patient profiles by taking into account the unique characteristics of their TME. Here, we explore some of the novel applications for AI in the crafting of interventional treatments.
One of the most promising directions for AI is the integration of multi-omics data to inform treatment decisions. The seminal work of Chen and Snyder [86] highlighted the potential use of omics data to obtain a holistic view of the patient’s biology. AI can synthesize information from genomics, transcriptomics, proteomics, and metabolomics to construct comprehensive models of the TME. This multi-faceted approach enables the identification of patient-specific molecular signatures that can predict response to specific interventional therapies.
The expanding field of spatial multi-omics is another area where AI is set to make a significant contribution. By combining high-dimensional data with spatial information, AI can map the complex organization of the TME and identify loci that are amenable to targeted intervention. A study on the use of spatial transcriptomics combined with AI analysis to map the cellular heterogeneity of tumors was described recently, providing insights into the spatial distribution of therapeutic targets [87].
In the drug development field, AI can accelerate the discovery of novel agents that modulate the TME to improve the efficacy of interventional treatments. Stokes et al. [88] reported on the use of DL to identify a new antibiotic compound through the analysis of bacterial gene expression profiles. A similar approach could be employed to discover agents that are able to reshape the TME, thereby increasing the effectiveness of interventional oncology treatments.
AI algorithms are also being developed to predict and monitor evolution of the TME in real-time during treatment, allowing for dynamic adjustments to therapy. The predictive modeling capabilities of AI can anticipate changes in the TME in response to treatment, enabling clinicians to modify the intervention strategy in order to avoid resistance or to enhance therapeutic efficacy.
In future, AI will become an indispensable tool in the personalization of interventional treatments. By harnessing the power of AI to interpret the wealth of biological data available, a new paradigm for cancer treatment arises whereby patients receive therapy that is precisely tailored to their individual TME, leading to improved outcomes and a higher quality of life.
A thorough understanding of the TME is crucial for the successful treatment of liver cancer, with AI being pivotal in uncovering its complexities. By facilitating analysis of the TME, AI has not only improved our understanding of tumor biology, but also enabled the discovery of novel therapeutic targets. This has the potential to enhance the specificity and efficacy of interventional therapies. The ability to target the TME effectively could lead to more durable responses and fewer systemic side effects, therefore fundamentally altering the prognosis for liver cancer patients.
ZX conceived the study, made tables and visualization, and wrote the original draft. WZ, JG, GT, and XC participated in literature collection, data interpretation, figure and table preparation, and critical revision of the manuscript. JS participated in searching references, manuscript review and editing. HC oversaw project administration, contributed to references searching, manuscript review and editing. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Not applicable.
Not applicable.
This research received no external funding.
The authors declare no conflict of interest.
During the preparation of this work the authors used ChatGpt in order to check spell and grammar. After using this tool, the authors reviewed and edited the content as needed and takes full responsibility for the content of the publication.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
