†These authors contributed equally.
Academic Editor: Graham Pawelec
Background: Dynamic contrast-enhanced (DCE) MRI is widely used to
assess vascular perfusion and permeability in cancer. In small animal
applications, conventional modeling of pharmacokinetic (PK) parameters from DCE
MRI images is complex and time consuming. This study is aimed at developing a
deep learning approach to fully automate the generation of kinetic parameter
maps, Ktrans (volume transfer coefficient) and Vp (blood plasma volume ratio), as
a potential surrogate to conventional PK modeling in mouse brain tumor models
based on DCE MRI. Methods: Using a 7T MRI, DCE MRI was conducted in U87
glioma xenografts growing orthotopically in nude mice. Vascular permeability
Ktrans and Vp maps were generated using the classical Tofts model as well as the
extended-Tofts model. These vascular permeability maps were then processed as
target images to a twenty-four layer convolutional neural network (CNN). The CNN
was trained on T
Glioblastoma multiforme (GBM) is the most common and lethal primary brain cancer. GBM is characterized by invasive tumor cell growth and extensive angiogenesis. Despite the development of highly angiogenic and leaky microvessels, many GBM cells grow by co-opting the pre-existing vessels, where the blood-brain barrier (BBB) may remain intact. Thus, the BBB disruption in GBM is markedly heterogeneous, which prevents therapeutic concentrations of chemotherapeutic agents from reaching the tumor in brain parenchyma [1, 2, 3, 4, 5].
MRI provides a noninvasive tool for acquiring information about the tumor
microenvironment. Advancements in MR techniques have increased noninvasive access
to a significant amount of useful information on cancer metabolism and tumor
heterogeneity, ranging from spatial scaled to functional or metabolic imaging.
Dynamic contrast-enhanced (DCE) MRI is a widely used technique for obtaining
information of vascular perfusion and permeability [6, 7, 8, 9, 10]. A series of
T
PK mathematical models require knowledge of the arterial input function (AIF),
which is an important variable for the modeling accuracy [16, 17, 18, 19, 20]. Lack of
accurate measurement of the AIF, also know was the vascular input function (VIF),
represents a major disadvantage of current PK models as a small deviation in the
AIF can markedly affect the estimation accuracy of kinetic parameters.
Measurements of the AIF is additionally more difficult if not impossible in small
animals in a case-by-case basis. Hence, a population-based averaged AIF from
literature assuming a consistent value is implemented here to avoid poor AIF
manual measurements leading to poor PK parameter accuracy. Population-based
averaged AIFs have additionally been shown to improve the reproducibility of PK
parameters [18, 20]. Moreover, PK models are solely based on fitting pixel
parameters to the CA concentration curves [21]. The pixel-wise model fitting is
computationally demanding for a whole MR scan with thousands of pixels and thus
is commonly time and memory consuming. Furthermore, manual identification of CA
arrival during the dynamic scans as well as additional MRI acquisitions for T
To address these limitations, we are seeking a machine learning approach for faster DCE parameter estimation without human bias. Machine learning has been increasingly used to power various research areas as it provides us with the ability to augment the knowledge and efforts of experts with a tool that can help to analyze data, and consequently make better predictions and decisions. Machine learning is commonly divided into two general forms, unsupervised and supervised learning. Unsupervised learning is concerned with understanding the internal structures of unlabeled data in a fully automated manner. Supervised learning requires training data to be manually labeled, where the algorithm learns from the labeled training data to predict the correct label for the unlabeled test data [22]. The model developed in this study is a supervised deep learning algorithm. Compared to the conventional machine learning methods, which require considerable research and design effort to capture higher-level features of the data, deep learning models learn data representations at various levels of abstraction automatically [23, 24]. One such advanced deep learning algorithm used for images is the convolutional neural network (CNN).
Recent studies have sought implementation of deep learning to facilitate
clinical PK parameter generation [25, 26, 27]. Nalepa et al. [26] have
developed a clinical end-to-end DCE processing pipeline for brain tumors.
However, this approach uses deep learning only for tumor segmentation and hence
still requires conventional PK modeling. Choi et al. [25] implemented
deep learning to improve the reliability of the AIF generation in astrocytomas to
improve PK parameter accuracy; however, similar to Nalepa et al. [26],
this approach still requires conventional PK modeling. In an effort to develop a
possible surrogate to conventional PK modeling, Ulas et al. [27] have
demonstrated the feasibility of implementation of deep learning for automatic PK
parameter generation directly from T
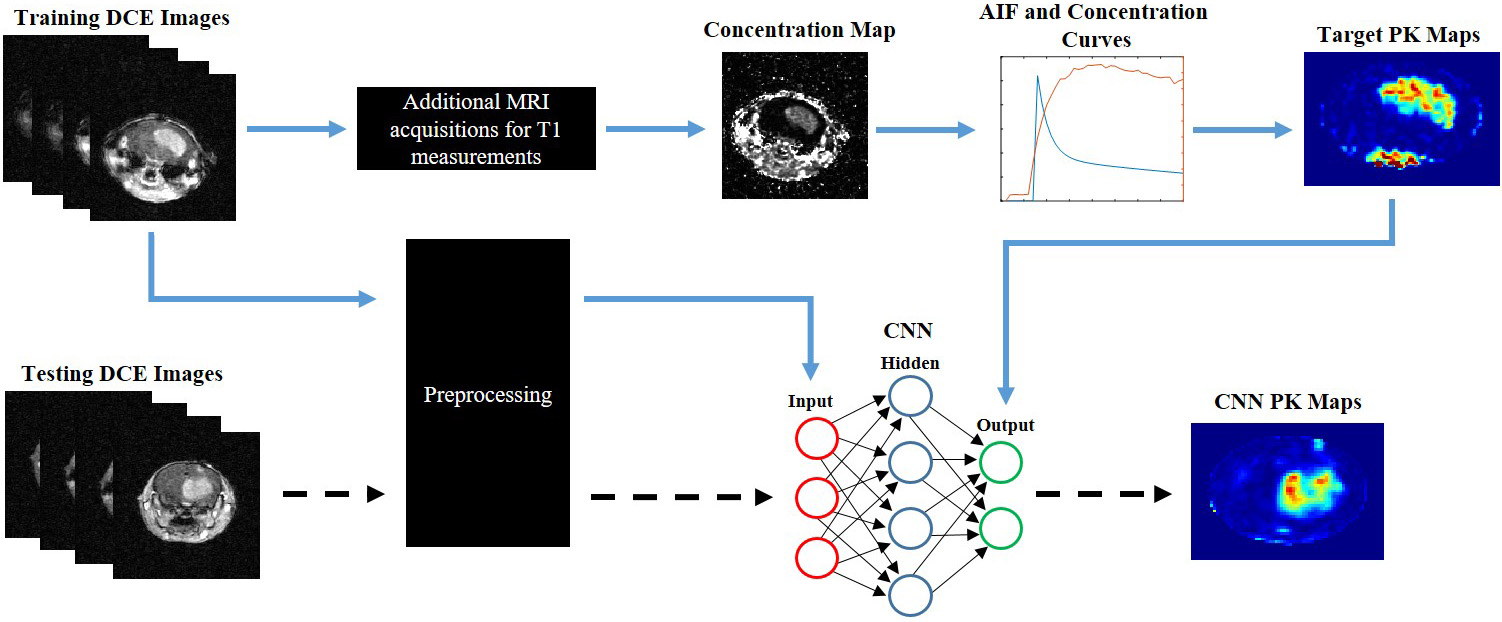 Fig. 1.
Fig. 1.Pipeline and workflow of this project. Conventional PK
modeling is applied to DCE dynamic images to generate target PK maps. Source DCE
dynamic images and target PK maps are fed into the 24-layer CNN for training
(blue arrows). An independent testing DCE dynamic imaging dataset can be fed
directly into the CNN to generate CNN PK maps following training of the neural
network (dashed arrows). Preprocessing of the DCE dynamic images involved
reshaping, segmentation, and batch normalization. The neural network can directly
predict the PK maps with the DCE time series alone. In contrast, conventional PK
modeling requires manual CA arrival identification, additional MRI acquisitions
for T
All animal procedures performed were approved by the Wake Forest University
Institutional Animal Care and Use Committee. Human glioblastoma U87 cells were
cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS, 1%
L-Glutamine, and 1% penicillin-streptomycin at 37
A breast cancer brain metastasis (BCBM) mouse model with intracardiac injection
of brain tropic breast cancer cells has been previously established at the lab
[31, 32]. Briefly, MDA-MB-231-BR cells (kindly provided by Dr. Steeg, NCI) were
cultured in DMEM with 10% FBS, 1% L-Glutamine, and 1% penicillin-streptomycin
at 37
Two weeks after tumor implantation, MRI was conducted. All MR imaging was
performed on a Bruker 7T Biospec 70/30 USR scanner (Bruker Biospin, Rheinstetten,
Germany). The mice were anesthetized in a chamber with 3% isoflurane, placed in
a 30 cm horizontal bore magnet, and the tail vein was catheterized for bolus
injection of Gd-DTPA (Magnevist; Bayer Healthcare) using a 27G butterfly. Animal
respiration was monitored and maintained throughout MRI experimentation using a
respiratory bulb. Anatomical T2-w images were acquired using a Rapid Imaging with
Refocused Echoes (RARE) sequence (TR/TE: 2500/50 msec; Number of Scan Averages
(NSA): 8; Echo Train Length (ETL): 8, Field of View (FOV): 22 mm
The acquired images were extracted in both raw Bruker 2dseq and DICOM file
formats. An additional 3D Gaussian filter was used on DCE dynamic data. DCE
images and variable flip angle T
Co-registered T
Similarly, Y coordinates were calculated by dividing the SI by sine of each angle.
These coordinates were then linearly fitted to quantify the slope (Sp). The T
where Tr is the repetition time of the MR scan. These T
where T
where S
where a and b are the coefficients determined from a population average of the
AIF and c
where Ktrans is the transfer rate constant from the blood plasma to the EES, Kep
is the transfer rate constant from the EES to the blood plasma, and Vp is the
fractional volume of blood plasma. C
In this study, the parameters are considered as a mapping problem between the dynamic time series and parameter maps. The CNN is trained to map the concentration of CA to the output. The estimated parameters from either the Tofts model or the Ex-Tofts model are processed as target images to the 24-layer CNN (Supplementary Fig. 1). The first convolutional layer applies 2D filters to each channel individually to extract low-level features. This was then followed by the two parallel pathways, local and global. The global pathway consists of 3 dilated convolutional layers each followed by a ReLU activation layer. The layers are dilated by factors of 2, 4 and 8, which captures the increase in receptive field size. This preserves the global structure and profile of the image. The local pathway consists of 3 non-dilated layers each of which are also followed by a ReLU activation layer. ReLU is applied to initiate nonlinearity into the mapping.
Zero-padding is applied to every convolutional operation to keep the spatial
dimensions of the output the same as the input. The filter size of each
convolutional layer is 4
The raw data from DCE MRI for each GBM animal was reshaped to have the temporal
series as the third dimension and scan slice as the fourth dimension. Data sets
were resized to 128
The networks were trained with a learning rate of 10
A transfer study was then performed to assess if the GBM trained CNN could
accurately predict PK maps for alternative brain tumors. Here, a total of 20
brain lesions detected in the three BCBM mice were implemented for testing. The
DCE images of BCBM were preprocessed similar to the DCE images from the GBM
study: reshaping of dynamic images to 128
All image processing was performed in MATLAB using homemade scripts. Following
testing of the proposed CNN, an ensemble of all networks was performed for each
study. Intratumoral PK parameters from both the target PK parameter maps and the
respective CNN maps were isolated for pixel-by-pixel comparisons. To this end,
anatomical co-registered T
Statistical analysis was performed using Microsoft Excel (Microsoft Corporation, Redmond, WA, USA) and GraphPad Prism 9.0 (GraphPad Software, San Diego, CA, USA). Pixel-by-pixel comparisons of Ktrans and Vp values within tumor regions between the CNN and the PK models were conducted. Linear regression was applied to provide statistical correlation and significance. To compare the similarity between the CNN maps and the corresponding PK target maps, root mean squared error (RMSE) values of the brain tumor were calculated along with normalized RMSE (nRMSE) values based on target PK parameter standard distributions (SD). Bland-Altman plots were generated to assess general trends in the predictive capability of the proposed CNN.
The Tofts Ktrans, Ex-Tofts Ktrans, and Ex-Tofts Vp generated maps were used as
target maps to train the deep neural network. The time required to generate maps
by the trained CNN was less than a few seconds with no human interference or
additional data. The Tofts and Ex-Tofts model required additional MRI
acquisitions for T
| Target PK model | Metric | Network 1 | Network 2 | Network 3 | Network 4 | Network 5 | Network 6 |
| Tofts Ktrans | RMSE | 8.60 × 10 |
8.73 × 10 |
2.37 × 10 |
4.34 × 10 |
1.34 × 10 |
7.05 × 10 |
| nRMSE | 1.28 | 1.04 | 1.02 | 0.83 | 1.26 | 0.76 | |
| Ex-Tofts Ktrans | RMSE | 1.87 × 10 |
2.24 × 10 |
1.50 × 10 |
1.88 × 10 |
1.84 × 10 |
2.31 × 10 |
| nRMSE | 0.80 | 0.95 | 0.86 | 0.94 | 0.97 | 0.93 | |
| Ex-Tofts Vp | RMSE | 1.72 × 10 |
2.36 × 10 |
1.58 × 10 |
1.88 × 10 |
2.04 × 10 |
2.27 × 10 |
| nRMSE | 0.86 | 0.81 | 1.04 | 0.90 | 0.81 | 1.38 |
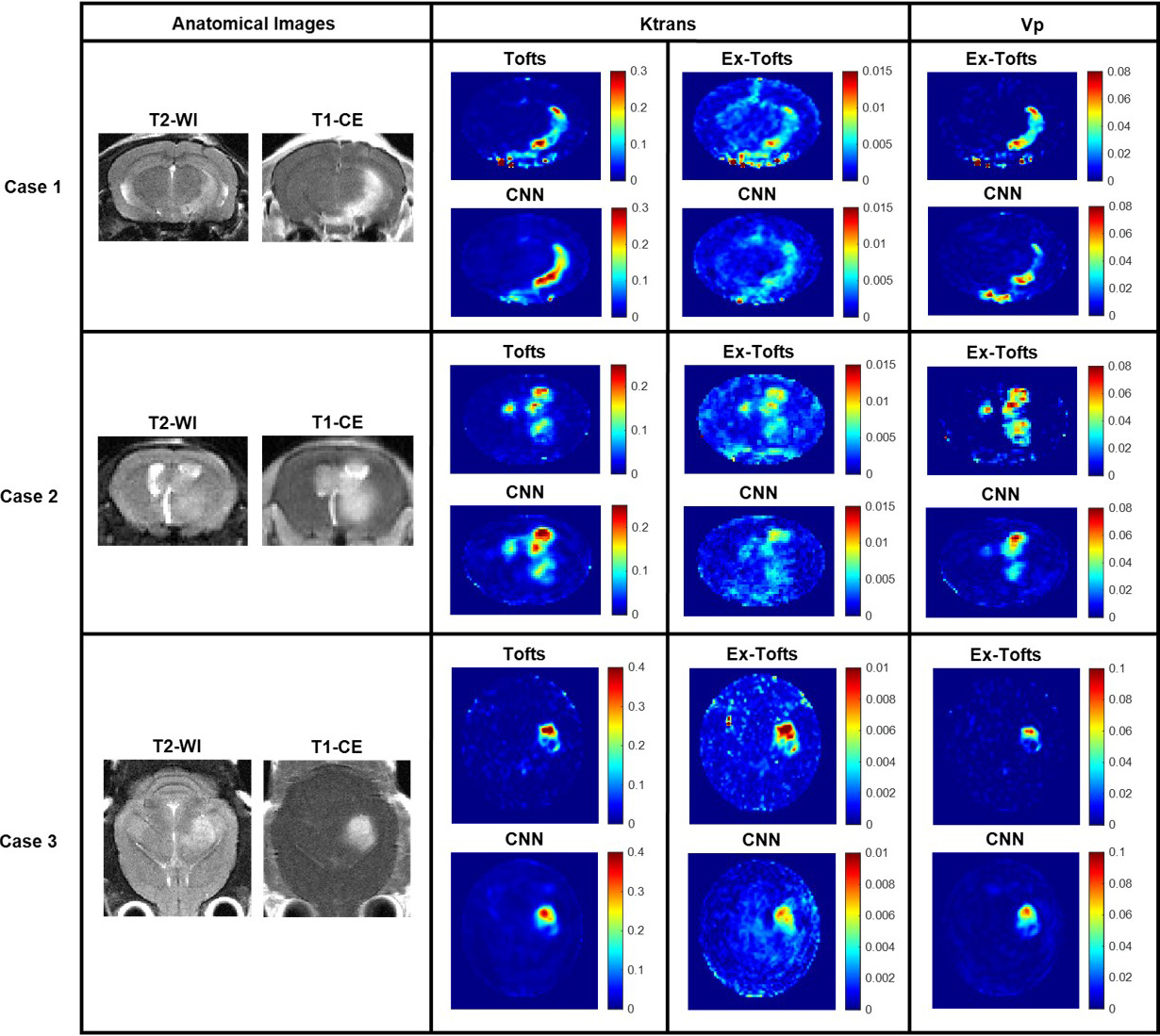 Fig. 2.
Fig. 2.GBM study. Three cases from the GBM study with permeable
lesions. Anatomical T2-w and T
To investigate if the CNN approach can decipher intratumoral heterogeneity, we
conducted pixel-by-pixel comparisons between the CNN and the target PK models.
Only the tumor region was considered following an ensemble of all networks for
the GBM study (n = 30 slices), containing a total of 7606 pixels. A significant
linear correlation (p
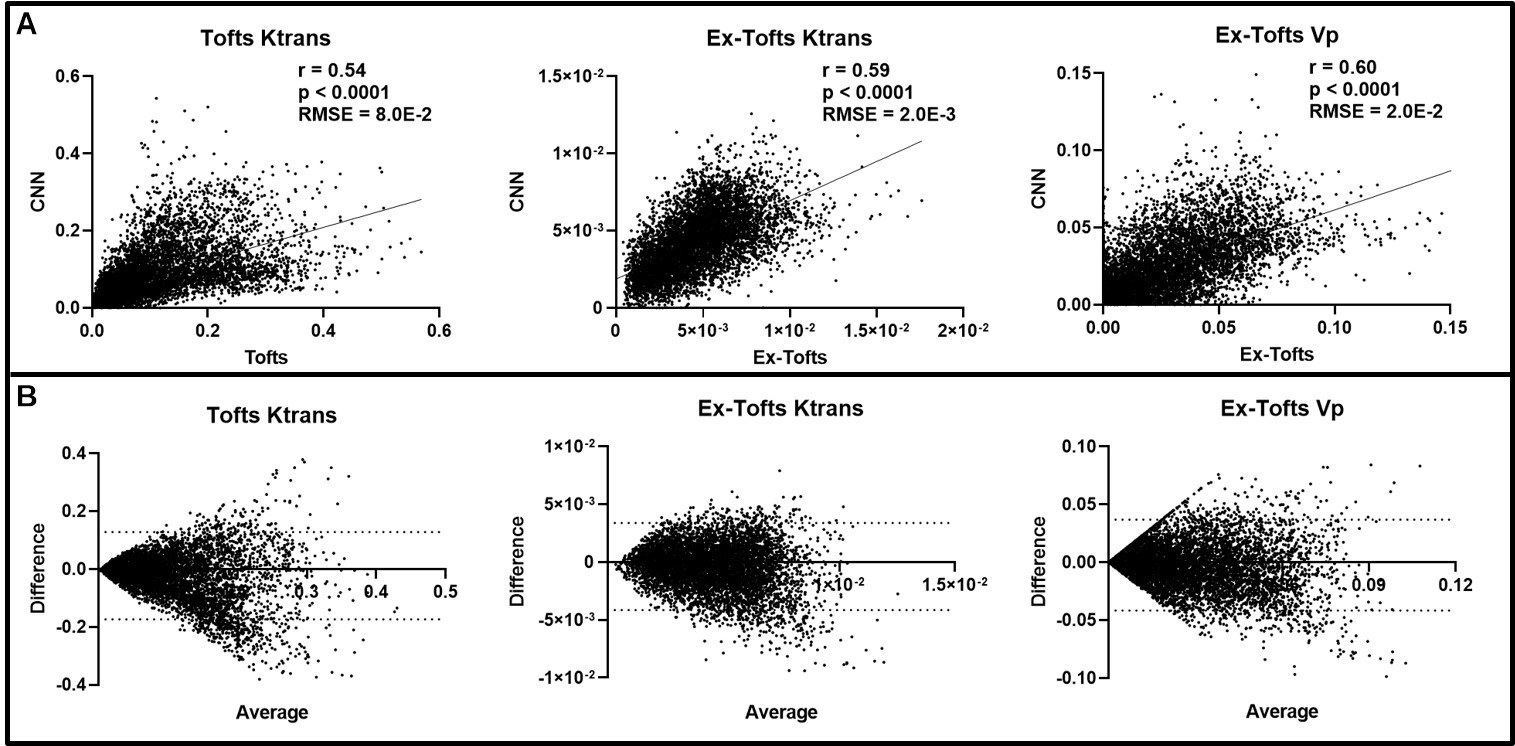 Fig. 3.
Fig. 3.Intratumoral ensemble analysis of GBM. (A) Pixel-by-pixel data
(n = 7606) of intratumoral regions were plotted, revealing a significant linear
correlation for Ktrans and Vp between CNN and target PK models (p
Following training and testing the proposed CNN with GBM, we performed a
transfer study with brain metastasis to assess the potential of the network for
alternative brain tumors. A total of 20 BCBM lesions were identified across the
15 dynamic slices used in this study. Unlike the glioma model, multiple brain
metastases including both contrast enhanced and non-enhanced lesions were
commonly seen in the BCBM model, representing an excellent phenotype for testing
the CNN approach for translation. Three networks, one for each target PK
parameter, were trained solely with GBM datasets followed by testing with BCBM.
Fig. 4 depicts three representative cases from individual BCBM mice. A small
lesion was identified for Case 1 on anatomical images that exhibited increased
vascular permeability on the target Tofts and Ex-Tofts PK maps. The CNN PK maps
retained the small size and shape of the BCBM lesion while learning each
corresponding PK parameter intratumoral values. Anatomical images for Case 2
revealed two tumors, one of which had only minimal enhancement on the T
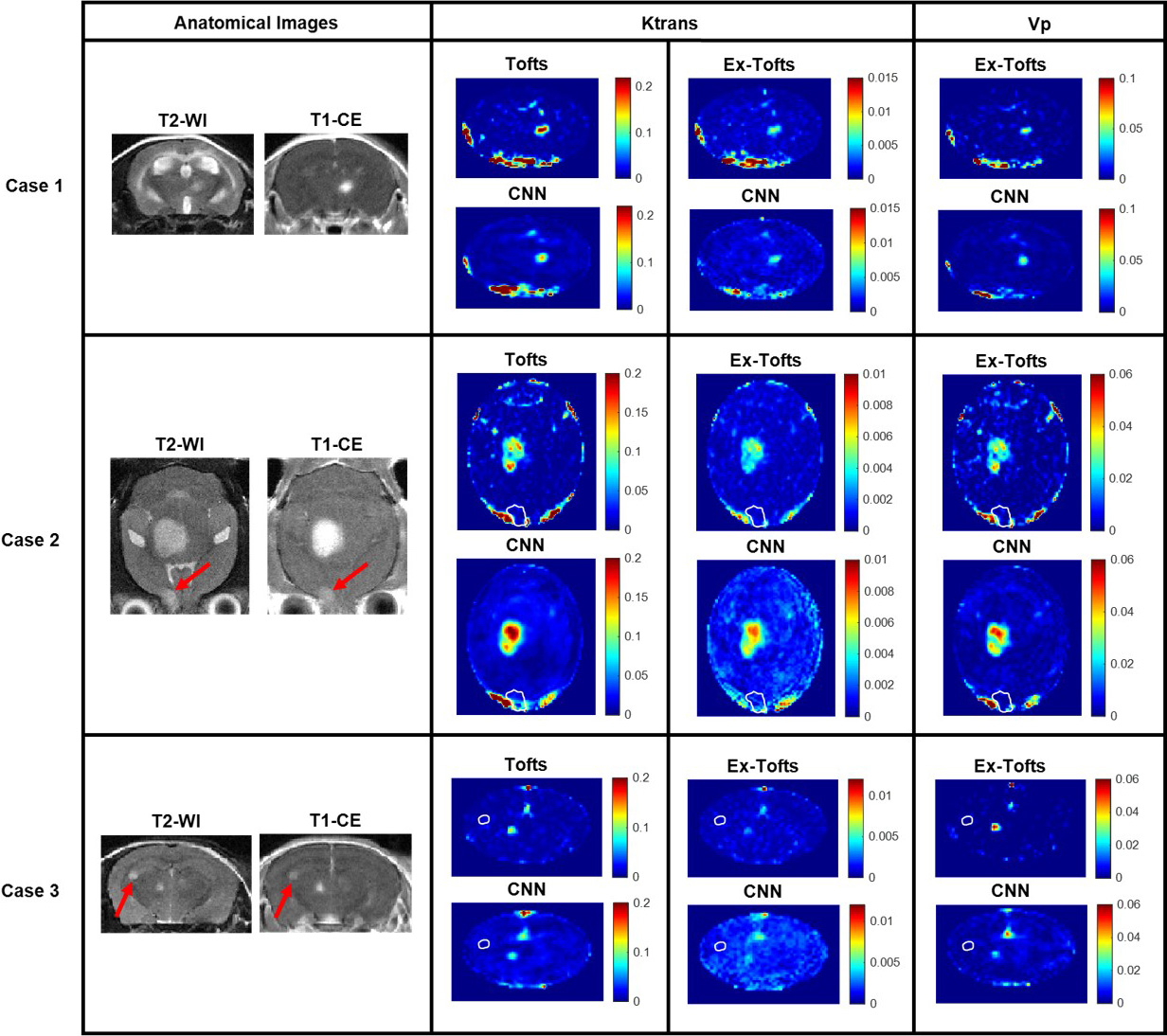 Fig. 4.
Fig. 4.Brain metastasis transfer study. MR imaging investigation of the brain metastasis transfer study revealed lesions with differing size, shape, and permeability. Tofts Ktrans, Ex-Tofts Ktrans, and Ex-Tofts Vp target maps as well as their corresponding CNN PK maps were depicted. The T2-w detectable edge of the lesions in Case 2 and 3 that exhibited lower permeability (arrows) were superimposed as a white contour on the PK maps.
Like the above glioma analysis, we conducted pixel-by-pixel comparisons for the
BCBM. There were a total of 20 lesions identified across the DCE dynamic images
(n = 15 slices), 14 of which were enhancing while six of them were non-enhancing,
indicating an intact BBB. Intratumoral pixel-by-pixel comparison revealed
significant linear correlation (p
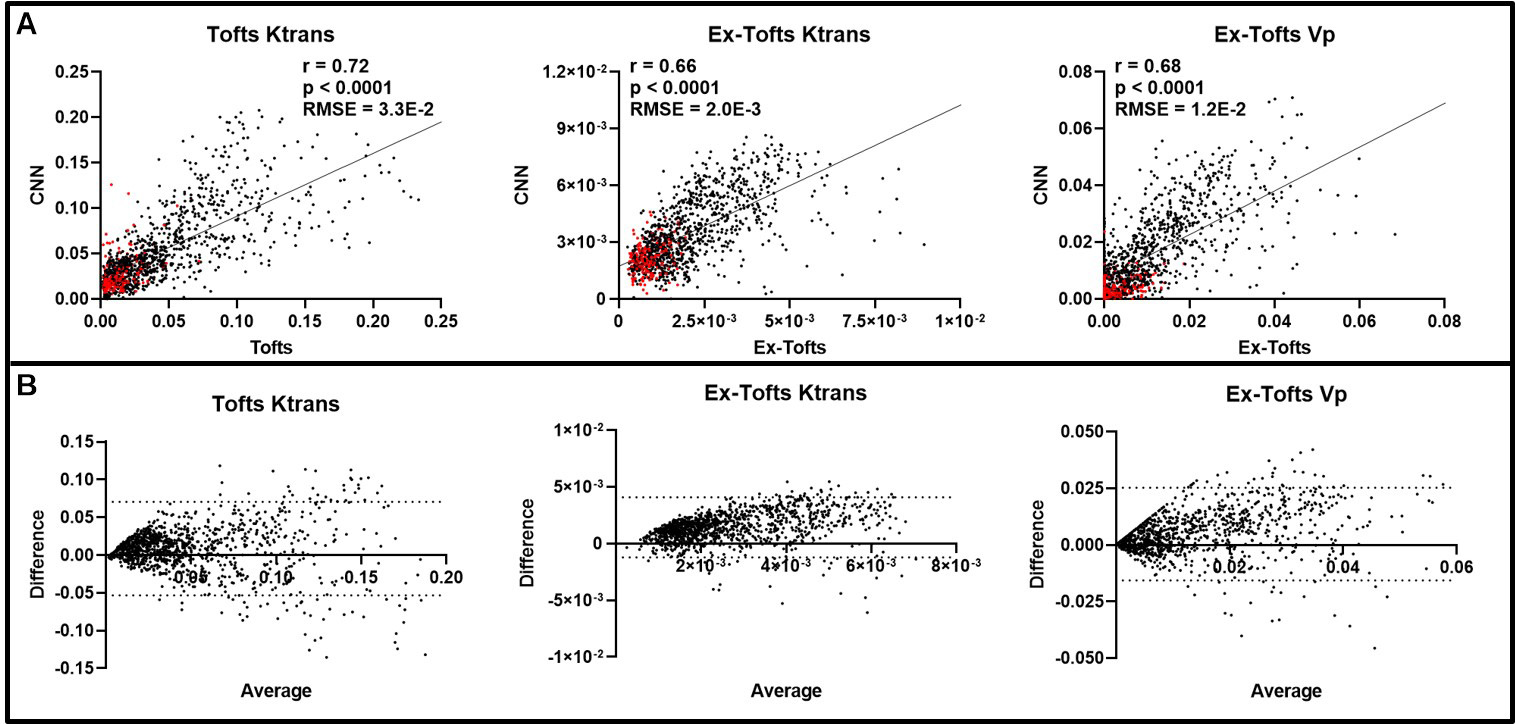 Fig. 5.
Fig. 5.Intratumoral ensemble analysis of brain metastases. (A)
Pixel-by-pixel data (n = 1148) of intratumoral regions were plotted that revealed
significant linear correlations between the CNN and target PK parameters
(p
In this study, we have successfully developed a deep learning-based CNN model to
generate the PK parameters for DCE MRI. CNNs are often implemented when working
with images and are the most commonly used networks for pattern recognition tasks
[36]. Hence, as generation of PK parameters is considered as a mapping problem to
train a deep neural network to recognize patterns of increased heterogeneous
intratumoral PK parameters from source DCE images, a CNN was chosen as an ideal
network for this study. The specific CNN methodology as used in this study has
previously been shown to yield accurate PK parameters directly from DCE images in
different brain regions of stroke patients without conventional PK modeling.
Hence, we hypothesized this CNN could be translated to small animal brain tumor
research studies. Generative adversarial networks (GANs) have been gaining
popularity in medical image processing as a more sophisticated deep learning
method in which two networks compete against each other and hence might serve as
a potential network for this application in future studies [36, 37].
Nevertheless, our results show a good match for both Ktrans and Vp maps generated
between the target PK parameter maps and the respective CNN PK parameter maps for
brain tumors. Importantly, the CNN PK maps were shown to successfully
recapitulate intratumoral heterogeneity and increased permeability in accordance
with their target PK model parameter maps. The CNN approach to directly generate
the PK maps from DCE dynamic images is time efficient (within a few seconds) and
requires no human interference or additional data, whereas conventional PK
modeling is much more time consuming and requires additional information
including CA arrival, T
K-fold cross validation is commonly implemented in deep learning applications to
assess the performance of the neural network through cycling training and testing
datasets [27, 38, 39]. This methodology ensures that the network can be
transferred to alternative testing cases of variable phenotypes and that the
network is avoiding overfitting of the training data through increasing the
amount of testing data. Here we trained and tested a total of 21 networks, 18 for
the GBM study and 3 for the brain metastasis transfer study. In the k-fold cross
validation GBM study, similar RMSE and nRMSE values were found for each
respective PK parameter (Tofts Ktrans, Ex-Tofts Ktrans, and Ex-Tofts Vp) across
each network demonstrating the power of the deep learning approach to decipher
intertumor variation for each testing case with high accuracy (Table 1). In
particular, the target Ex-Tofts Ktrans and CNN produced consistently low RMSE
values below the target parameter SD (nRMSE
In line with the GBM study, significant linear correlations (p
In general, deep learning and machine learning algorithms require huge amounts of data to perform efficiently upon training [40]. With limited availability of animal data in this study, there is a higher risk of overfitting the data, which can cause poor generalization and contribute to crude predictions. To solve such complications, the algorithm requires additional data to converge to a general case. Recent studies have shown the impact of augmented data in solving overfitting problems [41, 42]. However, this approach of data augmentation was computationally costly and complex on this project as the DCE data trained was a complex time-dependent series of images. Hence, we proceeded with no additional data augmented to train the network. Despite a small number of training data for both the GBM study (n = 25 slices) and the brain metastasis transfer study (n = 30 slices), our results indicate that the proposed neural network can still efficiently recapitulate the tumor size, shape, and PK permeability parameters in both primary and metastatic brain tumors while avoiding overfitting of the data as evidenced through its translatability to alternative brain tumor phenotypes. This is likely attributable to the fact that each of these training ‘slices’ involved multiple complex dynamic images with thousands of pixels per image for the network to learn from.
In summary, we have successfully deployed the Tofts model and the Ex-Tofts model as well as a deep learning-based CNN to study vascular permeability in small animal brain tumors. Results and observations from the CNN infer that this deep learning approach could perform as precisely as the conventional PK models. Importantly, the CNN approach requires limited data without human intervention and can be completed with much less time. Future studies by training and testing the system with more data and including additional glioma models with differential phenotypes of tumor aggressiveness and angiogenesis will be needed to further establish its utility. Furthermore, implementation of an accurate AIF measurement in these small animals in a case-by-case basis prior to CNN training might enhance the accuracy of the produced conventional PK and CNN maps by taking into account individual variation. We anticipate that the proposed deep learning-based CNN will serve as a useful surrogate for small animal brain tumor research of vascular PK parameters.
AIF, arterial input function; BBB, blood-brain barrier; BCBM, breast cancer
brain metastasis; BTB, blood-tumor barrier; CA, contrast agent; CNN,
convolutional neural network; CSF, cerebrospinal fluid; DCE MRI, dynamic
contrast-enhanced MRI; DMEM, Dulbecco’s modified Eagle’s medium; EES,
extravascular extracellular space; ETL, echo train length; Ex-Tofts, extended
Tofts; FA, flip angle; FOV, field of view; GBM, glioblastoma multiforme; Kep,
flux rate constant; Ktrans, volume transfer coefficient; nRMSE, normalized root
mean squared error; NSA, number of scan averages; PK, pharmacokinetic; RARE,
rapid imaging with refocused echoes; RMSE, root mean squared error; SD, standard
distribution; SI, signal intensity; Sp, slope; ST, slice thickness; T
CAA, DMS, LW, and DZ conceived and designed the research study. CAA, DMS, WNC, YL, and LW performed the research. WNC performed small animal imaging. CAA and DMS developed the network and analyzed the data. YL, LW, and DZ helped supervise the project and provided critical feedback. CAA, DMS, WNC, and DZ wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
All animal procedures performed were approved by the Wake Forest University Institutional Animal Care and Use Committee (A21-124 and A21-160).
We are grateful to Jared Weis, Biomedical Engineering at Wake Forest, Umit Topaloglu, Cancer Biology at Wake Forest, and Hannah Dailey, Mechanical Engineering and Mechanics at Lehigh University, for valuable input and collegial support.
This research has been supported in part by NIH/NCI R01 CA194578 and Wake Forest Comprehensive Cancer Center under Grant P30 CA01219740.
The authors declare no conflict of interest.
