Background: The world faces the challenge posed by the interaction between hosts and Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) with potential role for arthropod vectors, and the effect of SARS-CoV-2 variants on acquired immunity, vaccine efficacy and coronavirus disease-19 (COVID-19) pandemic control. Proposal: The characterization of the role played by animal hosts and host-virus interactions is essential to address this challenge. Zoonotic (animal-to-human) and reverse zoonotic (human-to-animal) routes may be involved in virus transmission with a possible still unconfirmed role for arthropod vectors. Herein we propose to consider the risks posed by the possible role of arthropod vectors in COVID-19 and that immunity against SARS-CoV-2 may increase the risk for zoonotic virus transmission. These risks should be considered when evaluating vaccine efficacy and monitoring animal SARS-CoV-2 variants. Conclusion: Virus surveillance, epidemiology, sequencing and evaluation of susceptibility to antibodies and other protective immune mechanisms from vaccinated individuals should be improved. A One Health approach such as the one applied by our group SaBio is necessary for a more effective control of COVID-19 and prevention of future pandemics.
Vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are a major achievement for the global control of the coronavirus disease 19 pandemic (COVID-19). Recently, identified SARS-CoV-2 variants have raised concern about fast-spreading of the virus and their potential effect on limiting vaccine efficacy [1, 2, 3]. Although current evidence suggest that vaccines remain effective at least partially against these variants of concern (VOC), the role played by animal hosts is also essential to address this question [4]. SARS-CoV-2 variants produced in low-susceptible and chronically infected hosts may be more effective in evading immunity and producing high viral loads, thus increasing the risk for zoonotic virus transmission [4].
Currently, the most accepted hypothesis is that the COVID-19 pandemic emerged by SARS-CoV-2 transmission from insectivorous bats through other intermediate (bridge) mammals to humans [5]. Natural infections with SARS-CoV-2 have identified several potential animal hosts with evidence of reverse zoonotic (human-to-animal) and sporadic zoonotic (animal-to-human) virus transmission [4, 6, 7]. Based on these results and models for SARS-CoV-2 Spike (S)-angiotensin I converting enzyme (ACE) host receptor interactions, animals with close-to-human S-ACE interactions (e.g., great apes or ruminants) may constitute effective hosts for maintenance, evolution and zoonotic transmission of virus variants highly infectious in humans. Other animal species susceptible to SARS-CoV-2 but with low S-ACE interaction capacity (e.g., cats or pigs) would be susceptible to reverse zoonotic virus transmission with low risk for human infection [8]. Additionally, modelling interactions between SARS-CoV-2 S-protein and arthropod ACE receptor suggested a possible role for vectors in coronavirus transmission even if low viremia reduces the risk for transmission [9].
Several factors affect the selection/appearance of new virus variants. As other coronaviruses, SARS-CoV-2 acquire amino acid substitutions as a result of the proofreading RNA polymerase [3]. Additionally, these viruses selectively delete small fragments of its RNA that are not corrected by RNA polymerase and may translate into protein modifications [2]. These modifications occur continuously in a recurring pattern of evolution and contribute to virus adaptation to host factors (i.e., receptors) and evasion of immune response [1, 2, 3]. Based on these mechanisms, the virus carrying the H69/V70 deletion appeared in Denmark as a virus variant that originated on mink farms and passed to humans, with the capacity to better adapt to minks while evading human antibody response [10]. Additionally, virus variants with various mutations have been isolated from animal hosts [11].
A One Health approach is required to control the COVID-19 pandemic and other infectious diseases with global incidence and for prevention of future pandemics. As defined by the Centers for Disease Control and Prevention (CDC; https://www.cdc.gov/onehealth/basics/index.html), “One Health is a collaborative, multisectoral, and transdisciplinary approach working at the local, regional, national and global levels, with the goal of achieving optimal health outcomes recognizing the interconnection between people, animals, plants, and their shared environment” and the World Health Organization (WHO; https://www.who.int/news-room/q-a-detail/one-health) “One Health is an approach to designing and implementing programmes, policies, legislation and research in which multiple sectors communicate and work together to achieve better public health outcomes”. The importance of One Health is reinforced by factors that increase risks of infectious diseases such as (a) growth and expansion of human populations, (b) increased people contact with wild and domestic animals, (c) climate change, (d) deforestation and intensive farming practices, and (e) international travel and trade of people, animals and animal products. For example, the impact of climate change on factors such as vegetation canopy and phenology, host density and abundance of reservoir species affect tick ecology and the spread of tick vectors and transmitted pathogens [12].
In this paper, we propose to consider the risks posed by the possible role of arthropod vectors in COVID-19 and in zoonotic virus transmission, and the need to use a One Health approach for a more effective control of COVID-19 and prevention of future pandemics.
Arthropods such as mosquitoes, flies, mites, fleas, lice and ticks can act as vectors of viruses and other pathogens via active biological transmission but also through passive mechanical transmission [9, 13, 14, 15, 16] (Table 1, Ref. [17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30]). Biological transmission occurs when vectors transmit pathogens that multiply within their bodies [31, 32]. Mechanical transmission is mediated by vector transfer of pathogens from contaminated exoskeleton, feet, mouth parts or internal organs to host body or sustenance [31].
| Pathogens | Arthropod vectors | Hosts | References |
| Lumpy skin disease virus, LSDV | Amblyomma hebraeum | Wild ungulates | [17] |
| Domestic ruminants | |||
| Rickettsia felis | Ctenocephalides felis | Cat | [18] |
| Dog | |||
| Opossum | |||
| Raccoon | |||
| Rodent | |||
| Human | |||
| Anaplasma marginale | Stomoxys calcitrans | Cattle | [19, 20, 21] |
| Human immunodeficiency virus 1, HIV-1 | Ornithodoros moubata | Human | [22] |
| African Swine Fever Virus, ASFV | Stomoxys calcitrans | Pig | [23] |
| Porcine reproductive and respiratory syndrome virus, PRRS | Musca domestica | Pig | [24] |
| Avian influenza virus H5N1 | Musca domestica | Chicken | [25] |
| Equine infectious anemia (EIA) RNA virus | Tabanus sp. | Horse | [26] |
| Stomoxys calcitrans | Donkey | ||
| Mule | |||
| Shigella spp. | Musca domestica | Human | [27] |
| Enterovirus C poliovirus and Coxsackievirus | Musca domestica | Human | [28] |
| Hepatitis A virus | Blattella germanica | Human | [28] |
| Turkey coronavirus | Musca domestica | Turkey | [29] |
| SARS-CoV-2 | Musca domestica | Human | [30] |
| Companion animals |
Animal species susceptible to SARS-CoV-2 with a role as coronavirus hosts and in transmission of the coronavirus [5, 6, 7, 33] are also infested or in contact with arthropod vectors (Table 1). Coronaviruses have been previously identified in ticks Ixodes uriae [34] and in unfed cat flea Ctenocephalides felis [35]. The application of experimentally validated models for SARS-CoV-2 S-ACE host receptor interactions support that not only vertebrate hosts, but also arthropod ACE may have the capacity to interact with SARS-CoV-2 S-protein [8, 9, 36]. Additionally, SARS-CoV-2 co-receptor integrins [37, 38] have been described in tick salivary glands and cement [39, 40] and in cat flea exoproteome [35, 41]. Cement is secreted by ticks as a complex protein polymerization substance with antimicrobial properties and a role in host attachment and ectoparasite feeding [40]. Recent experiments showed that mosquitoes are not competent vectors for SARS-CoV-2 [42, 43], results that agree with a predicted lower SARS-CoV-2 S-protein: ACE complex stability in insects [9]. Based on these results, it is possible that SARS-CoV-2 may be acquired by arthropod ectoparasites through feeding on infected hosts and may persist not only in contaminated mouthparts or exoskeleton [30], but also inside the vector through interactions with ACE and integrins for transmission to susceptible hosts by blood-meal regurgitation during secondary feeding or after transstadial transmission or inherited virus RNA [9, 35, 44, 45].
Another proposed connection between arthropods and COVID-19 is the immune
response to the glycan Gal
Altogether, these findings support a possible role for arthropods at the host-coronavirus interface with potential implications as low-susceptible non-mammalian hosts and biological and/or passive vectors for SARS-CoV-2, possibilities that need to be evaluated in future research [9, 30, 35, 65, 66, 67].
Some virus variants arising in low-susceptible hosts with modifications in S and other proteins could increase fitness and evade or reduce the efficacy of human immune response to vaccination and infection with currently circulating virus variants. Additionally, the role of integrins as co-receptors for SARS-CoV-2 cell attachment [38] and a putative implication of arthropod vectors [35] may also affect animal host susceptibility to infection with the selection of new virus variants [37]. Virus protein modifications could be related to genetic or post-translational modifications (e.g., glycosylation or ADP-ribosylation) [46, 68, 69]. Currently, these SARS-CoV-2 variants potentially arising in low-susceptible hosts do not constitute a major risk for humans. Nevertheless, in a growing immunized population after infection or vaccination, variants produced in low-susceptible hosts may be more effective in evading immunity, thus increasing the risk for zoonotic virus transmission. For example, a virus variant with high fitness in a low-susceptible host may not be efficiently transmitted to humans as compared with currently circulating variants. However, a genetic and/or post-translational modification in a virus variant arising from a low-susceptible host could acquire the capacity to evade human immune response and thus establish infection in individuals protected against currently circulating virus variants (Fig. 1). The evolutionary history of coronaviruses suggests that SARS-CoV-2 infectivity derived from adaptation in bats and not humans [69]. Furthermore, these new virus variants may be also susceptible to reverse zoonotic transmission to animal hosts for maintenance and appearance of new variants [4]. Despite the controversy regarding the origin of SARS-CoV-2 [70], understanding the role of low-susceptible animal hosts such as ferrets in coronavirus infection, adaptation and arising of new virus variants is important to control COVID-19 and other emerging infectious diseases [71, 72, 73, 74, 75].
 Fig. 1.
Fig. 1.Increased risk for zoonotic virus transmission. SARS-CoV-2 variants produced in low-susceptible and chronically infected hosts may be more effective in evading immunity and with higher human-to-human transmission. SARS-CoV-2 high and low susceptible hosts are only shown as representative species.
Based on the questions related to the appearance of new virus VOC such as Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1) and Delta (B.1.617.2) (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/) [76], the ECDC [77] recommends avoiding prolonged situations with high proportion of the population partially susceptible to virus infection, surveillance of zoonotic SARS-CoV-2 transmission and monitorization and isolation of individuals with long-lasting infection.
A global One Health approach is required for the prevention and control of
infectious diseases with a transdisciplinary research between ecology,
biotechnology and human/animal health. This approach requires the international
collaboration between scientific, industrial and health partners. As an example,
our group SaBio (from “Sanidad & Biotecnología” which translates into
“Health & Biotechnology”) applies this approach for the study of infectious
diseases such as those caused by ticks and tick-borne pathogens or mycobacteria
e.g., [78, 79]. With the emergence of COVID-19, the challenge posed by this
pandemic should be approached with a global One Health perspective by combining
research on environmental and epidemiological factors, virus animal hosts and
transmission, and human immune response [6, 9, 35, 36, 46, 48, 75, 80, 81, 82, 83, 84] (Fig. 2). These results advance research for the development of interventions for the
control of current COVID-19 and future pandemics including SARS-CoV-2 diagnostic
and environmental monitoring, epidemiological studies with case mapping, contact
tracing, potential role of animal hosts and arthropod vectors, palliative
treatments and prognostic biomarkers, and personalized medicine including the use
of
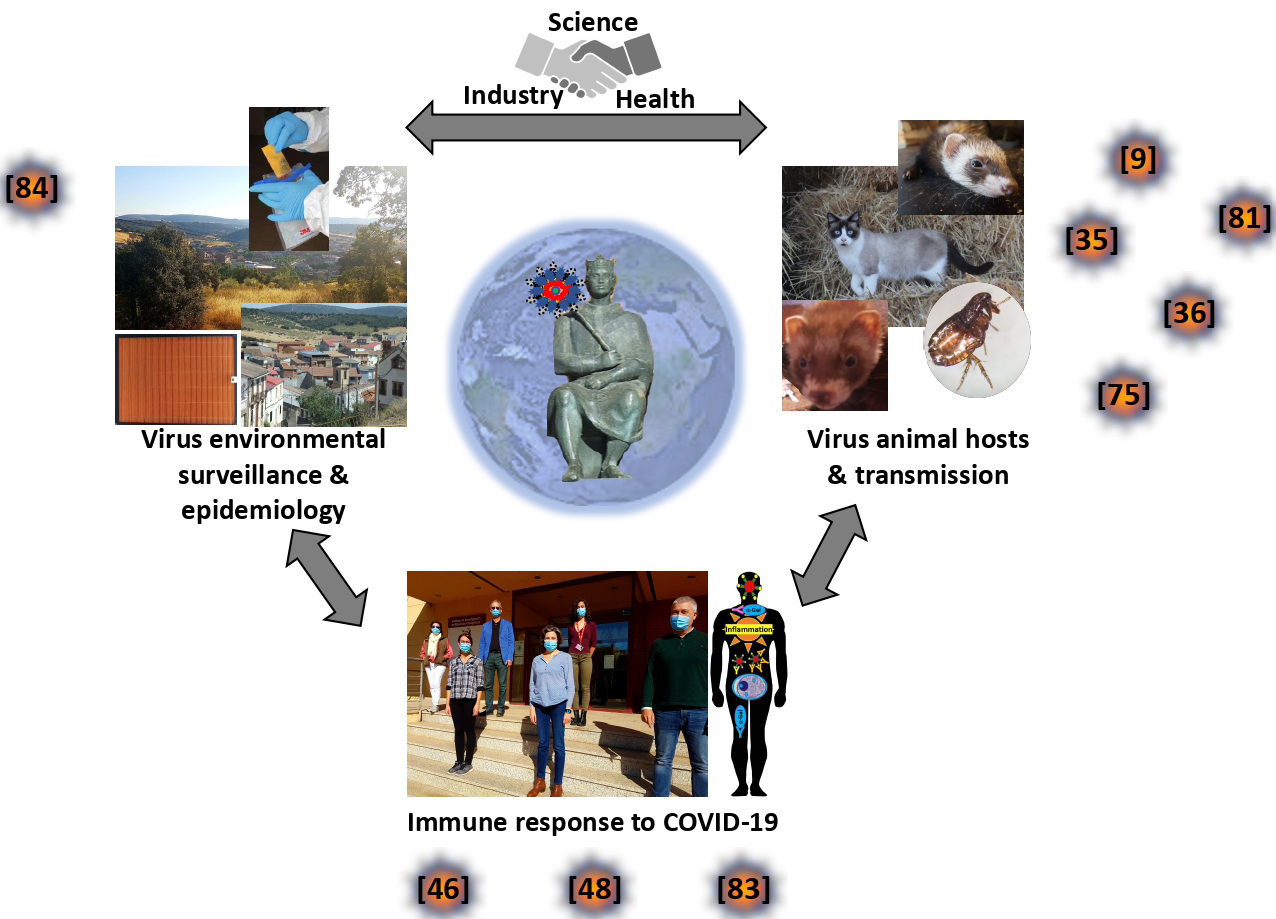 Fig. 2.
Fig. 2.SaBio on COVID-19: A global One Health approach. Our group, SaBio, applies a global One Health approach in research for the control of infectious diseases. Regarding COVID-19, we have applied this approach by developing research in three areas: (a) SARS-CoV-2 environmental surveillance and epidemiology, (b) virus animal hosts and transmission, and (c) immune response to COVID-19. The results (illustrated in publications [9, 35, 36, 46, 48, 75, 81, 83, 84]) have contributed to the characterization of disease epidemiology and immune response mechanisms with a possible impact on the development of prevention, diagnostic, prognostic and therapeutic interventions. Images courtesy of the authors.
As highlighted in this paper, host and virus derived factors are the key drivers of the COVID-19 pandemic [8, 35, 85, 86], and non-human hosts may gain protagonism in the near future. The information about viral host ranges and associations between known viruses and susceptible hosts is limited but key to prevent future pandemics [87]. In addition to their role in the generation of new virus variants, relaxing measures in response to pandemic control may also increase human-to-animal contact in both urban and rural settings. These risks are underestimated and should be considered when evaluating vaccine efficacy in relation to the potential role of animal reservoirs and zoonotic virus transmission. SARS-CoV-2 surveillance at the human-animal interface, sequencing and evaluation of susceptibility to antibodies from vaccinated individuals should be improved [67, 71, 72, 73, 74, 75, 76, 77, 85, 86, 87, 88, 89]. The direct and indirect impact of COVID-19 on animal health should also be considered [6, 90, 91]. A One Health approach searching a balanced interaction of humanity with nature and a more holistic approach to disease control is necessary for a more effective prevention of future pandemics [92].
Conceptualization, JF and CG; investigation, JF, IGFM and CG; writing—original draft preparation, JF; writing—review and editing, JF, IGFM and CG; visualization, JF; All authors have read and agreed to the published version of the manuscript.
Not applicable.
We thank members of SaBio and collaborators worldwide for their contributions to COVID-19 research and acknowledge the University of Castilla La Mancha (UCLM, Spain) for support to Group SaBio.
This research received no external funding.
The authors declare no conflict of interest.
VOC, variants of concern; S; Spike; ACE, angiotensin I converting enzyme 2;
