Epigenetic regulation in animals induces rapid and long-lasting effects on gene expression in response to environmental changes that frequently affect animal behavior. In the last decade, accumulating studies have revealed how epigenetic regulation affects the behavior of animals, such as learning and memory, mating and courtship, the circadian sleep-wake cycle, and foraging/starvation-induced hyperactivity. In each section of this review, we discuss what we have learned from studies with mammals, mostly mouse models. We then highlight studies with Drosophila models to compare data with mouse models. Finally, we discuss several unanswered questions and future developments in this field.
Gene expression may be regulated without any changes in actual DNA sequences. Epigenetic regulation is achieved by several processes involved in DNA methylation, histone modifications, and nucleosome structure modifications (1). DNA methylation is generally added onto CpG dinucleotides, and the methylation of CpG-rich regions is responsible for gene silencing in most cases (2). Histone modifications and their function in gene regulation were initially reported in 1964 (3), and more than 100 residues on various histone tails have so far been shown to be post-translationally modified by processes including acetylation, phosphorylation, methylation, ubiquitylation, SUMOylation, and ADP-ribosylation, which affect gene expression through alterations in the chromatin structure (4, 5). Another main epigenetic process is chromatin structural modifications through the regulation of nucleosome positioning. Nucleosomes consist of histone octamers wrapped up with 146 base-pair DNA and function as a barrier to transcription. Therefore, the precise positioning of nucleosomes around genes is critical for their expression (6). Nucleosome positioning may be influenced by DNA methylation, histone modifications, and untranslated RNAs, such as microRNAs (7–9).
Previous studies revealed that epigenetic regulation is critical for a wide variety of biological processes, such as imprinting, inactivation of the X chromosome, reprogramming, and aging (10–13). In the last decade, the functions of epigenetic regulation in animal behavior have attracted increasing attention because epigenetic regulation induces rapid and long-lasting effects on gene expression in response to environmental changes. Accumulating evidence has clarified how epigenetic regulation affects the behavior of animals, such as learning, mating, the sleep-wake cycle, and foraging, particularly in Drosophila melanogaster (14) and mammals. In this article, we review advances in our understanding of the relationship between epigenetic regulation and behavior in mammals and D. melanogaster.
The ability to memorize and learn is essential for behavioral adaptation to environmental changes. Previous studies revealed that epigenetic markers are temporarily regulated in the post-mitotic neurons of adult rodents, Drosophila, honeybees, and Aplysia during the process of learning and memory (15). In mammals, memory formation requires the coordinated expression of neuronal genes for synaptic restructuring through epigenetic regulation (15). Temporal epigenetic modifications may mediate memory consolidation through the regulation of gene expression within a few hours of learning. In contrast, sustained changes in epigenetic modifications in cortical brain regions underlie the maintenance of memory over prolonged periods of time (15).
Hippocampal memory formation and consolidation are regulated by specific histone modifiers, such as the histone acetyltransferase (HAT) CREB-binding protein (CBP) (16, 17), histone deacetylase HDAC2 (18), HDAC3 (19), HDAC4 (20, 21), histone demethylase JMJD2B (22), histone methylase G9a/EHMT2 and GLP (G9a related protein)/EHMT1 (23, 24). CBP, a coactivator of transcription, functions as a platform for the recruitment of other required components of the transcriptional machinery and also as a HAT that alters the chromatin structure. Transgenic mice expressing an inhibitory truncated CBP protein in forebrain neurons (CBP transgenic mice) have been established (16). Studies with CBP transgenic mice revealed that CBP plays a role in specific forms of hippocampal synaptic plasticity and long-term memory formation that is dependent on the hippocampus (16). Other studies with transgenic mice expressing CBP in which HAT activity is eliminated also revealed that the HAT activity of CBP is a critical component of memory consolidation (17).
The increased acetylation of the histone tail induced by inhibitors of HDACs is known to facilitate learning and memory in mice. Transgenic mice overexpressing HDAC2, but not HDAC1 exhibit decreased dendric spine density, synapse numbers, synaptic plasticity, and memory formation (18). In contrast, HDAC2 knockout mice show increased synapse numbers and memory facilitation (18). Furthermore, a chromatin immunoprecipitation assay (ChIP assay) revealed a relationship between HDAC2 and the promoters of genes involved in synaptic plasticity and memory formation (18). Therefore, HDAC2 appears to play a role in regulating synaptic plasticity and long-lasting changes in neural circuits that result in the negative regulation of learning and memory. Other studies revealed that the deletion of HDAC3 in mice improved age-related impairments in long-term memory and synaptic plasticity (19). Furthermore, the lack of HDAC3 restored the experience-induced expression of the circadian gene Per1 in the dorsal hippocampus (19). The hippocampal expression of PER1 is important for the formation of long-term memory. Thus, the hippocampal overexpression of Per1 improves age-related memory impairments, and the regulation of PER1 may be a mechanism by which the lack of HDAC3 improves memory and synaptic plasticity in aged mice (19). These findings demonstrated that HDAC3 negatively regulates long-term memory, synaptic plasticity, and the experience-induced expression of Per1 in the aging hippocampus. The expression of a truncated form of HDAC4 impaired spatial memory in mice and repressed a set of genes required for synaptic plasticity (20). A brain-specific knockout of HDAC4 also impaired long-term memory in mice (21). HDAC4 binds the transcription factor MEF2 to repress its transcriptional activity. Other studies revealed a role for MEF2 family members in learning and memory (25).
JMJD2B is a histone demethylase that regulates gene expression through the demethylation of tri-methylated histone H3 lysine 9 (H3K9me3). JMJD2B is expressed in the central nervous system (CNS) from embryos through to adults with markedly high expression levels in the hippocampus (22). Neuron-specific JMJD2B-deficient mice showed an increase in total spine numbers, but decreases in mature spine numbers in the hippocampus (22). JMJD2B-deficient mice also showed hyperactive behavior, sustained hyperactivity in a new environment, defects in working memory, and spontaneous epileptic-like seizures (22). Therefore, JMJD2B positively regulates spine maturation and long-term memory.
G9a and GLP forms complex which dominantly regulates H3K9 mono- and demethylation (H3K9me2) at euchromatic region. It has been reported that G9a/GLP mediated H3K9me2 in the hippocampus and entorhinal cortex (EC) regulates the long-term memory (LTM) consolidation of contextual conditioned fear (23). Interestingly, pharmacological inhibition of G9a/GLP activity in the EC altered H3K9me2 level in hippocampal area CA1, which suggests that G9a/GLP functions in mediating cellular and molecular connectivity between these two brain regions during memory consolidation (23). Furthermore, a recent study has reported that pharmacological inhibition of G9a/GLP activity reinforces early Long-term potentiation (LTP) and promotes synaptic tagging and capture (STC) in the EC (24). LTP and STC are considered as the critical models for the cellular basis of LTM (26, 27).
A previous study reported that DNA methylation in the ventral tegmental area (VTA) regulated reward-related memories (28). Reward-related memories are critical for adaptive behavior and evolutionary fitness. Reward learning requires dopaminergic neurons located in the VTA, encoding relationships between predictive cues and future rewards. The formation of reward-related associative memories in rats up-regulates key plasticity genes in the VTA, which correlate with memory strength associated with gene-specific DNA methylation changes (28).
Three H3K9-specific histone methyltransferases have been identified in Drosophila: Drosophila G9a (dG9a), Su(var)3-9, and SetDB1 (DmSETDB1). dG9a shares approximately equal homology to human G9a/EHMT2 and GLP/EHMT1. dG9a is ubiquitously expressed in various tissues, including the testis (29) and CNS, and dG9a null mutants are viable under laboratory conditions. As well as the mouse model, the significance of the G9a/GLP-mediated epigenetic regulation of learning and memory has been reported in Drosophila model. Neurodevelopmental and behavioral analyses on dG9a null mutants revealed that dG9a positively regulated peripheral dendrite development, larval locomotor behavior, non-associative learning, and courtship memory. The requirement for dG9a in memory was predominantly mapped in the mushroom body neurons of adult brains. The re-expression of dG9a in adults restored memory, suggesting that cognitive defects are reversible in dG9a mutants. The genome-wide profiling of H3K9 dimethylation by ChIP-sequencing (ChIP-seq) analyses revealed that the loss of H3K9 dimethylation in dG9a mutants occurred at 5% of the euchromatic genome and was enriched at distinct classes of genes. These genes are involved in neuronal and behavioral processes. Thus, dG9a regulates dendrite branching as well as non-associative learning and courtship memory (30).
Drosophila carries the following five HDACs: Rpd3, HDAC3, HDAC4, HDAC6, and HDAC11. Rpd3 is a homologue of yeast Rpd3 and shares approximately equal homology to human HDAC1 and HDAC2. Rpd3 is abundantly expressed throughout the adult brain and localizes in neuronal nuclei. Although the modulation of Rpd3 levels predominantly in the adult mushroom body, an important structure for memory formation, exerts no apparent effect on immediate recall or one-hour memory, 24-hour long-term memory was severely impaired (31). Not only the RNAi-mediated knockdown of Rpd3, but also its overexpression induced defects in long-term courtship memory, indicating that the proper level of Rpd3 is important for normal long-term memory (31). The proper level of Rpd3 may be required to maintain the proper transcriptional levels of the plasticity-related genes needed for long-term memory.
The role of HDAC4 in long-term memory was also examined in Drosophila. The overexpression of HDAC4 in the adult mushroom body resulted in defects in long-term courtship memory, but exerted no effect on short-term memory. The overexpression of an HDAC4-H968A mutant without catalytic activity also abolished long-term memory, suggesting that deacetylase activity is not required for this mode of action (32). The overexpression of HDAC4 induced the redistribution of MEF2 from a relatively uniform distribution in the nucleus into punctate nuclear structures colocalizing with HDAC4, suggesting that the repressive effects of HDAC4 on long-term memory are mediated by an interaction with MEF2 (32). Similar to the case of Rpd3, not only the overexpression of HDAC4, but also its RNAi-mediated knockdown impaired long-term memory (32), suggesting the proper level of HDAC4 critical for normal long-term memory.
A recent study using the PILGRAM (the Platform for Interactive Learning by Genomics Results Mining) machine learning analysis revealed that Grunge/Atrophin, a transcriptional repressor, histone deacetylase, is closely related to learning and memory genes (33). Grunge plays a positive role in the modulation of memory retention in the adult mushroom body (33). It is also involved in memory retention through the regulation of neuronal activity and not by altering neuronal development (33).
Several HATs have been identified in Drosophila. Hat1 (KAT1, lysine acetyltransferase 1) is a B-type HAT involved in generating acetylated H4K5 (H4K5ac) and H4K12ac marks on newly synthesized histones. These marks play roles in transporting histones to the nucleus. These marks are then rapidly removed after the integration of histones into nucleosomes. MOF (KAT8), chameau (KAT7), and Tip60 (KAT5) belong to the MYST HAT family. dGCN5 (KAT2) and Elongator Protein 3 (Elp3) (HAT9) are members of the GNAT HAT family, while nejire (KAT3) is a single Drosophila CBP/p300 homologue (34). Tip60 is expressed throughout the adult brain, including the mushroom body. Tip60 functions in the mushroom body to promote short-term memory (35). The targeted loss of the HAT activity of Tip60 through the expression of a dominant negative HAT defective form of Tip60 in the mushroom body induced an abnormal morphology in axonal lobes, while the overexpression of Tip60 exerted no morphological defect. However, the loss and gain of Tip60 levels in the mushroom body both induced defects in short-term memory (35). ChIP-seq analyses revealed that genes involved in cognitive processes were misregulated in the Tip60 mutant fly brain (35). Increases in Tip60 levels in the mushroom body were shown to rescue both learning and memory defects under amyloid precursor protein (APP)-induced neurodegenerative conditions (35), suggesting the potential of HAT activators as a therapeutic target for neurodegenerative disorders. Collectively, HATs may activate early memory genes to establish memory formation, while HDACs appear to inactivate memory extinction genes in order to promote memory retention.
Mating behavior requires rapid decision-making, including sexual and species discrimination, based on external information and internal states, such as epigenetic states (36). Limited information is currently available on the epigenetic regulation of mating and courtship behavior. Sexually naïve female mice acquire sexual receptivity experientially. Repeated experience with sexually active males and simultaneous treatments with estradiol and progesterone were found to gradually increase the sexual receptivity of female mice. Ovarian hormones activate their target nuclear steroid receptors, estrogen receptor-α and the progesterone receptor, to induce female sexual receptivity. Nuclear receptors also recruit the coactivators of transcription, including HAT, thereby facilitating histone acetylation to ovarian hormone-responsive genes. The treatment of female mice with the HDAC inhibitor, sodium butyrate enhanced the experiential acquisition of receptivity (37). Moreover, the actions of sodium butyrate on receptivity acquisition depended on the activated estrogen receptor-α and progesterone (37). Since sodium butyrate is a general class I and class II HDAC inhibitor, it is not yet known which HDAC is involved in this process.
The socially monogamous prairie vole (Microtus ochrogaster) has emerged as a unique model for investigating the neuronal bases of social attachment (pair bonds) that is initiated by the formation of selective affiliation behaviors toward the partner, the so-called partner preference. The formation of partner preference is regulated by a number of neurotransmitters, including oxytocin, vasopressin, and dopamine. The HDAC inhibitors, sodium butyrate and trichostatin A were previously shown to facilitate partner preference formation in female prairie voles (38). This was associated with the up-regulation of oxytocin receptors and vasopressin V1a receptors with an increase in histone acetylation (H3K14ac) at their respective promoter regions (38). Mating-induced partner preference similarly enhanced the up-regulation of oxytocin receptor and vasopressin V1a receptor genes with an increase in H3K14ac at their promoter regions, indicating that trichostatin A and mating both facilitate partner preference via epigenetic regulation (38).
The male courtship behavior of D. melanogaster is an innate program without the involvement of learning. After finding a female, a male fly orients his body axis toward the female (orientation) and then starts to chase her (following) with or without prior touching of the female abdomen with the forelegs (tapping). During the chasing process, the male fly extends and vibrates his wings to generate specific sounds, called courtship songs (singing). The male fly then accesses the female from behind, licks the genitalia (licking), and tries to mount her back (attempted copulation) and connect his genitalia with hers (copulation). Copulation typically continues for 15–25 min. Wild-type females never display courtship behavior and instead show either acceptance or rejection to a courting male.
The functional roles of epigenetic factors on sex-specific behaviors have been elucidated in studies on the fruitless (fru) gene (39). Male courtship requires the products of the fru gene, which is differentially spliced between males and females (40). The bonus (bon) gene genetically interacts with fru (41). The bon gene encodes a Drosophila homologue of mammalian transcriptional intermediary factor 1 (TIF1). TIF1 has been shown to participate in chromatin regulation by forming protein complexes with HDAC1 and heterochromatin protein 1 (HP1) in mammals. Co-immunoprecipitation assays revealed that Rpd3 (a Drosophila homologue of HDAC1 and HDAC2), Su(var)205 (a Drosophila homologue of HP1a), and Bon formed a complex with Fru in vivo (41). Fru functions appear to be facilitated by Rpd3 and impeded by Su(var)205. Biochemical analyses revealed that Rpd3 competed with Su(var)205 in forming a complex with Fru (41). In addition, an immunostaining assay of polytene chromosomes showed that Fru-containing complexes bound to multiple sites on the genome, suggesting that Fru proteins orchestrate the transcription of a large number of downstream genes in controlling male-specific behaviors (41).
The Drosophila brain contains approximately 150 clock neurons that may be divided into seven clusters depending on their anatomical locations and functional properties. They are the small and large ventral lateral neurons (sLNvs and lLNvs), dorsal lateral neurons (LNds), lateral posterior neurons, and three dorsal neuron clusters (DN1–3). Four sLNvs are referred to as morning (M) cells, while the fifth sLNv and LNds are called evening (E) cells. These two clusters of clock neurons—M cell and E cell oscillators—are mainly responsible for activity bursts at dawn and dusk. In contrast, male–female pairs of flies follow a distinct pattern: low activity at dusk, followed by male courtship activity during the night, named the “male sex drive rhythm” (MSDR). Male flies lacking the expression of salt-inducible kinase 3 (SIK3) in M cells show a short period of MSDR, but a long period of single-fly locomotor rhythm (42). Immunocytochemical analyses revealed that the circadian nucleocytoplasmic shuttling of HDAC4 was disrupted in these flies (42). Therefore, SIK3–HDAC4 signaling in M cells involves MSDR by regulating molecular oscillations in DN1 neurons.
A previous study reported that the D. melanogaster methyl-CpG-binding domain (MBD)-containing protein, dMBD-R2 contributed to reproductive isolation (43). Male flies with a reduction in dMBD-R2, specifically in octopamine neurons, show courtship toward diverged interspecies D. virilis and D. yakuba females and a decrease in conspecific mating success (43). Conspecific male-male courtship was found to be increased between dMBD-R2-deficient males, while aggression was reduced (43). Therefore, Drosophila MBD-containing proteins are required within the octopamine neural circuitry to inhibit interspecies and conspecific male-male courtship.
Circadian clocks are intrinsic, time-tracking systems that enable organisms to adopt their behaviors, such as feeding and the sleep-wake cycle, and physiology, including hormonal levels and body temperature, to the environmental changes associated with the 24-hour day-night cycle (44). The conserved molecular mechanisms of circadian clocks are featured by regulation with the transcriptional negative feedback loop. CLOCK and BMAL1 are positive regulators of the mammalian clock that form a complex to promote the expression of target genes, such as cryptochromes (CRY1 and CRY2) and periods (PER1, PER3, and PER3), which repress the transcriptional activity of the CLOCK/BMAL1 complex (44). Several epigenetic factors, such as the histone deacetylase SIRT1, HDAC3, histone methyltransferase MLL1, and demethylase JumonjiC (JmjC), and ARID domain-containing histone lysine demethylase 1a (JARID1a) have been reported to regulate the expression of these clock genes (45). The HDAC activity of SIRT1 is regulated in a circadian manner in the mouse liver and controls circadian histone H3 acetylation as well as the acetylation of BMAL1 (46). SIRT proteins catalyze the protein deacetylation reaction consuming NAD+ as a substrate. Since the NAD+/NADH ratio is a direct indicator of the cellular energy status (47), SIRT1 transduces signals originating from cellular metabolic conditions to the circadian clock through the acetylation of histone and clock proteins (46, 48). A previous study reported that the interaction of HDAC3 and nuclear receptor corepressor 1 is required for the regulation of the circadian clock and physiology in the mouse liver (49). MLL1, one of the H3K4-specific methyltransferases, functions as a key regulator for circadian transcription and cyclic H3K4 trimethylation by forming a complex with CLOCK/BMAL1 (50). JARID1a also forms a complex with CLOCK/BMAL1, and the complex is recruited to the Per2 promoter to activate Per2 expression (51).
In addition to histone modifications, recent studies revealed that the profiles of cytosine modifications, mainly comprising 5-methyl- and 5-hydroxymethylcytosines, are changing in a circadian manner in the liver and lung (52). These oscillating modified cytosines are associated with the aging effect on the amplitude of circadian gene oscillations (52). Furthermore, another study demonstrated that DNA methylation profiles at circadian genes are associated with obesity, metabolic disturbances, and carbohydrate intake (53).
In D. melanogaster, the roles of JmjC domain-containing proteins on the circadian rhythm were comprehensively examined utilizing 11 viable mutants of the following JmjC domain-containing genes: lid, KDM2, KDM3, KDM4A, KDM4B, JMJD4, JMJD5, JMJD7, PSR, NO66, and HSPBAP1 (54). Two out of the 11 viable mutants (lid and KDM3 mutants) exhibited high levels of arrhythmicity, while two others (KDM2 and JMJD5 mutants) showed the subtle shortening of the circadian period length (54). Furthermore, many of these 11 mutants significantly affected sleep and locomotion activity (54). The JMJD5 mutant exhibited a reduction in daytime sleep as well as the promotion of daytime activity (54). The JMJD7 mutant also showed a reduction in daytime sleep (54). The NO66 mutant showed a reduction in sleep and increase in locomotion activity both in daytime and nighttime (54). The KDM4B mutant showed a marked increase in daytime and nighttime sleep (54). These findings indicate that most genes encoding JmjC domain-containing proteins play a role in tuning circadian behaviors in a non-overlapping manner.
A recent study reported that the loss of Elp3, one of the histone acetyltransferases, during neurodevelopment induced the hyperactive phenotype and sleep loss in the adult stage (55). The knockdown of Elp3 resulted in the misregulation of genes involved in sleep and hyperactivity (55). Furthermore, the Elp3 mutant showed significant increases in synaptic bouton numbers and axonal lengths as well as branching in the larval neuromuscular junction (55). These findings suggested that gene regulation by Elp3 controls the hyperactive and sleep phenotypes in the adult stage of Drosophila by modulating the development of synaptic boutons and axonal lengths of the larval neuromuscular junction (55). It is important to note in Drosophila that the majority of the peripheral neurons of the adult fly originate from larval peripheral neurons. Moreover, the peripheral neurons of Drosophila are glutamatergic neurons that are similar to most neurons in the CNS.
Foraging behavior has been extensively examined, particularly in D. melanogaster (56). There are two foraging strategies: “rovers” and “sitters”. “Rovers” traverse a large area while feeding, whereas “sitters” stay in a small area (57). This rover-sitter behavioral difference was found to be induced by allelic variations in a single gene called foraging (for), which encodes a cyclic guanosine monophosphate-dependent protein kinase (58, 59). Ina A et al recently reported that one of the histone methyltransferases, dG9a, interacted with for to induce rover-sitter behavioral differences (60). The for gene is transcribed to at least 21 different transcripts, which may be clustered into four classes depending on four promoters, pr1-4 (61). Ina A et al showed that the rover-sitter behavioral difference was induced by an expression difference in the transcripts from pr4, and that dG9a was required for the rover-sitter expression difference of the transcripts from pr4 (60). Ina A et al also demonstrated that the rover-sitter expression difference of the transcripts from pr4 was highly dependent on dG9a, particularly in the brain and ovaries (60).
Epigenetic regulation plays an important role in the acquisition of starvation resistance. For example, Rpd3 accumulates in the nucleolus in the early phase of starvation to up-regulate rRNA synthesis, and increases stress tolerance proteins to acquire starvation stress resistance (62). Regarding starvation-induced behavioral changes, starved flies enhance their locomotion activity to increase the probability of finding desirable foods (63, 64). This starvation-induced hyperactivity requires octopamine, the insect counterpart of vertebrate norepinephrine, and neurons expressing octopamine (65). We recently reported that dG9a functions as a key regulator in the decision of behavioral strategies under starvation in D. melanogaster (66). dG9a null mutant flies were found to be viable, but sensitive to starvation and dG9a saved energy through the recycling of cellular components by regulating the expression of genes required for autophagy (67) (Figure 1). Based on these studies, we performed RNA-sequence (RNA-seq) analyses using adult flies of the dG9a null mutant and wild type after starvation to identify which genes are regulated by dG9a under starvation stress.
 Figure 1.
Figure 1.
Schematic model of the role of dG9a in acquisition of starvation stress tolerance. The Figure shows that dG9a increase the stress tolerance in the cellular level (right) and in the behavioral level (left). In the cellular level, dG9a promotes the autophagy-induced energy recycling through regulating the gene expression of Atg8a, one of the essential genes to proceed the autophagy. In behavioral level, dG9a is responsible for starvation-induced increase in the sensitivity to sucrose to get desirable food by controlling the expression of Gustatory receptors (Grs) genes. Furthermore, dG9a is responsible for suppressing the locomotion activity under starvation stress by regulating the Drosophila insulin-like receptors (dilps).
RNA-seq with gene ontology analyses demonstrated that the expression of genes encoding gustatory receptors (Grs) and odorant binding proteins were altered in dG9a mutants in starvation (66). We found that the depletion of dG9a extensively up-regulated the expression of Gr64a, Gr64b/Gr64c, and Gr64d/Gr64e, which are required to sense sweetness (66, 68). Consistent with these findings, proboscis extension reflex tests, one of the well-established behavioral assays for evaluating gustatory sensitivity in Drosophila (69, 70), revealed that dG9a depletion significantly increased sensitivity to sucrose under starved conditions (66). Since dG9a expression decreases under starvation conditions, these findings suggest that dG9a is responsible for an increase in sensitivity to sucrose in order to survive starvation conditions (66).
As well as the promotion of taste sensitivity, starved flies also enhance their locomotion activity to increase the possibility of finding desirable foods (63, 64). The locomotion activity test utilizing the Drosophila Activity Monitoring system (71) showed that dG9a depletion increased starvation-induced hyperactivity, which suggested that dG9a is responsible for suppressing locomotion activity under starvation (66). Furthermore, we revealed that dG9a regulated the expression of Drosophila insulin-like peptides (dilps). A previous study reported that insulin and glucagon signaling exerted opposing effects on the regulation of starvation-induced hyperactivity (72). These findings indicate that dG9a is responsible for suppressing locomotion activity under starvation stress by regulating the insulin signaling pathway (66).
In the natural world, animals are frequently exposed to starvation stresses, while foraging requires the cost of food-seeking energy and associated threat of predation as well as environmental changes with migration (73–75). An alternative strategy to survive under starvation is saving energy without moving, such as the hibernation associated with seasonal fluctuations in food availability, which is adopted by a wide variety of organisms, including Drosophila (76, 77). Wild-type flies may save energy without moving at the early stage of starvation and then become active to seek food with increasing risks along with a reduction in dG9a expression at the late stage of starvation. Therefore, dG9a functions as a key regulator for flies to make decisions on these strategies along the time course of starvation and has an adaptive advantage for surviving starvation (Figure 1). The refeeding of wild-type flies after starvation may restore hyperactivity and increased sensitivity to sucrose as well as the expression level of dG9a (66), which indicates that regulation by dG9a is reversible and highly dependent on food intake.
In addition to dG9a, we investigated the relationship between starvation-induced hyperactivity and other epigenetic factors. We found that the fat body-specific knockdown of Tip60 promoted starvation-induced hyperactivity (Figure 2). These findings indicate that Tip60 functions in the fat body as a critical regulator of starvation-induced hyperactivity. We utilized three different Tip60 RNAi strains and significant increases in locomotion activity were observed in all strains, particularly during the light phase (Figure 2). These findings indicate that the promotion of starvation-induced hyperactivity is not induced by background mutations or the off-target effects of RNAi strains. In Drosophila, the fat body expresses DILP6, one of the eight insulin-like peptides (DILP1-8), extensively during the larval and adult stages (78–81). DILP6 expression in the adult fat body reduces dilp2 and dilp5 mRNA expression levels in the brain, and the secretion of DILP2 into the hemolymph (82). As described above, insulin signaling regulates starvation-induced hyperactivity (72). Although the DILPs that play a role in the regulation of starvation-induced hyperactivity have not yet been identified, Tip60 may control starvation-induced hyperactivity by regulating the expression of DILPs in the fat body.
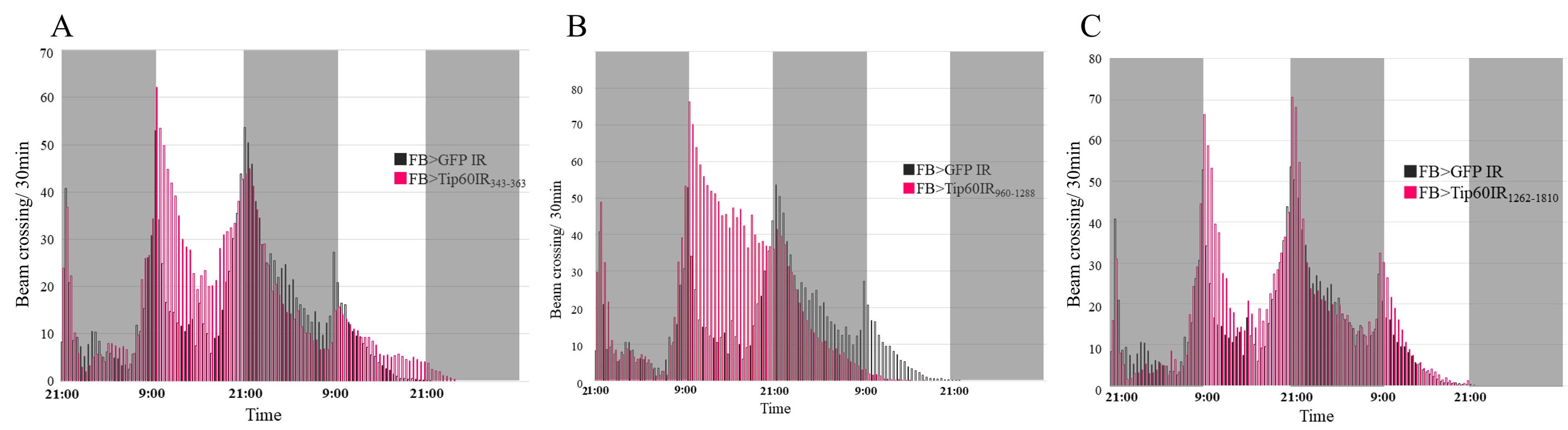 Figure 2.
Figure 2.
Tip60 functions in the fat body as a critical regulator of the starvation-induced hyperactivity. The beam crossing activity of the control fly (w/Y; FB-GAL4/+; UAS-GFP IR/+) and the Tip60 knockdown fly; (A) w/Y; FB-GAL4/+; UAS-TIP60 IR343-363/+ (B) UAS-TIP60 IR960-1288/Y; FB-GAL4/+; + (C) w/Y; FB-GAL4/UAS-TIP60 IR1262-1810; +. Gray and white background in the graph represents 12 h dark periods and 12 h light periods, respectively. n=30. Fly strains were kept on 12 h light/dark cycles for at least 5 generations before performing locomotion activity tests. Individual 3-5 day-old adult male fly was placed in glass tube and its locomotion activity was continuously recorded by DAM2 Drosophila Activity Monitor (TriKinetics). One end of glass tubes was filled with cotton wool soaked with water and the other was sealed with PARAFILM® (Bemis). These instruments were put in the incubator at 25 ºC and kept on 12 h light/dark cycles.
The importance of epigenetic regulation in animal behavior is becoming more apparent. In addition to the behaviors described above, epigenetic regulation has been identified as critical for extensive behaviors, such as drug addiction, fear, and mental disorders, including autism spectrum disorder (ASD) (83–87). Regarding the relationship between epigenetic regulation and mental disorders, recent studies showed that missense variants for genes encoding H3K9-specific methyltransferases GLP/EHMT1 and G9a/EHMT2 were ASD-associated genes (88, 89). It is well-known that ASD is affected by environmental stresses, including starvation stress. Therefore, Drosophila models for ASD (90) and dG9a mutants may contribute to studies on the potential roles of dG9a in the pathogenesis of ASD. Furthermore, recent studies have shown that epigenetic regulators are associated with intellectual disorders, such as Kleefstra syndrome (KS), as well as hereditary motor and sensory neuropathies, including Charcot-Marie-Tooth disease (CMT) (91, 92). KS is a rare genetic disorder that is characterized by intellectual disability, childhood hypotonia, and distinctive facial features (91). The haploinefficiency of GLP/EHMT1 was shown to be associated with KS (93), and a lysine-specific methyltransferase 2B recessive variant induced a KS-like intellectual disability phenotype (94). In a mouse model of human CMT, HDAC6 inhibitors were shown to reverse the loss of axonal transport, which is induced by mutations in the 27-kDa small heat-shock protein gene (HSPB1) (95). These findings strongly suggest that epigenetic regulators have potential as therapeutic targets for these mental/intellectual/neurodegenerative disorders.
One of the features of epigenetic regulation is that it may be inherited by the next generations. Previous studies demonstrated that epigenetic regulation mediates the trans- and inter- generational transmission of behaviors, such as drug addiction, sexual behaviors, and hedonic behaviors (96–99). Time is needed to examine trans- and inter- generational inheritance, and, thus, model organisms that have a short life span, such as D. melanogaster, are useful. To effectively examine the extensive relationship between epigenetic regulation and animal behaviors, it will become important to utilize the advantages of several model organisms.
We thank the Bloomington Drosophila Stock Center and Vienna Drosophila Genetic Resource Center for the fly lines. This research was partially supported by the Practical Research Project for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development (AMED), the JSPS Core-to-Core Program, Asia-Africa Science Platforms, the JSPS Program for Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers (Grant No. S2802), JSPS KAKENHI Grant Number 16K07346, and Award from American Heart Association- Postdoctoral Fellowship.
