- Academic Editor
Background: Fall armyworm (Spodoptera frugiperda) is a highly destructive maize pest that significantly threatens agricultural productivity. Existing control methods, such as chemical insecticides and entomopathogens, lack effectiveness, necessitating alternative approaches. Methods: Gut-associated bacteria were isolated from the gut samples of fall armyworm and screened based on their chitinase and protease-producing ability before characterization through 16S rRNA gene sequence analysis. The efficient chitinase-producing Bacillus licheniformis FGE4 and Enterobacter cloacae FGE18 were chosen to test the biocontrol efficacy. As their respective cell suspensions and extracted crude chitinase enzyme, these two isolates were applied topically on the larvae, supplemented with their feed, and analyzed for their quantitative food use efficiency and survivability. Results: Twenty-one high chitinase and protease-producing bacterial isolates were chosen. Five genera were identified by 16S rRNA gene sequencing: Enterobacter, Enterococcus, Bacillus, Pantoea, and Kocuria. In the biocontrol efficacy test, the consumption index and relative growth rate were lowered in larvae treated with Enterobacter cloacae FGE18 by topical application and feed supplementation. Similarly, topical treatment of Bacillus licheniformis FGE4 to larvae decreased consumption index, relative growth rate, conversion efficiency of ingested food, and digested food values. Conclusion: The presence of gut bacteria with high chitinase activity negatively affects insect health. Utilizing gut-derived bacterial isolates with specific insecticidal traits offers a promising avenue to control fall armyworms. This research suggests a potential strategy for future pest management.
The fall armyworm (FAW), Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae), is an important pest to numerous crops, including maize (Zea mays), cotton (Gossypium hirsutum), and sorghum (Sorghum bicolor). The production of maize in India rose from 29 million tonnes in 2018–2019 to 35.91 million tonnes in 2022–2023. Due to its industrial usage, maize is a low-input and high-profit crop. However, the invasive FAW, Spodoptera frugiperda, represents a new threat to maize cultivation [1]. Annual output losses from FAW ranged from 8.3 million to 20.6 million tons [2]. Chemical insecticides are frequently used to manage FAW in maize, which has the potential to build resistance over many generations and negatively impact natural enemies [3]. FAW has been demonstrated to be susceptible to entomopathogens such as nuclear polyhedroviruses (SfMNPV), Metarrhizium rileyi, and Nomuraea rileyii and were reported to cause larval infection and mortality [4, 5, 6].
To combat entomopathogens, insects produce reactive oxygen species (ROS), gut phagocytosis, antimicrobial proteins (AMPs), and phenoloxidase (PO) [7]. The immune systems of insects make it difficult for entomopathogens to survive and function effectively in the insect’s gut [8]. Mumcuoglu et al. [9] revealed that the indigenous gut bacteria could be used for control measures because the exogenous bacteria fed to the insects were killed during the passage through the gut. The indigenous microbiota may quickly adjust to changes in the intestinal environment [10]. Hence, insect gut bacteria would be the better way to implement insect pest management.
Interestingly, insect gut microorganisms are crucial to their biology because they establish symbiotic relationships and cause host insect disease [11]. Zhang et al. [12] reported that Enterobacter hormaechei promoted housefly larvae growth by inhibiting harmful Pseudomonas aeruginosa, Providencia stuartii, and Providencia vermicola and improved the reproduction of beneficial bacteria. Enterococcus, Comamonas, and Elizabethkingia were reported to be responsible for most functional alterations in S. frugiperda microbiota [13]. Rozadilla et al. [14] described that archaea and bacteria from the S. frugiperda gut play a significant role in the nutritional requirement of fifth-instar larvae. The gut microorganisms of S. frugiperda fluctuate throughout developmental stages and show vertical transmission of bacteria, while nutrition and the environment might influence gut bacteria [15]. Gut bacteria produce iron-chelating compounds termed siderophores to collect iron from the host insect for bacterial development and proliferation [16]. These siderophores protect the host insects from entomopathogens [17]. According to Krishnamoorthy et al. [8], Bacillus sp. in papaya mealybugs helped detoxify profenophos and chlorpyrifos pesticides used for mealybug management.
A protein–carbohydrate matrix with a chitin concentration ranging from 3 to 13% v/v forms the insect’s peritrophic membrane [18] and is crucial for the insect’s food uptake, growth, and development. The development of insects is influenced by the changes caused by the peritrophic membrane’s chitin and protein compositions [19]. Since it is difficult for insects to develop resistance to microbial enzymes, an environmentally benign tactic is to use gut bacterial enzymes to break down the insect’s structural component by taking advantage of its chitinous morphological structure [20]. Chitinases produced by gut bacteria are employed to degrade the insect’s cuticle partially and they can reduce insect growth by decreasing feeding rate and body weight, leading to insect mortality [21, 22]. It was found that the chitinase-producing Serratia marcescens caused the highest mortality in the treated larvae of Spodoptera littura and was suggested as a biocontrol agent against Spodoptera littura [23]. Harrison et al. [24] demonstrated that the protease enzyme can act as insecticidal when overexpressed. The protease toxic activity can occur in various areas of the insect body, such as the midgut, hemocoel, and cuticle. The present study aimed to isolate and select gut-associated FAW bacteria based on their chitinase and protease activities. In addition, attempts were made to control FAW in vitro by providing chitinase-producing bacteria in the diet and topical application in crude enzyme form and cell suspension. We also examined the harmful effects on the growth and development of their insect hosts.
FAW used in this study was obtained from laboratory-grown and infected maize
field populations. Nearly forty larvae were collected. The larval collection was
performed between November 2021 and January 2022 in the maize fields, which had
not been exposed to any of the pesticides at Tamil Nadu Agricultural University,
Coimbatore, India (11.0123° N, 76.9355° E) and Dharapuram
(10.7343° N, 77.51861° E), Tamil Nadu, India. The
laboratory-reared FAW populations were obtained from the Department of Plant
Biotechnology and Molecular Biology, Tamil Nadu Agricultural University,
Coimbatore, India. The larvae were raised in a laboratory for nearly 190
generations using an artificial feed prescribed by “CIMMYT” [25] at 25
Since fourth (IV) and fifth (V) instar larvae inflict substantial damage upon
their hosts, these instar larvae were selected to isolate gut-associated
bacteria. To remove the transitory microbiomes, 25 larval instars were selected
from the laboratory, reared and field-caught populations, and left to fast for a
whole day. Subsequently, the larvae were surface disinfected for five minutes
using 70% ethanol and washed three to five times with sterile distilled water
[26]. Under sterile conditions, the larvae were dissected, and gut samples were
collected in 0.1 M phosphate buffer (pH 7.0). The gut samples were homogenized in
a sterile pestle and mortar, serially diluted, and spread in eleven different
growth media-containing Petri plates. The used growth media were Corn Meal agar,
Czapek Dox agar, De Man, Rogosa, and Sharpe (MRS) agar, Eosin-methylene blue
(EMB) agar, Endo agar, Luria–Bertani agar, MacConkey agar, nutrient agar,
Reasoner’s 2A (R2A) agar, Tryptose soy agar, and yeast extract peptone dextrose
(YPD) agar (HiMedia, Mumbai, India). The plates containing the gut suspensions
were incubated for 72 hours at 28
Chitin degradation was quantified using the 3,5-dinitrosalicylic
acid (DNS) assay. Briefly, bacterial isolates (1
Skim milk agar plates were used to assess the protease enzyme activity of FAW
gut-associated bacterial isolates qualitatively [28]. The bacterial isolates (1
Siderophore-producing bacterial isolates were qualitatively assessed using Chrom
Azurol S (CAS) agar plates, as described by Dutta et al. [29]. The
succinate medium was prepared separately (pH 6.5), then CAS indicator solution
was added to the medium, adjusted to pH 7.0, and autoclaved. Next, the bacterial
cultures (1
For molecular identification, the genomic DNA of bacterial isolates with high
chitinase and protease activity was extracted using the CTAB method. The 16S rRNA
gene was amplified from the isolated DNA using the universal primers 27 F
(5
Nitrogen-fixing ability, zinc, silica, and phosphate solubilization efficiency
were assessed to identify the role of gut-associated bacteria in host insect
nutrition. The nitrogen-fixing ability of gut bacterial isolates was assessed by
growing the isolates in a nitrogen-free bromothymol blue malic acid medium (Nfb).
The color change of the medium from green to blue indicated that the isolates
could fix nitrogen [8]. The zinc and silica solubilization efficiency was
detected by spotting the isolates (10 µL, 1
The efficient chitinase-producing Bacillus licheniformis FGE4 and
Enterobacter cloacae FGE18 isolates from the gut of FAW were chosen to
test their effect on the nutritional indices of host insects as a consequence of
chitinase. Hence, both isolates were added as food supplements and topically
applied to the larvae. To extract the crude chitinase enzyme, the bacterial
isolates were cultured in a liquid nutrient medium supplemented with 0.3%
colloidal chitin. The cells were separated by centrifugation (10,000 rpm at 4
°C for 5 min), and the enzyme-containing supernatant was then purified
with a membrane filter before being utilized in the bioassay. The cell suspension
was prepared by washing the pellets in 0.05 mol phosphate buffer. The phosphate
buffer was prepared by mixing sodium phosphate dibasic stock (0.5 M) and sodium
phosphate monobasic stock (0.5 M), and the pH was adjusted to 7.0. After being
washed twice with the buffer, the cell pellets were suspended in sterilized
distilled water and were used for experiments. The following were the test
solutions (treatments) used per 2 grams of CMMYT diet for conducting bioassay:
(i) 1 mL of the cell suspension (10
ANOVA was used to analyze the data, and the General Linear Models Tukey’s HSD test was used to compare the means. The square root and arcsine transformations were used for the data transformation of numbers and percentages. IBM SPSS (SPSS, 2013, IBM Corp., Armonk, NY, USA) was used for all data analysis.
A total of 111 morphologically distinct bacteria were isolated from the IV and V instars of the field-caught and laboratory-reared (with artificial diet) FAW populations. Of the 111 isolates, 38 and 29 were IV and V instars, respectively, from the laboratory-reared FAW population; a further 20 and 24 isolates were recovered from IV and V instar larvae, respectively, from the field-caught FAW population. No bacterial colonies were detected in the gut suspensions from the IV and V instar artificial diet-reared larval populations or V instar field-caught FAW populations in the Corn Meal agar plates. In the MRS medium, bacterial colonies were only visible 48 hours after incubation. The maximum numbers of bacteria were found in the gut suspension of IV instar field-caught larvae from the NA medium. In contrast, the lowest numbers were found in the IV instar artificial diet-reared larvae from the Endo agar medium. Out of all the isolates from the gut samples of the larvae raised on an artificial diet, the TSA medium revealed the largest bacterial population. In contrast, the Endo agar medium revealed the lowest population.
The 16S rRNA gene analysis revealed that the isolates recovered from S. frugiperda belonging to Firmicutes (also known as Bacillota) contain three different genera, with Bacillus being the predominant one. Additionally, our results reported three different genera of Gammaproteobacteria. The nucleotide sequences of the recovered bacterial isolates were subjected to homology searches in DNA databases, which revealed that the sequences of the FGE1, FGE4, FGE5, FGE8, FGE10, FGE17, FGE19, FGE20, FGE21 isolates showed a 95% to 98% similarity with the 16S rRNA gene sequences for the Bacillus species and FGE2, FGE13, FGE15, FGE16, and FGE18 (99%) were homologous with the Enterobacter species. Similarly, FGE6, FGE9, FGE11, and FGE14 showed 98% similarity with the Enterococcus species (Table 1).
| Isolates | Closest match | Similarity % | Length (bp) | NCBI accession number | Chitinase activity (µmol/min/mL) | Protease activity (%) | Siderophore production (%) |
| FGE1 | Bacillus amyloliquefaciens longA | 98.65 | 1505 | OP070959 | 1.78 |
ND | ND |
| FGE2 | Enterobacter cloacae TBMAX89 | 99.59 | 1472 | OP068371 | 1.72 |
34.62 |
50 |
| FGE3 | Klebsiella variicolaAHKv-S01 | 97.54 | 1450 | OP070061 | 1.54 |
ND | 78.57 |
| FGE4 | Bacillus licheniformis DS3 | 98.11 | 1457 | OP070050 | 2.1 |
ND | ND |
| FGE5 | Bacillus subtilis ANA4 | 98.75 | 1476 | OP070059 | 1.04 |
75 |
66.67 |
| FGE6 | Enterococcus mundtii UDFX4 | 98.62 | 1464 | OP081022 | 0.94 |
28 |
ND |
| FGE7 | Kocuria turfanensis NL52 | 98.78 | 1439 | OP070058 | 0.68 |
27.27 |
ND |
| FGE8 | Bacillus subtilis OH2377A | 95.47 | 1465 | OP070955 | 0.16 |
65.38 |
ND |
| FGE9 | Enterococcus mundtii15-1A | 98.38 | 1487 | OP081005 | 0.22 |
51.85 |
20 |
| FGE10 | Bacillus thuringiensis SRG2 | 95.13 | 1452 | OP070946 | 0.22 |
28.57 |
ND |
| FGE11 | Enterococcus sp. RJ-7 | 98.9 | 1500 | OP070961 | 1.08 |
ND | ND |
| FGE12 | Pantoea agglomerans P18 | 97.66 | 1469 | OP070952 | ND | 87.5 |
ND |
| FGE13 | Enterobacter hormaechei DS02Eh01 | 99.25 | 1441 | OP070948 | ND | 39.29 |
ND |
| FGE14 | Enterococcus durans 4434 | 98.82 | 1459 | OP070943 | ND | 16.67 |
ND |
| FGE15 | Enterobacter mori YIM Hb-3 | 98.19 | 1503 | OP070944 | 0.14 |
9.52 |
33.33 |
| FGE16 | Enterobacter asburiae A2563 | 99.14 | 1426 | OP070960 | 1.42 |
ND | ND |
| FGE 17 | Bacillus cereus P3B | 97.78 | 1525 | OP070957 | 0.68 |
26.06 |
ND |
| FGE 18 | Enterobacter cloacae R6-366 | 98.36 | 1461 | OP070949 | 2.3 |
51.85 |
ND |
| FGE19 | Bacillus halotolerans BBRIST011 | 96.00 | 1462 | OP070947 | 0.4 |
61.54 |
ND |
| FGE20 | Bacillus velezensis JF37 | 96.03 | 1475 | OP070950 | 0.94 |
60 |
ND |
| FGE21 | Bacillus pumilus SBMP2 | 98.48 | 1456 | OP070926 | ND | 54.55 |
ND |
The first column in the table represents the gut bacterial isolates associated with fall armyworm (FAW). FGE1–FGE3 represents isolates from the fourth instar
field-caught FAW population, FGE4–FGE12 represents isolates from the fifth
instar artificial-diet-reared FAW population, FGE13–FGE14 represent isolates
from the fifth instar field-caught FAW population, FGE15–FGE21 represents
isolates from fourth instar artificial-diet-reared FAW population. Values in each
column are the mean of three replications
Among 111 bacterial isolates, only 81 were chitinase positive, and 37 were
protease positive (Table 1). The maximum chitinolytic activity (2.3
Among the 21 screened gut bacterial isolates, 9 isolates (Klebsiella
variicola FGE3, Bacillus subtilis FGE5, Bacillus subtilis
FGE8, Pantoea agglomerans FGE12, Enterobacter hormaechei
FGE13, Enterobacter asburiae FGE16, Bacillus cereus FGE17,
Enterobacter cloacae FGE18, Bacillus pumilus FGE21) were found
to fix nitrogen in the medium (Table 2). Eleven isolates were found to solubilize
zinc, with the maximum solubilization shown by Enterobacter hormaechei
FGE13 (81.82
| Isolates | Nitrogen fixation | Solubilization (%) | ||
| Zinc | Silica | Phosphate | ||
| Bacillus amyloliquefaciens FGE1 | - | ND | ND | ND |
| Enterobacter cloacae FGE2 | - | ND | ND | ND |
| Klebsiella variicola FGE3 | + | 7.15 |
40 |
33.34 |
| Bacillus licheniformis FGE4 | - | 27.28 |
ND | ND |
| Bacillus subtilis FGE5 | + | ND | ND | ND |
| Enterococcus mundtii FGE6 | - | 33.34 |
ND | ND |
| Kocuria turfanensis FGE7 | - | ND | 20 |
ND |
| Bacillus subtilis FGE8 | + | ND | ND | ND |
| Enterococcus mundtii FGE9 | - | 20 |
ND | ND |
| Bacillus thuringiensis FGE10 | - | ND | ND | ND |
| Enterococcus sp. FGE11 | - | ND | ND | ND |
| Pantoea agglomerans FGE12 | + | 10 |
50 |
66.67 |
| Enterobacter hormaechei FGE13 | + | 81.82 |
81.82 |
87.5 |
| Enterococcus durans FGE14 | - | 44.45 |
10 |
ND |
| Enterobacter mori FGE15 | - | ND | ND | ND |
| Enterobacter asburiae FGE16 | + | 55.56 |
53.85 |
50 |
| Bacillus cereus FGE17 | + | 66.67 |
92.5 |
83.34 |
| Enterobacter cloacae FGE18 | + | 50 |
83.34 |
66.67 |
| Bacillus halotolerans FGE19 | - | ND | 23.08 |
ND |
| Bacillus velezensis FGE20 | - | ND | ND | ND |
| Bacillus pumilus FGE21 | + | 62.5 |
81.82 |
50 |
The first column in the table represents the gut bacterial isolates associated
with fall armyworm (FAW). FGE1–FGE3 represents isolates from the fourth instar
field–caught FAW population, FGE4–FGE12 represents isolates from the fifth
instar artificial-diet-reared FAW population, FGE13–FGE14 represents isolates
from the fifth instar field-caught FAW population, FGE15–FGE21 represents
isolates from the fourth instar artificial-diet-reared FAW population. Values in
each column represent the mean of three replications
There was a decrease in consumption rate (1.87
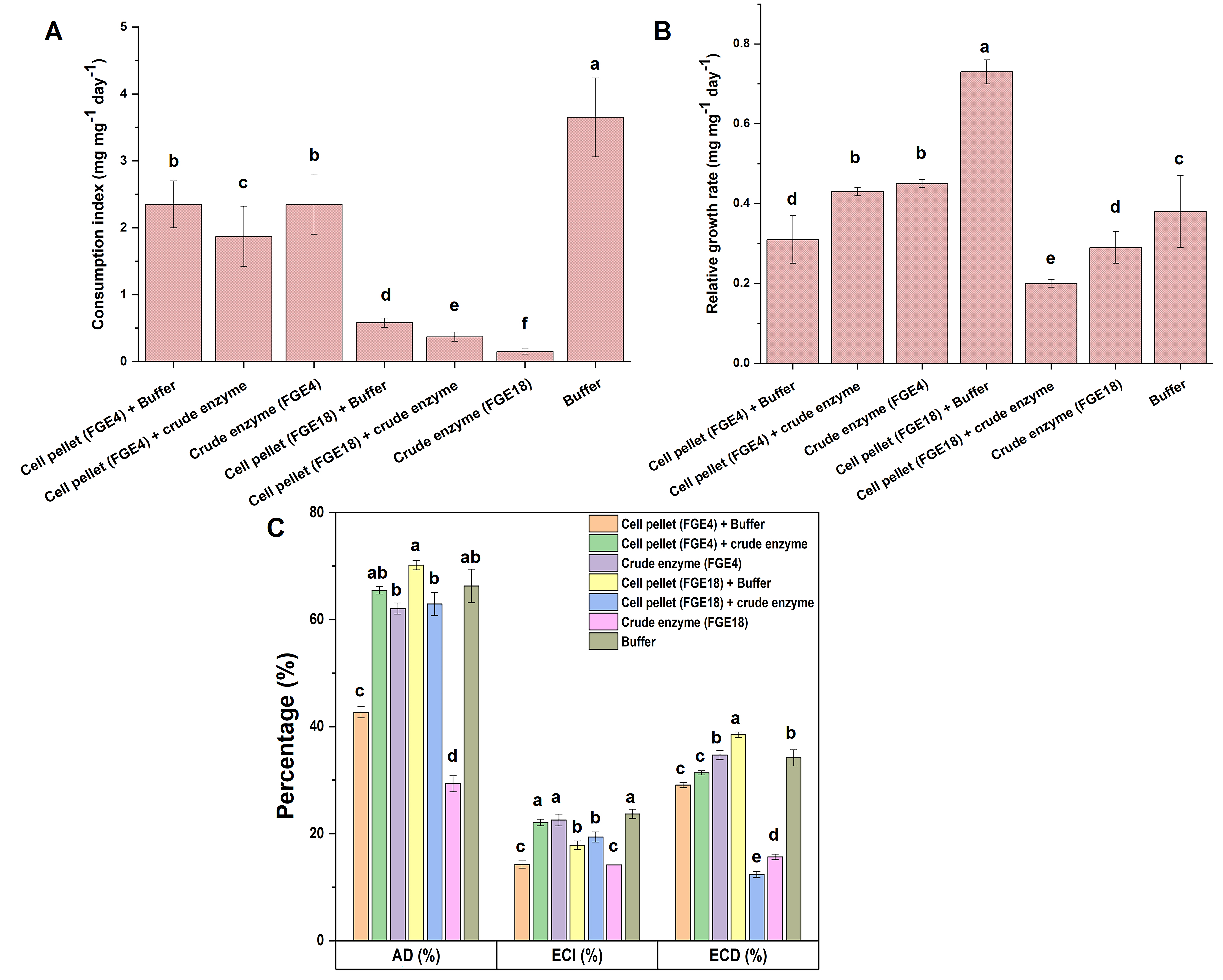 Fig. 1.
Fig. 1.Effect of chitinase on quantitative food use efficiency through topical application. Each panel represents a mean of three replicates, the error bar indicates standard error, and panels with the letter(s) a–f are significantly different at 0.05 levels (Tukey’s HSD test). (A) Consumption index, (B) relative growth rate, (C) approximate digestibility (AD), efficiency of conversion of ingested food (ECI), and efficiency of conversion of digested food (ECD).
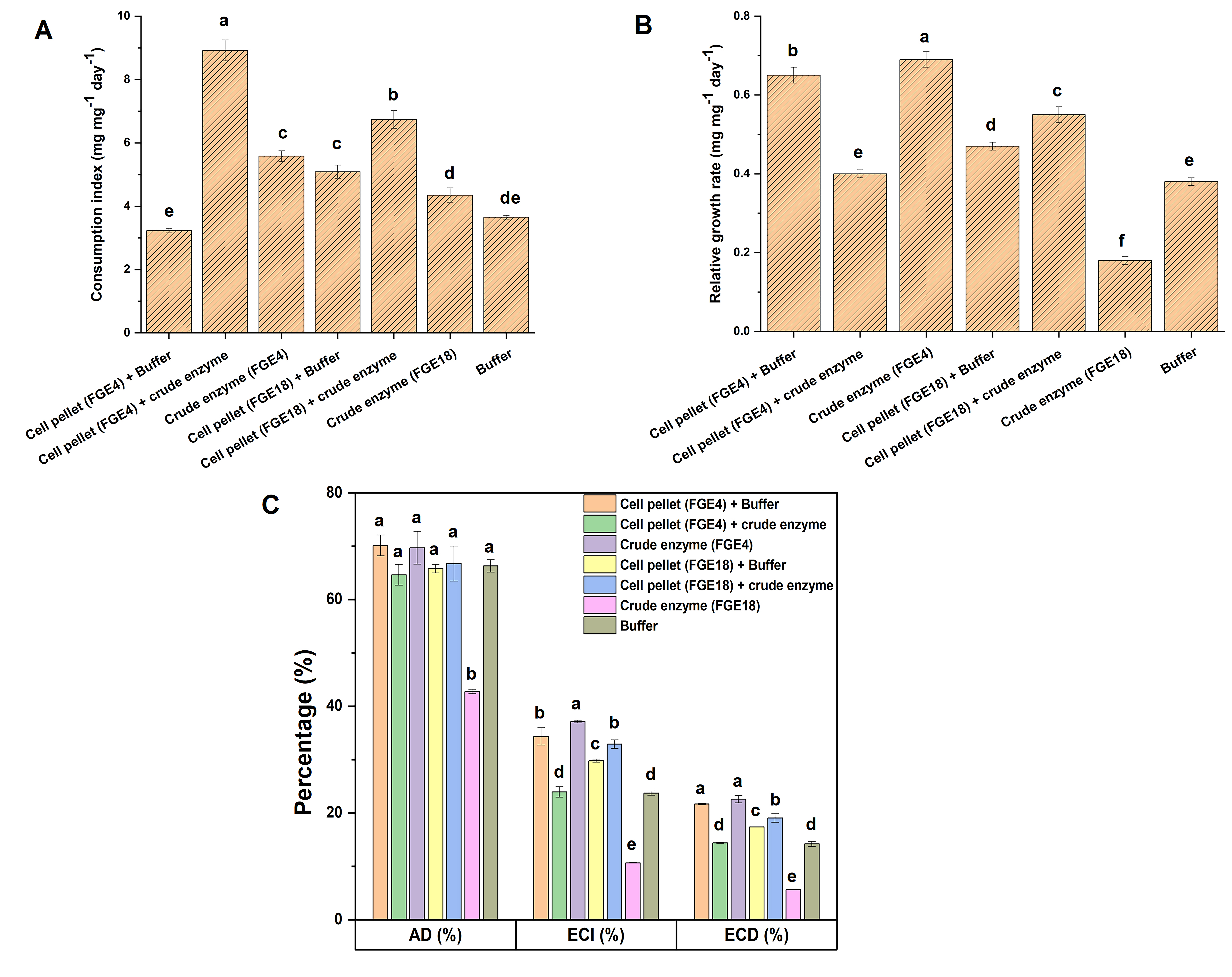 Fig. 2.
Fig. 2.Effect of chitinase on quantitative food use efficiency through feed supplementation. Each panel represents mean of three replicates, the error bar indicates standard error, and panels with the letter(s) a–f are significantly different at 0.05 levels (Tukey’s HSD test). (A) Consumption index, (B) relative growth rate, (C) approximate digestibility (AD), efficiency of conversion of ingested food (ECI), and efficiency of conversion of digested food (ECD).
Understanding the contribution of gut bacteria to host insect activities, such as antagonistic against invading pathogens, detoxification of pesticides, and host feeding, requires determining the species of bacteria and their probable involvement in the host insect gut environment. This study revealed the presence of different cultivable FAW gut bacteria, which were collected from field conditions and laboratory-reared populations. Based on the molecular characterization, the cultivable gut bacterial isolates belong to 18 different bacterial species, viz., Bacillus amyloliquefaciens, Bacillus licheniformis, Bacillus subtilis, Klebsiella variicola, Kocuria turfanensis, Enterococcus mundtii, Enterococcus durans, Bacillus thuringiensis, Enterococcus sp., Pantoea agglomerans, Enterobacter hormaechei, Enterobacter mori, Enterobacter asburiae, Enterobacter cloacae, Bacillus cereus, Bacillus halotolerans, Bacillus velezensis, and Bacillus pumilus. Similar reports were found in S. frugiperda-infested maize fields in Shaanxi Province, where Enterococcus and Enterobacteriaceae members, such as Enterobacter, Klebsiella, Pantoea, Escherichia, Rhodococcus, and Ralstonia, predominated in the guts of both adults and larval stages [15]. The genus Enterococcus was the most common in the FAW gut samples obtained from eastern parts of India, followed by Klebsiella sp. and Enterobacter sp., with a small proportion of Raoultella, Citrobacter, Leclercia, and Pantoea [32]. Acevedo et al. [33] also reported that S. frugiperda oral secretions contained Pantoea, Enterobacter, Raoultella, and Klebsiella. Indiragandhi et al. [17] reported the different bacterial phylotypes present in the insecticide-resistant, susceptible, field-caught population of Plutella xylostella. The absence of specific bacteria in the larval gut limits its pupation and successful adult emergence [34]. Hence, the cultivable bacteria isolated from the FAW might have a significant role in the biology of S. frugiperda larvae and even with adult development. The study conducted by Sivakumar et al. [35] reported that the gut bacterium B. pumilus associated with A. biguttula biguttula has a role in host insect nutrition and defense for the first time.
Chen et al. [36] suggested that Enterococcus may affect the metabolism level in the gut of S. frugiperda by aiding carbohydrate transport and energy production. Enterococcus spp., identified in this study, may contribute to the defense response to FAW. Research has demonstrated that members of Enterobacteriaceae are involved in the metabolism of sugar in larvae and digestion, defense, courtship, and reproduction [37]. In the present study, cultivable Enterobacteriaceae members were also isolated. The highest bacterial population was found in the NA medium and the lowest in the MacConkey agar medium among the field-caught FAW larval gut samples of IV and V instar larvae. Similar results were observed in the cultivable gut bacteria isolated from the diamondback moth, Plutella xylostella, where the highest bacterial population was observed in NA medium, and the lowest number of bacterial populations was observed in MacConkey agar [17]. The gut of the field-caught population may have the highest bacterial population level because of the increased nutritional availability from the natural host plants; however, the artificially reared population harbored a lower abundance of the bacterial population. The increased number of Firmicutes in the guts of S. frugiperda larvae might result from the larvae’s improved ability to absorb various nutrients [37].
The exoskeleton and peritrophic membrane of insects serve as a physicochemical defense and are composite materials predominantly composed of chitin and protein, with the latter also including trace amounts of lipids, catecholamine metabolites, minerals, and other minor constituents [38]. The pathogens or pests may be exposed to chitinases at unsuitable concentrations or stages of development to make them more susceptible to host defenses [38]. Consequently, bacterial isolates in the current study were screened based on their chitinolytic and proteolytic activity. Of the 111 gut bacterial isolates, 81 could produce chitinase, 37 were protease producers, and 5 were siderophore producers. The gut flora controls the thickness of the peritrophic membrane, which affects how nutrients pass through the insect gut [39]. Chitinase and protease-producing gut bacteria were found to enter the host gut through feeding and disturb the thickness of the peritrophic membrane, which led to a nutritional imbalance in the host insect and mortality [8, 22]. Similarly, the protease enzymes produced by Xenorhabdus nematophila could suppress the insect’s immune system [40]. Due to the low iron concentration in the gut environment, siderophore production is believed to be widespread in insect gut bacteria. The siderophores are produced and secreted by bacteria complexes with iron outside the bacterial cell, which they then deliver to the insect [41]. Some opportunistic pathogens that require iron for pathogenicity are found in the gut of insects. By generating siderophores, they chelate the iron from the consumed insect feed. As a result, an insect may die from iron toxicosis if it consumes too much iron [42].
Insect gut bacteria correlate with their insect partners in terms of nutrition [43]. The droplets of poplar and willow borer, Cryptorhynchus lapathi, contained bacterial enzymes involved in nitrogen and sulfur metabolism and were also involved in the biosynthesis of essential amino acids and vitamins [7]. The bacteria isolated from the gut samples in the present study showed nitrogen fixation, zinc, silica, and phosphate solubilization. The organisms involved in nutrient provisioning and insect physiology can be altered or eliminated to retard insect growth, which can be useful for pest control. Eliminating gut bacteria with antibiotics impaired the growth and development of adult host insects [34].
In the present study, the extracted crude chitinase enzyme and chitinase-producing bacteria were supplemented in their diet, fed to the larvae, and topically sprayed on the larvae. There was a 98.63% reduction in the consumption index of larvae treated with the crude chitinase enzyme plus chitinase-producing E. cloacae FGE18 through topical application, and the growth reduction was also observed in both feeds of supplemented and topically infected larvae (52.63% and 23.68% growth reduction, respectively). The genomic sequence of E. cloacae subsp. cloacae indicated the presence of four chitinases and two N-acetyl-glucosaminidases that may be involved in chitin breakdown. Furthermore, the E. cloacae genome contains genes that code for one CBM-33 lytic polysaccharide monooxygenase and one polysaccharide deacetylase, which may play an important role in the depolymerization of chitin [44]. Additionally, Liao et al. [45] claimed the role of E. cloacae insecticidal protein in killing the host insect, Galleria mellonella larvae, by destroying or inhibiting their host immune response. In the case of Bacillus licheniformis FGE4, decreased ECI and ECD values were found in the topically applied larval population. These results were similar to the experiments conducted using transgenic tobacco plants expressing the Manduca sexta chitinase gene to feed tobacco budworms [46], and the results showed a reduction in larval biomass and feeding damage. In our investigation, most larval parameters exhibited lower values when the larvae were treated topically with cell suspension and crude enzymes of both isolates. Similar results were obtained by Kim et al. [47], where they described that chitinolytic and proteolytic effects in the culture supernatant of Beauveria bassiana could cause death when topically sprayed against aphids (Aphis gossypii). Harrison and Bonning [24] demonstrated that cuticle-degrading proteases such as PR1A may be hazardous to insects when provided topically. The colonization of chitinase-producing bacteria and the production of chitinase in the gut may lead to damage in the peritrophic membrane of the insect gut and cause diffusion of nutrients, which were similar in the mode of action of permethrin insecticides and delta endotoxins of Bacillus thuringiensis [21]. In evidence of this, several researchers have demonstrated the insecticidal properties of the chitinases from diverse microorganisms. Additionally, chitinolytic bacteria that produce protease, siderophores, and secondary metabolites were discovered to be the best for controlling nematodes [48]. The growth of the tobacco caterpillar, Spodoptera litura, was hindered by purified chitinases from B. subtilis. A talc-based formulation of Pseudomonas fluroescens and chitin has been observed to decrease the incidence of the leaf folder Cnaphlocrocis medinalis in rice by 56.1% [49].
Our study concluded that gut-inhabiting bacteria with high chitinase activity can negatively influence the insect’s health and survivability. Hence, enhancing them would be an alternate strategy in insect pest management. Chitinase-producing Bacillus licheniformis FGE4 and Enterobacter cloacae FGE18 isolated from the gut of S. frugiperda larvae can be used against the FAW larvae. However, the application of these identified bacteria in the field conditions, evaluating their persistence in the field, and method of applications for effective management of insect pests needs to be thoroughly studied in the future to drive this approach more effectively for the benefit of the farming community.
FAW, Fall Armyworm; SfMNPV, Spodoptera frugiperda Multicapsid Nuclear polyhedrosis Virus; PO, Phenoloxidase; ROS, Reactive Oxygen Species; AMP, Antimicrobial proteins; CIMMYT, International Maize and Wheat Improvement centre; EMB agar, Eosin Methylene Blue agar; R2A agar, Reasoner’s 2A agar; TSA, Typtose Soy Agar; NA, Nutrient Agar; YPD, Yeast Extract Peptone Dextrose agar; MRS agar, de Man Rogosa and Sharpe agar; cfu, Colony Forming Unit; CAS, Chrom Azurol S; CTAB, Cetyl Trimethyl Ammonium Bromide; PCR, Polymerase Chain Reaction; NCBI, National Center for Biotechnology Information; CI, consumption index; RGR, relative growth rate; AD, approximate digestibility; ECI, the efficiency of the conversion of the ingested food; ECD, the efficiency of the conversion of the digested food.
The data utilized and/or examined in the present study can be obtained from the corresponding author upon a reasonable request.
NOG and RA designed the research study. RA contributed to the conception of the present work. TDW performed the research, analyzed the data and wrote the manuscript. PI and VB provided oversight, literature search and direction for the drafting of the manuscript. All authors have participated sufficiently in the work to take public responsibility for appropriate portions of the content and agreed to be accountable for all aspects of the work in ensuring that questions related to its accuracy or integrity. All authors read and approved the final manuscript. All authors contributed to editorial changes in the manuscript.
Not applicable.
The author places their sincere thanks for the facilities extended by the Department of Agricultural Microbiology, Department of Agricultural Entomology and Department of Plant Biotechnology, Tamil Nadu Agricultural University, Coimbatore.
This research received no external funding.
The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
