- Academic Editors
-
-
-
Background: Autism Spectrum Disorder (ASD) is a complex
neurodevelopment disease characterized by impaired social and cognitive
abilities. Despite its prevalence, reliable biomarkers for identifying
individuals with ASD are lacking. Recent studies have suggested that alterations
in the functional connectivity of the brain in ASD patients could serve as
potential indicators. However, previous research focused on static
functional-connectivity analysis, neglecting temporal dynamics and spatial
interactions. To address this gap, our study integrated dynamic functional
connectivity, local graph-theory indicators, and a feature-selection and ranking
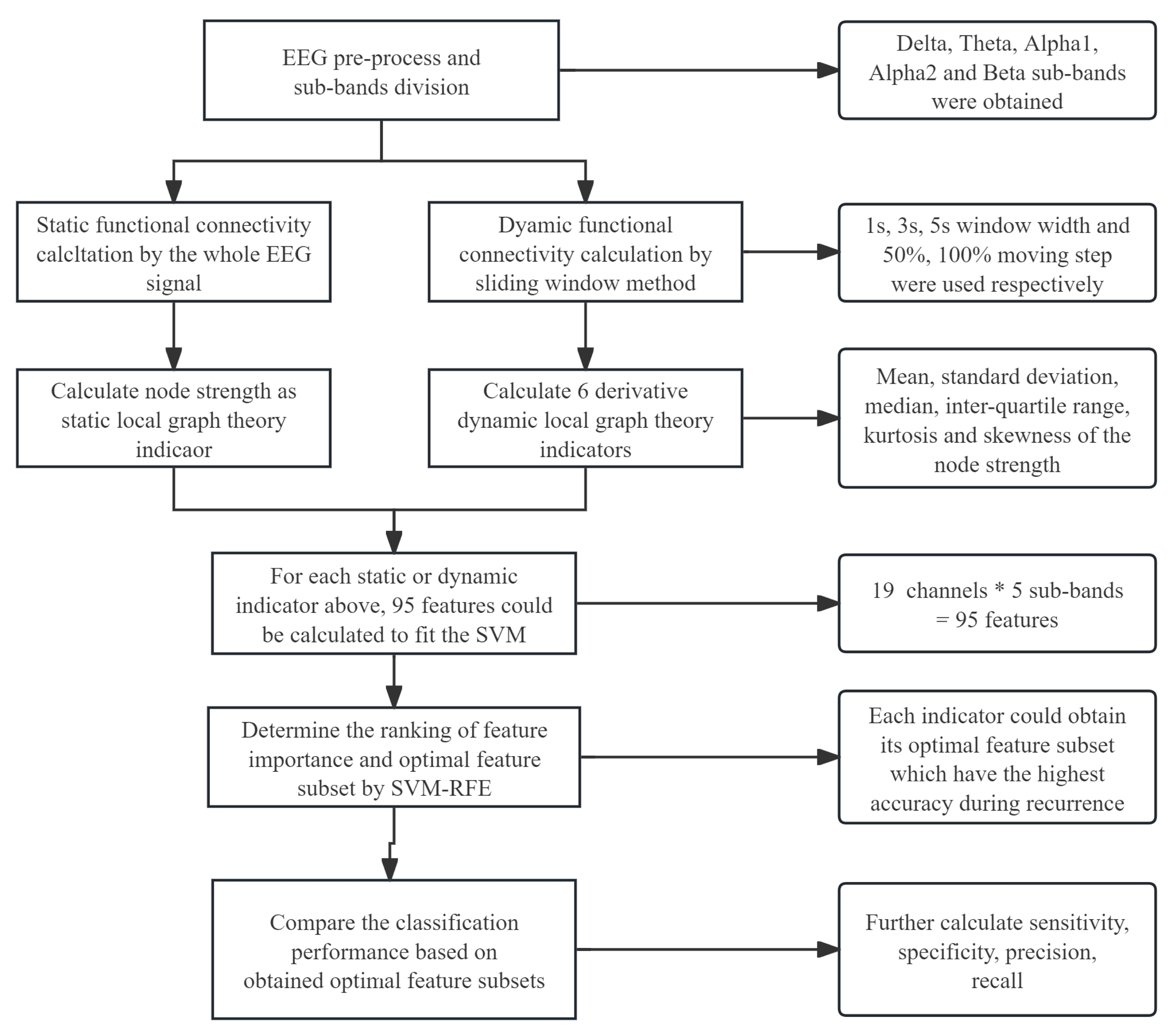
approach to identify biomarkers for ASD diagnosis. Methods: The
demographic information, as well as resting and sleeping electroencephalography
(EEG) data, were collected from 20 ASD patients and 25 controls. EEG data were
pre-processed and segmented into five sub-bands (Delta, Theta, Alpha-1, Alpha-2,
and Beta). Functional-connection matrices were created by calculating coherence,
and static-node-strength indicators were determined for each channel. A
sliding-window approach, with varying widths and moving steps, was used to scan
the EEG series; dynamic local graph-theory indicators were computed, including
mean, standard deviation, median, inter-quartile range, kurtosis, and skewness of
the node strength. This resulted in 95 features (5 sub-bands