-
- Academic Editors
-
-
-
Developmental language disorders (DLDs) are common neurodevelopmental conditions, affecting approximately 7–10% of children, with significant impacts on communication, academic achievement, and social integration. While genetic factors are known contributors, the underlying genomic architecture and biological pathways remain incompletely understood. This analysis explores key genomic biomarkers of DLD and investigates their functional interactions.
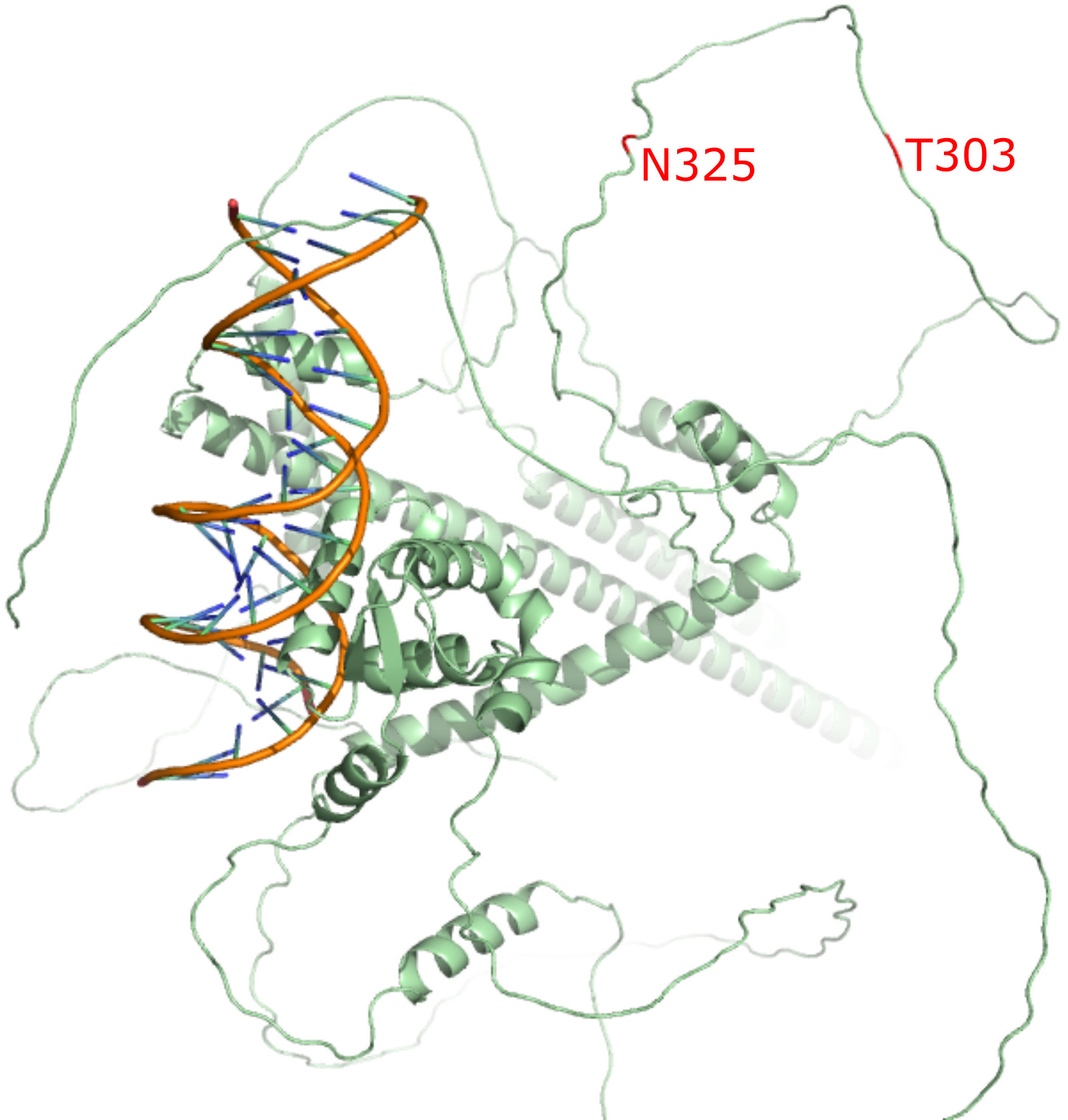
We conducted an integrative genomic analysis combining multiple data-driven approaches. Using the Open Targets platform, we compiled a set of genes associated with DLD-related phenotypes (based on evidence scores ≥0.3) and constructed a gene-phenotype network to visualize these associations. Protein-protein interaction mapping of the identified genes was performed using the STRING database to uncover interaction clusters and shared pathways. We then analyzed sequence and structural relationships among the encoded proteins, including pairwise sequence homology (BLAST alignments), 3D structural modeling, and multimeric interaction prediction using AlphaFold 3.
Our analysis identified 89 genes linked to 14 DLD-related phenotypic terms, with strong clustering around delayed speech. Several genes (e.g., GRN, MAPT, FOXP2, FOXP1, AP4E1) showed particularly high-confidence associations. Structural analysis of encoded proteins revealed unexpected similarity between functionally related but sequence-divergent pairs (e.g., WDR45 and GNB1). AlphaFold 3 modeling predicted a potential interaction between DCDC2 and KIAA0319, suggesting a plausible structural mechanism for their co-involvement in dyslexia.
DLDs emerge from diverse genetic contributors but converge on shared neurodevelopmental pathways. Structural modeling enhances genomic insights by uncovering hidden relationships and candidate interactions, paving the way for more precise genetic screening and functional studies in language disorders.