- Academic Editor
Background: Multiple radiomics models have been proposed for
grading glioma using different algorithms, features, and sequences of magnetic
resonance imaging. The research seeks to assess the present overall performance
of radiomics for grading glioma. Methods: A systematic literature review
of the databases Ovid MEDLINE PubMed, and Ovid EMBASE for publications published
on radiomics for glioma grading between 2012 and 2023 was performed. The
systematic review was carried out following the criteria of Preferred Reporting
Items for Systematic Reviews and Meta-Analysis. Results: In the
meta-analysis, a total of 7654 patients from 40 articles, were assessed.
R-package mada was used for modeling the joint estimates of specificity (SPE) and
sensitivity (SEN). Pooled event rates across studies were performed with a
random-effects meta-analysis. The heterogeneity of SPE and SEN were based on the
With the 5th edition of the World Health Organization (WHO) Classification of Tumors of the Central Nervous System (CNS) published in 2021 [1], a major role has been assigned to molecular patterns for the differential diagnosis of gliomas [2]. These innovations have been followed by improvements in therapeutic strategies, with more targeted and focused therapies [3]. The gold standard for diagnosis of gliomas remains biopsy, although it puts the patients at inevitable risk of procedure-related complications [4]. Over the last 10 years, artificial intelligence (AI) applied to diagnostic imaging progressively gained more popularity [5]. Radiomics, through deep learning (DL) and machine learning (ML) techniques, refers to the extraction of mineable data from medical imaging, boosting its diagnostic capability. Since its first appearance in 2012 as a computer-aided detection and diagnosis system [6], radiomics has grown massively infiltrating the diverse fields of neuro-oncology [4]. Clinical applications of these noninvasive radiomics-based models range from screening and characterization to monitoring and prediction [7]. This has clinical implications for any CNS tumor, from gliomas to meningioma and pituitary tumors [8, 9, 10].
The grading of glioma has been one of the main focuses of radiomics [11]. Nowadays, multiple radiomics-based models have been proposed for grading glioma, using different features, models, and sequences of magnetic resonance imaging (MRI). Each carried out its diagnostic accuracy, thus, the actual performance of radiomics is dependent on the specific model and is not standardized [12]. Given the heterogeneity and rapid expansion of this technique, we aimed to better define its overall potential in grading gliomas. Therefore, based on the current literature we conducted a meta-analysis and systematic review to evaluate the current performance state of radiomics for the grading of brain gliomas.
The Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) criteria were followed in conducting the systematic review [13]. An expert librarian created and carried out a thorough literature search of the Ovid MEDLINE, PubMed, and Ovid EMBASE databases with advice from the authors. Three terms were utilized in “AND” combinations: “radiomics”, “glioma”, and “grading”. Only publications published between 2012 and 2023 were included in the search.
Two Authors (L.D.M. and F.P.) assessed the study inclusion criteria during the review process. The following inclusion criteria included: (1) Written in the language of English; (2) case series including based on 10 patients or more; (3) studies reporting exclusively histologically proven brain gliomas; (4) studies that included the WHO grade; (5) studies based on radiomics; (6) studies that reported the performance data of radiomics for glioma grade prediction.
Three investigators (L.D.M., F.P., and E.A) independently collected data from systematic reviews that were eligible using piloted forms. Each systematic review was then validated by a different investigator, and consensus was used when confronted with disagreements by. Study characteristics were obtained from each eligible systematic, based on the baseline information that included: year of publication, number of patients, WHO grade, and MRI protocol. As for the radiomics model, we collected information about: the AI sub-category (i.e., ML or DL), the classification algorithm (i.e., logistic regression (LR), gradient boost (GB), naïve bayes (NB), support vector machine (SVM), multilayer perceptron (MLP), elastic net regression (ENR), linear discriminant analysis (LDA), nearest neighbors (NN), random forest (RF), convolutional neural network (CNN), and others), the selected features (i.e., textural, geometrical or morphological, voxel intensities-based, others), the employed MRI modalities (i.e., T1-weighted [T1W], T2-weighted [T2W], others), and the application of cross-validation analysis (i.e., yes or no).
We analyzed the clarity of reporting in eligible systematic reviews with the PRISMA checklist. The checklist includes 27 items that are used to assess the reporting quality. The PRISMA checklist is shown in Supplementary Material [13].
Our primary outcomes were the specificity (SPE), sensitivity (SEN), and Summary Receiver Operating Characteristics (SROC) curve of radiomics for predicting the WHO grade of gliomas of the brain. Bivariate analyses by discrimination task between low-grade gliomas (LGGs; WHO grade 1 and 2) vs. high-grade gliomas (HGGs; WHO grade 3 and 4), and WHO grade 4 gliomas vs. WHO grade 3 gliomas were conducted.
The impact of the following variables on the performance of the proposed radiomics models was evaluated as secondary outcomes: year of development, cohort size, AI sub-category, validated classifiers, selected features, MRI modalities, and cross-validation strategy. These variables were also studied in quantitative terms to picture the current trends of radiomics models for glioma grading.
To evaluate the methodologic quality of the studies that were part of our meta-analysis, we adjusted the Newcastle-Ottawa Scale (NOS) [14]. The purpose of this tool is to be used in comparative investigations. However, as our investigations lacked a control group, we evaluated the methodologic quality of the data using a subset of the scale’s items, paying particular attention to the following queries: (1) Was a random sample used in the study, or did all patients or consecutive patients participate? (2) Was the research prospective or retrospective? (3) Did the clinical follow-up provide enough information to determine every outcome? (4) Were the results published? (5) Were the criteria for inclusion and exclusion well-defined [15]?
Data from primary studies were reported in a 2
After duplicates were eliminated, 390 papers were found. 103 articles were found for full-text study following the analysis of the titles and abstracts. Forty papers were evaluated for eligibility. The following criteria led to the exclusion of the remaining 63 articles: (1) studies not reporting data on radiomics performance for glioma grading (34 articles), (2) studies not reporting the WHO grade (13 articles), (3) studies reporting on AI-based models other than radiomics (8 articles), (4) improper study design (5 articles), and (5) language other than English (3 articles). The Authors of three included articles [17, 18, 19] provided missing performance data and the results were integrated into the data abstraction process. For each of the patient groups under consideration, at least one or more outcome measures were available for all of the studies that were part of the analysis. The PRISMA statement’s flow chart is seen in Fig. 1.
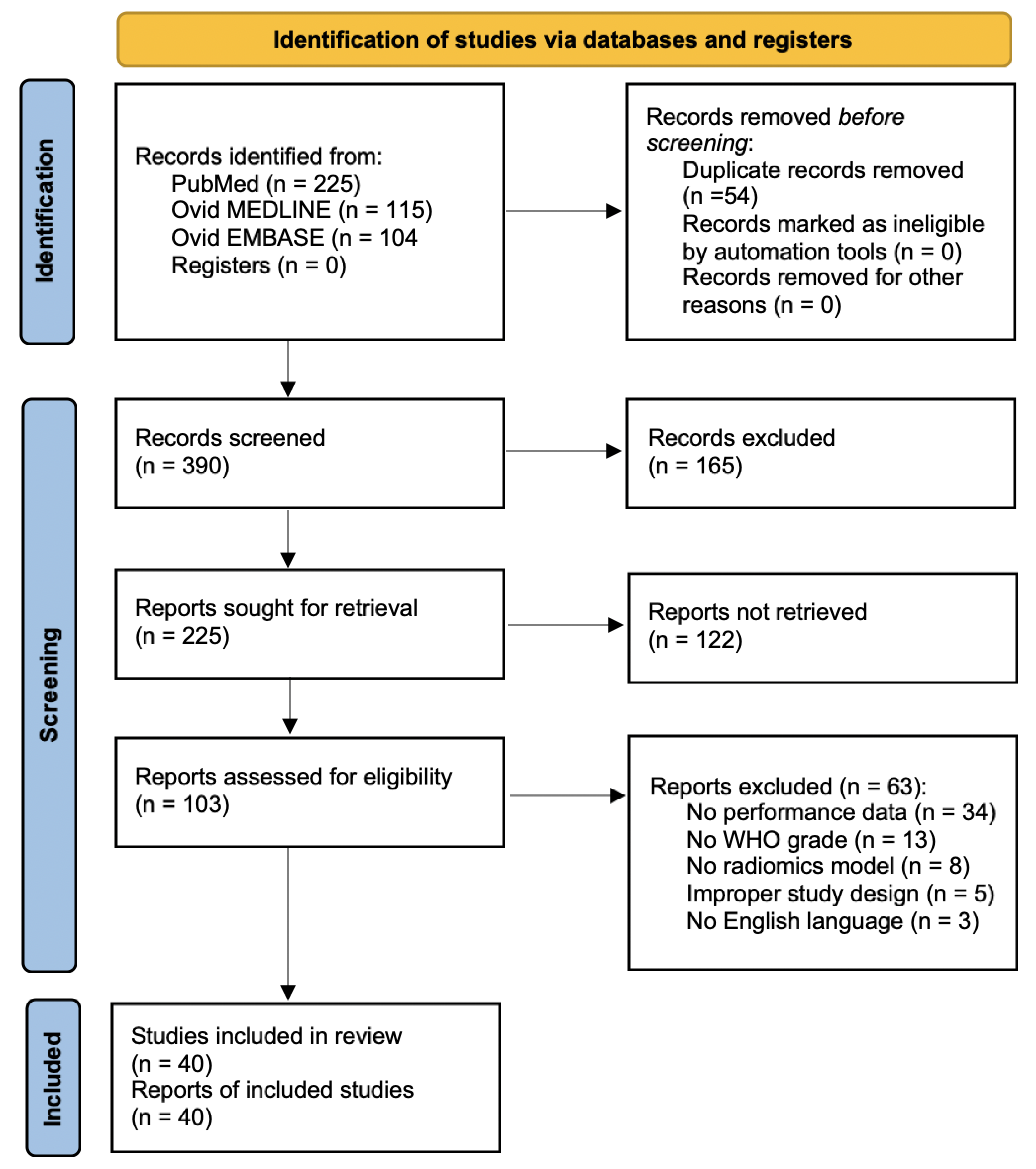 Fig. 1.
Fig. 1.PRISMA flow diagram showing the search process of the literature. Abbreviations: PRISMA, preferred reporting items for systematic reviews and meta-analysis; WHO, World Health Organization.
A total of 7654 patients were included in this study. Most studies were published in 2022 (25%), followed by 2021 (20%), and 2020 (17.5%). The smallest study included 26 patients, while the largest 572. Differentiation in LGG and HGG was reported for 6951 patients (90.8%), of which 4537 had HGGs (65.3%). Among those with HGGs, discrimination between WHO grade 4 and WHO grade 3 was reported for 2742 patients (60.4%), of which 1781 had grade 4 gliomas (65%). Each study included different MRI sequences and the most common was the contrast-enhanced T1W (CE-T1W; 92.5%), followed by T2W (T2W; 77.5%), T2W fluid-attenuated inversion recovery (T2-FLAIR; 70%), and T1W (65%). Other MRI modalities were diffusion-weighted imaging (DWI; 20%), perfusion-weighted imaging (PWI; 15%), and proton magnetic resonance spectroscopy (1H-MRS; 5%).
A number of 31 studies reported on ML (77.5%), 3 articles reported on DL (7.5%), and 6 were hybrid studies (15%). As for the classifiers, LR was the most adopted (55%) followed by SVM (40%), RF (30%), CNN (17.5%), GB (7.5%), and others. Table 1 (Ref. [17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56]) shows a summary of the studies included.
| Author, Journal, Year | Patients (n.) | WHO Grade (n.) | MRI Protocol | Method | Classifiers | |||
| LGG | HGG | |||||||
| 3 | 4 | |||||||
| Chen, Int J Biomed Imaging, 2018 [20] | 274 | 54 | 220 | NA | NA | T1W, CE-T1W, T2W, T2-FLAIR | ML & DL | SVM, CNN |
| Cheng, IEEE J Biomed Health Inform, 2022 [21] | 438 | 119 | 319 | NA | NA | T1W, CE-T1W, T2W, T2-FLAIR | DL | CNN |
| Cheng, IEEE/ACM Trans Comput Biol Bioinform, 2022 [22] | 350 | 92 | 258 | NA | NA | T1W, CE-T1W, T2W, T2-FLAIR | ML | LR, SVM, RF, GB |
| Cho, Annu Int Conf IEEE Eng Med Biol Soc, 2017 [23] | 108 | 54 | 54 | NA | NA | T1W, CE-T1W, T2W, T2-FLAIR | ML | LR |
| Cho, PeerJ, 2018 [19] | 285 | 75 | 210 | NA | NA | T1W, CE-T1W, T2W, T2-FLAIR | ML | LR, SVM, RF |
| Ding, Quant Imaging Med Surg, 2022 [24] | 50 | NA | NA | NA | 25 | CE-T1W | ML & DL | RF, CNN |
| Ditmer, J Neurooncol, 2018 [25] | 94 | 14 | 80 | NA | NA | CE-T1W | ML | LR |
| Gao, Front Oncol, 2020 [26] | 369 | 147 | 222 | 116 | 106 | CE-T1W | ML | LR, SVM, RF |
| Gihr, Front Oncol, 2020 [27] | 26 | 26 | 0 | 0 | 0 | T2W, DWI | ML | LR |
| Guo, Diagn Interv Radiol, 2021 [28] | 152 | 47 | 105 | 39 | 66 | T1W, CE-T1W, T2W, T2-FLAIR | ML | LR |
| Gutta, Am J Neuroradiol, 2021 [29] | 220 | 59 | 161 | 46 | 115 | T1W, CE-T1W, T2W, T2-FLAIR | DL | CNN |
| Hashido, J Comput Assist Tomogr, 2021 [30] | 52 | 18 | 34 | NA | NA | T1W, CE-T1W, T2W, T2-FLAIR, DWI, PWI | ML | LR, SVM, RF |
| Hashido, Sci Rep, 2020 [31] | 46 | 15 | 31 | 4 | 27 | T1W, CE-T1W, T2W, T2-FLAIR, PWI | ML | LR |
| Hu, Comput Biol Med, 2021 [32] | 505 | 233 | 272 | 107 | 165 | T1W, CE-T1W, T2W, T2-FLAIR | ML | SVM |
| Huang, J Comput Assist Tomogr, 2021 [33] | 59 | 13 | 46 | NA | NA | T1W, CE-T1W, T2W, T2-FLAIR | ML | LR |
| Kobayashi, Sci Rep, 2021 [34] | 355 | NA | NA | NA | 259 | T1W, CE-T1W, T2W, T2-FLAIR | ML & DL | LR, CNN |
| Li, Cancers, 2022 [35] | 212 | 105 | 107 | 0 | 107 | T1W, CE-T1W, T2W, T2-FLAIR | ML | SVM |
| Lin, Med Phys, 2022 [36] | 100 | 50 | 50 | NA | NA | T1W, CE-T1W, T2W, DWI, 1H-MRS | ML | LR |
| Liu, Neuroradiology, 2022 [37] | 182 | 63 | 119 | NA | NA | T1W, CE-T1W, T2W, T2-FLAIR, DWI | ML | LR |
| Lu, Clin Cancer Res, 2018 [38] | 214 | NA | NA | NA | 106 | CE-T1W, T2-FLAIR, DWI | ML | SVM |
| Nakamoto, Sci Rep, 2020 [39] | 157 | NA | 157 | 55 | 102 | CE-T1W, T2W | ML | LR, SVM, RF, NB, NN |
| Ning, Ann Transl Med, 2021 [40] | 334 | 112 | 222 | 117 | 105 | CE-T1W, T2-FLAIR | ML & DL | SVM, CNN |
| Park, Korean J Radiol, 2019 [41] | 314 | 213 | 101 | 101 | NA | CE-T1W, T2W, T2-FLAIR | ML | RF, GB, ENR, LDA |
| Reza, J Med Imaging, 2019 [42] | 285 | 75 | 210 | NA | NA | CE-T1W, T2W, T2-FLAIR | ML | SVM, RF, GB |
| Skogen, European Journal of Radiology, 2016 [17] | 95 | 27 | 68 | 34 | 34 | T1W, CE-T1W, T2W, T2-FLAIR | ML | LR |
| Su, Am J Transl Res, 2021 [43] | 139 | 69 | 70 | 36 | 34 | T1W, CE-T1W, T2W, T2-FLAIR, DWI | ML | LR |
| Su, Eur Radiol, 2019 [44] | 217 | 95 | 122 | 61 | 61 | T1W, CE-T1W, T2W, T2-FLAIR, DWI, PWI | ML | LR |
| Sudre, BMC Med Inform Decis Mak, 2020 [45] | 333 | 101 | 232 | 74 | 158 | PWI | ML | RF |
| Takahashi, Int J Radiat Oncol Biol Phys, 2019 [46] | 55 | 14 | 41 | 12 | 29 | T1W, CE-T1W, T2W, T2-FLAIR, DWI | ML | LR, SVM |
| Tian, J Magn Reson Imaging, 2018 [47] | 153 | 42 | 111 | 33 | 78 | T1W, CE-T1W, T2W, DWI, PWI | ML | SVM |
| Vamvakas, Phys Med, 2019 [18] | 40 | 20 | 20 | NA | NA | T1W, CE-T1W, T2W, T2-FLAIR, DWI, PWI, 1H-MRS | ML | SVM |
| van der Voort, Neuro Oncol, 2023 [48] | 238 | 47 | 191 | 59 | 132 | T1W, CE-T1W, T2W, T2-FLAIR | DL | CNN |
| Wang, J Magn Reson Imaging, 2019 [49] | 85 | 34 | 51 | NA | NA | CE-T1W, T2W, DWI | ML | LR |
| Xie, J Magn Reson Imaging, 2018 [50] | 42 | 15 | 27 | 13 | 14 | T1W, CE-T1W, T2W, T2-FLAIR | ML | NA |
| Xu, Front Oncol, 2022 [51] | 572 | 190 | 382 | NA | NA | T1W, CE-T1W, T2W | ML & DL | RF |
| Xu, Quant Imaging Med Surg, 2022 [52] | 129 | 62 | 67 | NA | NA | CE-T1W, T2 FLAIR, DWI | ML | LR, SVM, RF, NB, NN |
| Zhang, J Digit Imaging, 2020 [53] | 108 | 43 | 65 | NA | NA | DWI | ML & DL | SVM |
| Zhao, BMC Neurol, 2020 [54] | 69 | 36 | 33 | 33 | NA | CE-T1W, T2-FLAIR | ML | RF |
| Zhou, Int J Clin Pract, 2022 [55] | 114 | 35 | 79 | 21 | 58 | T1W, CE-T1W, T2W | ML | LR |
| Zhou, Neuro Oncol, 2017 [56] | 84 | NA | NA | NA | 0 | T1W, CE-T1W, T2W, T2-FLAIR | ML | LR |
Abbreviations: NA, not available; WHO, world health organization; LGG, low-grade glioma; HGG, high-grade glioma; MRI, magnetic resonance imaging; T1W, T1-weighted; CE-T1W, contrast-enhanced T1W; T2W, T2-weighted; T2-FLAIR, T2W-fluid-attenuated inversion recover; DWI, diffusion-weighted imaging; PWI, perfusion-weighted imaging; 1H-MRS, proton magnetic resonance spectroscopy; ML, machine learning; DL, deep learning; SVM, support vector machine; CNN, convolutional neural network; LR, logistic regression; RF, random forest; GB, gradient boost; NB, naïve bayes; NN, nearest neighbors; ENR, elastic net regression; LDA, linear discriminant analysis.
The performance of radiomics for LGGs vs. HGGs and WHO grade 3 vs. 4 categorizations was reported for a total of 3290 patients and 704 patients, respectively. Overall SPE and SEN for differentiation between LGGs and HGGs were 91% (95% CI = 0.86–0.94) and 84% (95% CI = 0.78–0.89), respectively. With regards to the discrimination task between WHO grade 4 and WHO grade 3, the overall SEN was 89% (95% CI = 0.82–0.94) and the overall SPE was 81% (95% CI = 0.66–0.91), respectively. Fig. 2 shows the univariate forest plots of the analysis for discrimination between LGGs vs. HGGs and grade 3 vs. grade 4 gliomas. Fig. 3 provides the respective SROC curves for the different binary categorization tasks considered in our analysis. In the canonical receiver operating characteristic (ROC) curve, each data point belongs to a single study in which several different diagnostic thresholds are employed to categorize between two classes of interest (e.g., cases and non-cases). In a single study, changing the threshold results indeed in the plot of the true positive rate (TPR) against the false positive rate (FPR) at each threshold setting. Conversely, in a meta-analysis, the units of analysis are separate and single studies. Hence, the SROC curve aims to represent the relationship between TPR and FPR across studies, recognizing they may have used diverse thresholds. As above mentioned, to fit such curves, we put into effect the R package mada, which, besides the SROC, provides two further figures of merit: (i) the confidence region of the summary estimate, which provides a measure of the ROC-based performance heterogeneity among the included studies; (ii) the prediction region for a new hypothetical study, which is the zone in the false positive rate vs. sensitivity space, in which it is likely to expect a new study.
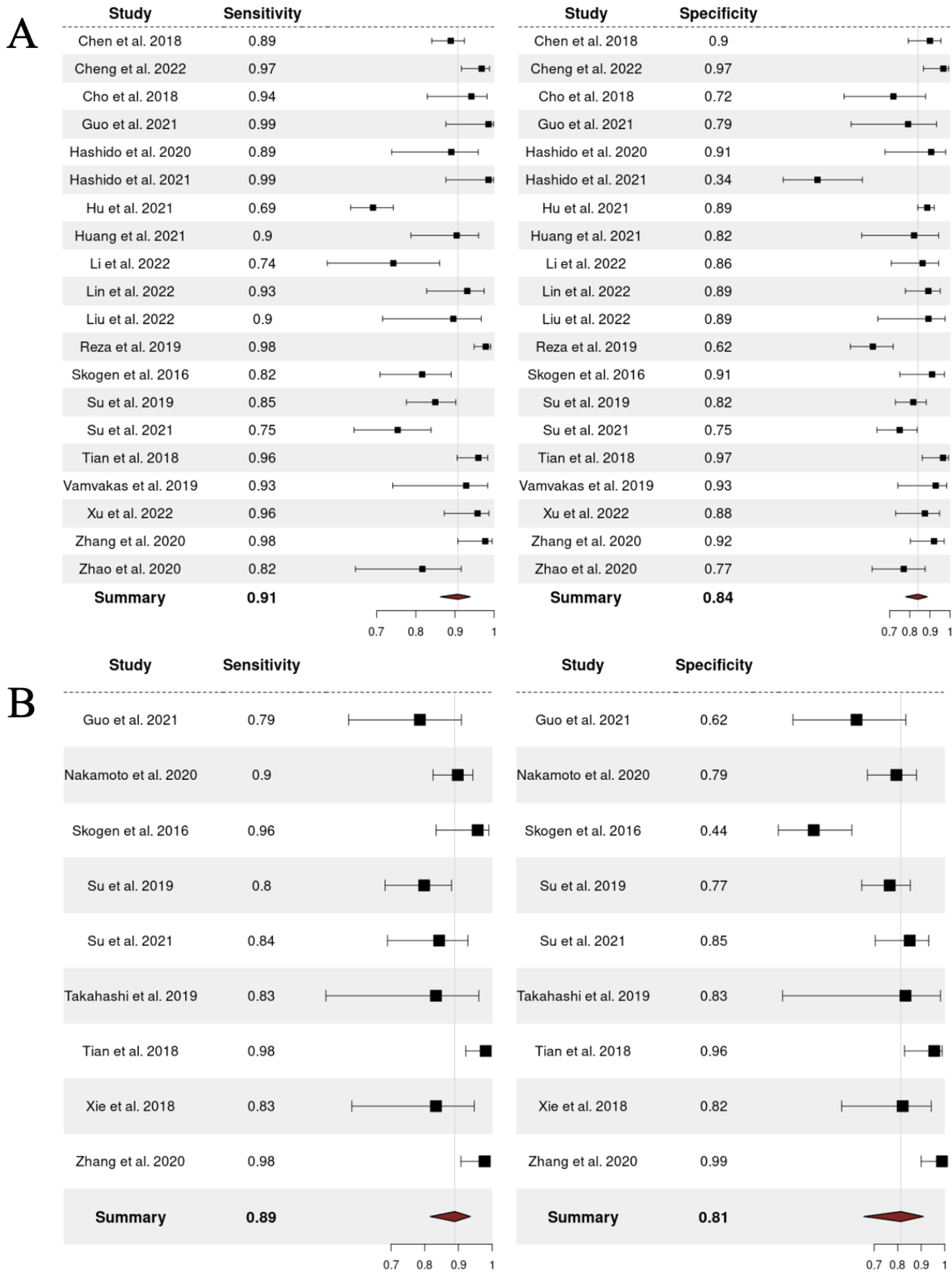 Fig. 2.
Fig. 2.Forest plots for discrimination between LGGs vs. HGGs (A) and grade 3 vs. grade 4 gliomas (B). The red diamonds represent the overall results.
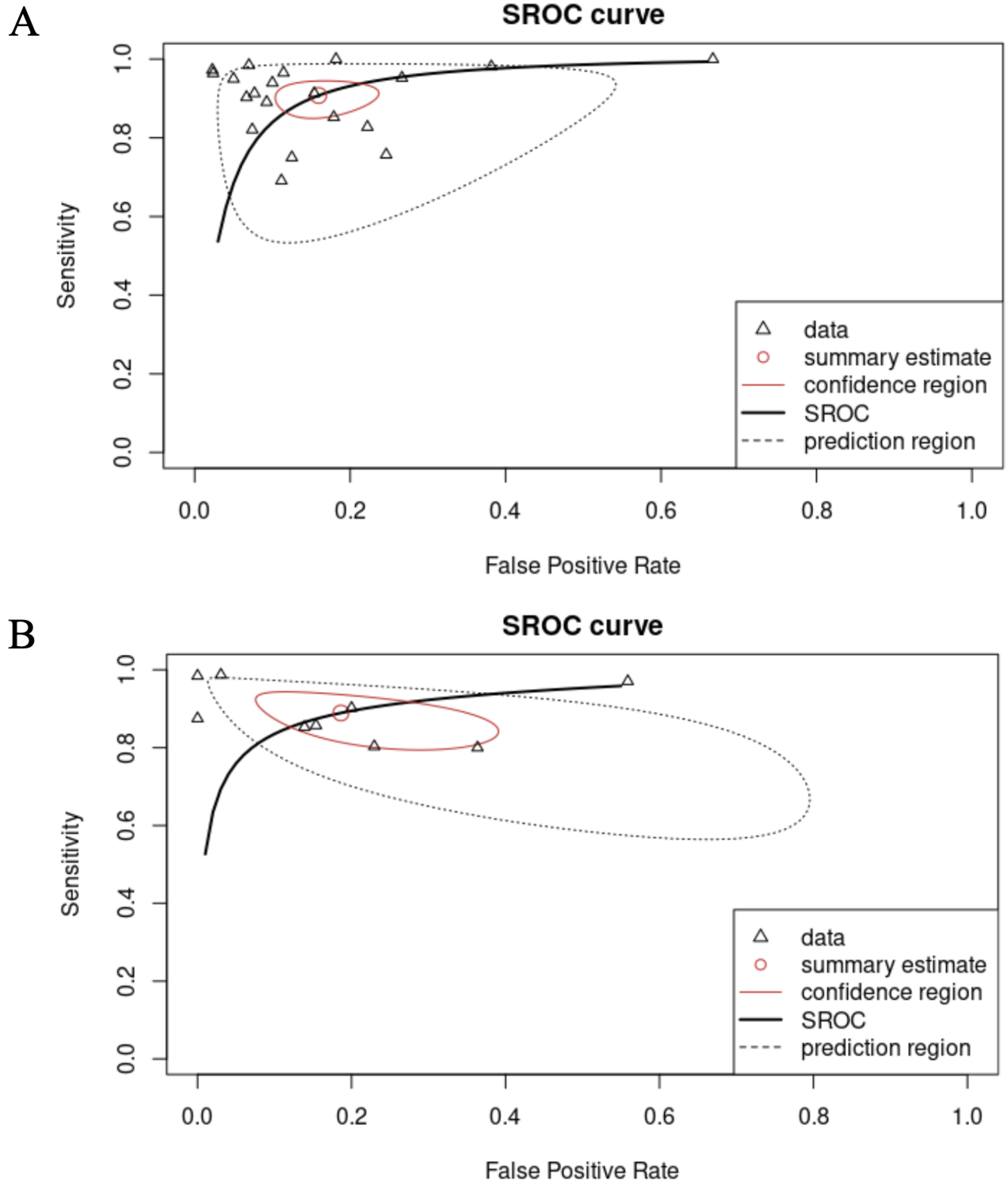 Fig. 3.
Fig. 3.SROC curves for the differentiation between LGGs vs. HGGs (A) and grade 3 vs. grade 4 gliomas (B). Each curve provides: (i) the summary estimate of the considered study (see the red circle); (ii) the confidence region of the summary estimate (see the red ellipse-like trace), which furnishes a measure of the performance heterogeneity among the included studies; (iii) the prediction region for a new hypothetical study, where it is likely to expect a new study (see the dashed line). Abbreviations: SROC, summary receiver operating characteristic.
Our subgroup analysis did not evidence variables to significantly impact the performance of the models. Nonetheless, when looking at the most performing classifiers, we observed a better trend for non-linear algorithms such as SVM and CNN, as shown in Fig. 4.
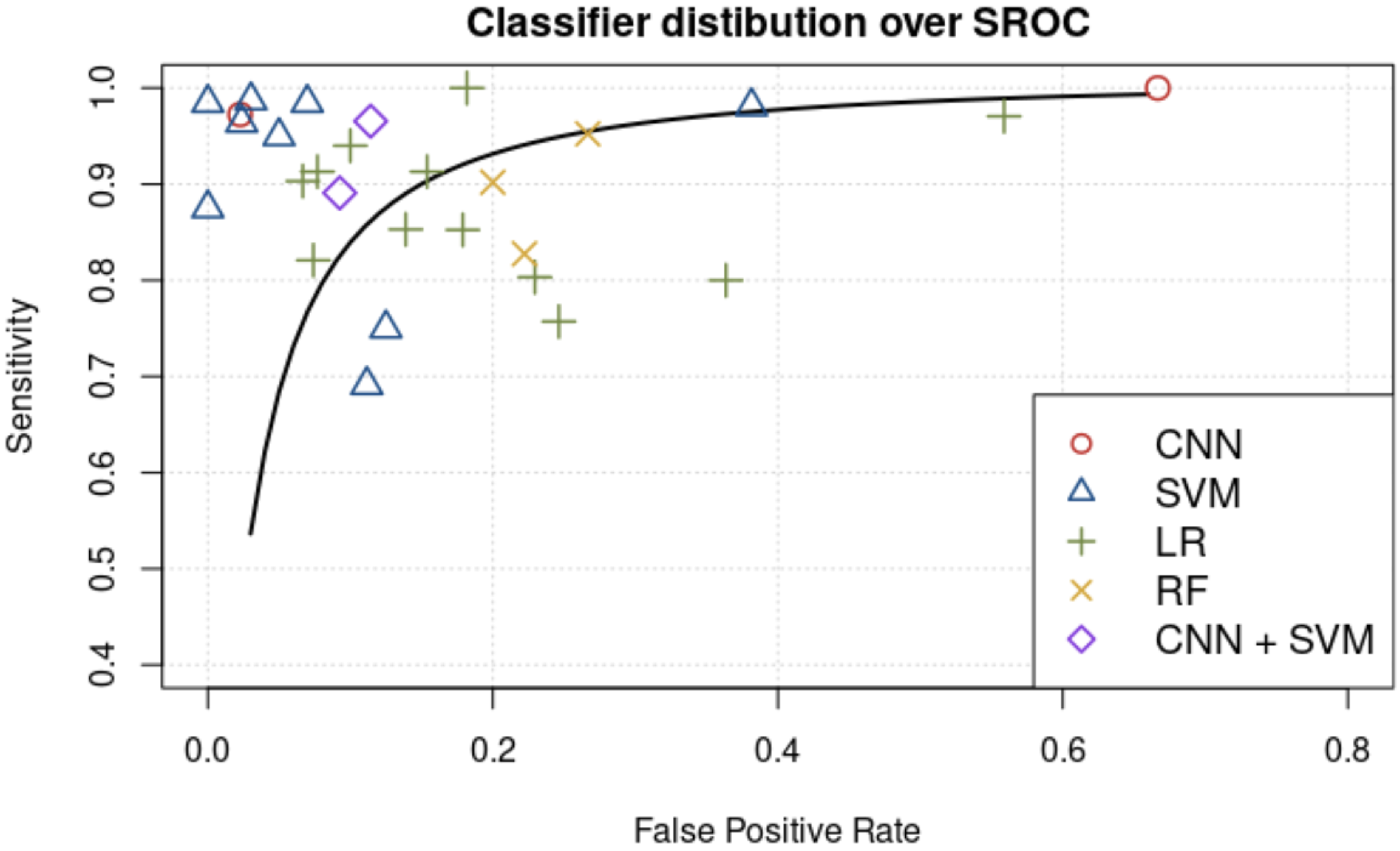 Fig. 4.
Fig. 4.SROC curve for the two classification tasks considered together. From this graph, we can assess that the non-linear classifiers tend to perform better with respect to simpler solutions such as logistic regression classifiers. This can be evinced from the position of the non-linear classifiers (CNN, SVM, RF, and CNN + SVM) which are more towards the top-left corner of the graph, with respect to the linear (LR).
We furthermore investigated the following variables, which may significantly impact the model performance: (i) the selected features, which are the final data representation, in a numerical form, fed to the AI algorithm to perform the given classification task; (ii) the MRI modalities from which the input images are obtained. Regarding features, the most used were the textural (28.6%), followed by the deep (20.4%) and voxel intensities features (12.2%). Fig. 5A provides a visual representation of the features distribution among the studies included in our review. In this figure, each square corresponds to a single radiomic model. Note that in a single study, more than one radiomic model may be described, each one tailoring a different and specific classification task. Concerning the employed MRI modality, the CE-T1W was the most reported MRI sequence (36.5%), followed by T2-FLAIR (15.6%), and DWI (14.6%). Fig. 5B shows the distribution of the MRI modalities among the included studies. A majority of studies (58.1%) used a cross-validated analysis.
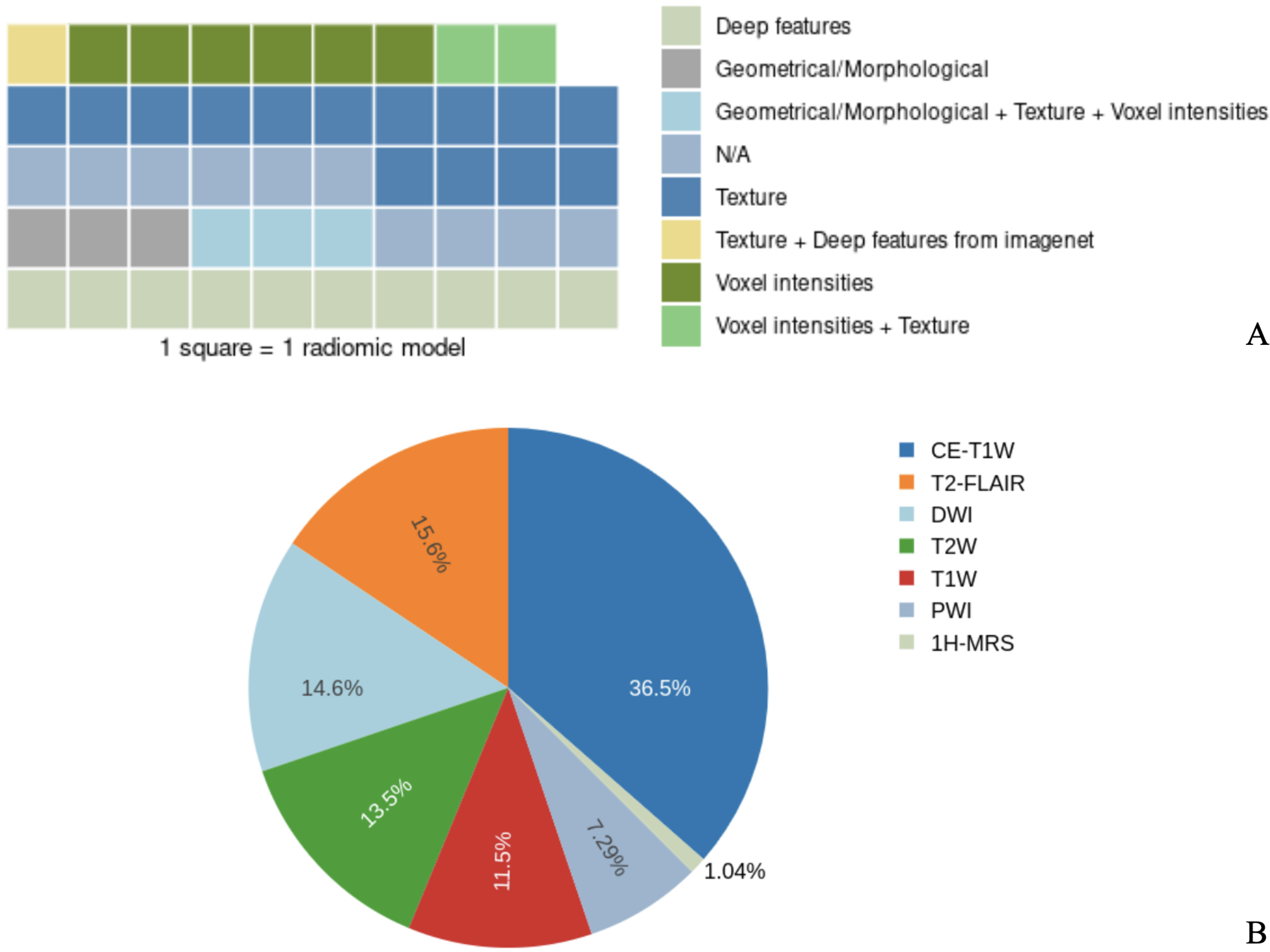 Fig. 5.
Fig. 5.Radiomics features. (A) A visual representation of the radiomics features distribution among the studies included in our review. Each square corresponds to the features fed as input to a single radiomic model. Note that in a single study, more than one radiomic model may be described, each one tailoring a different and specific classification task. A final remark must be made concerning the so-called “deep features from Imagenet”. This is indeed a widely used data representation in the context of deep learning, as detailed in the corresponding studies. (B) Pie chart showing the distribution of the MRI modalities among the included studies. Note that a single radiomic model may be fed with data obtained from different MRI modalities.
The
We provided an overview of the performance of the current radiomics models for glioma grading prediction. The overall performance resulted higher for the HGGs vs. LGGs discrimination task than the WHO grade 3 vs. 4 task, both in terms of SPE and SEN. The FPR was higher than the false negative rate (FNR) for both differentiation tasks indicating a greater capability to rule out HGGs or WHO grade 4 gliomas rather than identifying these entities. The studied variables did not impact significantly the performance, but we observed a better trend for non-linear classifiers such as SVM and CNN.
As shown in Fig. 6, there has been an outstanding growth of expertise in AI developments and laboratories, which has brought on a significantly higher number of studies published over the years.
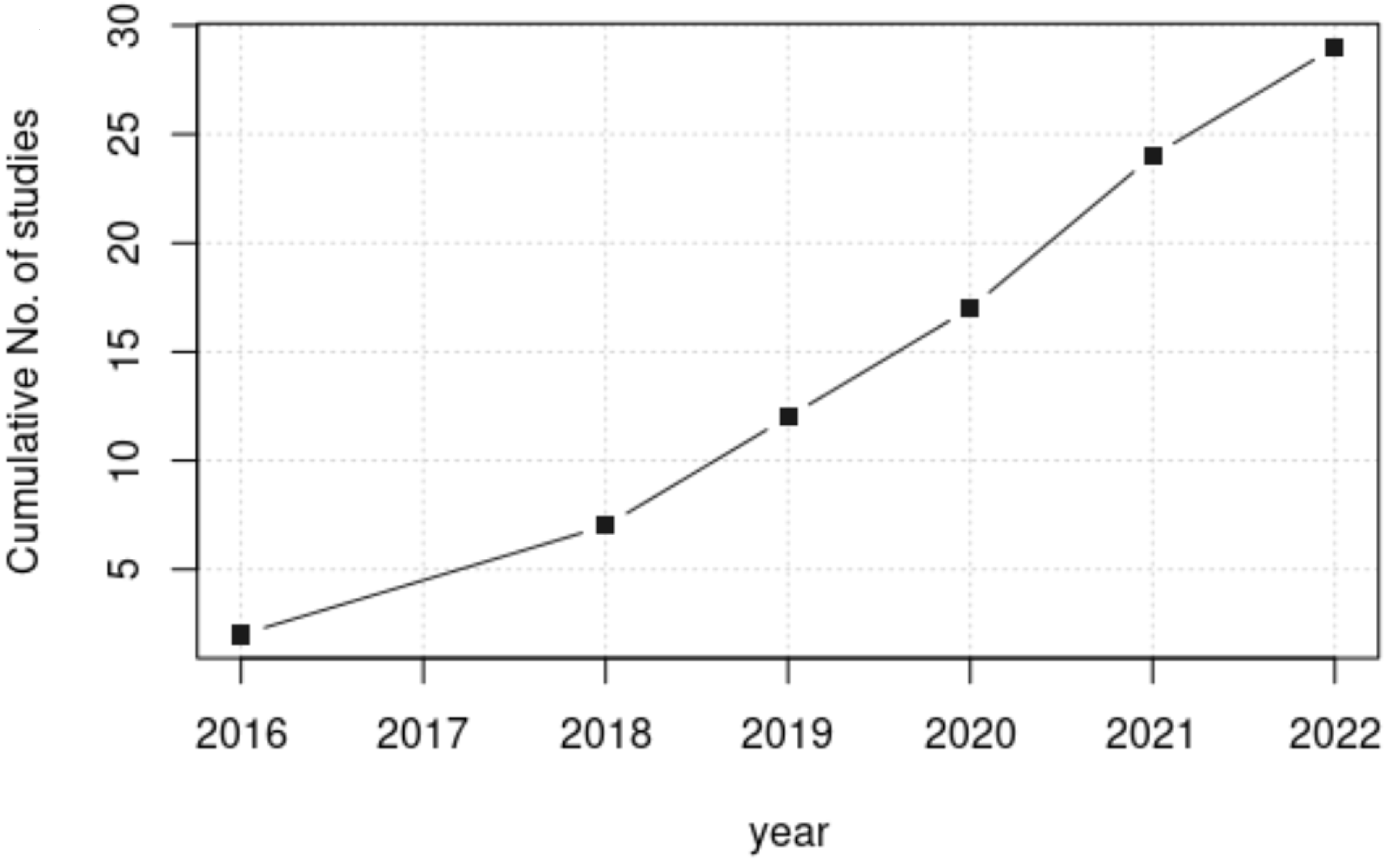 Fig. 6.
Fig. 6.Cumulative number of radiomics studies included in our study over years.
Radiomics has captured the interest of neuroradiologists and neuro-oncologists, who progressively upheld the research field [12, 57]. On the one hand, novel strategies for feature extraction and segmentation methods have been developed that reverberated the impact of radiomics on many fields of neuro-oncology and neuroradiology, multiplying the possible clinical applications [4, 7, 9, 11]. On the other hand, the concept of DL, as a method of data representational learning able to learn from end to end by itself without requiring any handcrafted features or human-based data representation, originated as an innovative branch of radiomics [58]. Even though a minority of the articles focused on DL, many hybrid studies were included that imbricate the ML classifiers with novel DL algorithms, accomplishing fully automatic detection processes. All studies included in our meta-analysis extracted features (handcrafted or DL-based) from MRI sequences, but other imaging modalities have been used for different purposes, particularly positron emission tomography in radiogenomics [59, 60].
Our data confirm the paramount role of radiomics in predicting the grade of cerebral gliomas, as suggested by previous authors [61]. The overall performance reflects the great predictivity of the current models included in the meta-analysis.
We found a higher prediction capability of radiomics for the HGGs vs. LGGs differentiation task than the WHO grade 3 vs. grade 4 task. In accordance with the current literature, the main prognostic impact of glioma grading is provided by the LGGs vs. HGGs differentiation [62, 63, 64]. Even though the definitive diagnosis of gliomas is histopathological, these data strongly support the potential role of radiomics for an initial diagnostic orientation in the definition of grade glioma. This may be particularly important for those patients with doubtful neuroradiological imaging where there is no clear orientation toward a diagnosis of LGG vs. HGG [65, 66, 67, 68].
When looking at the SROC curves, we observed a smaller and better-performing prediction region in the LGGs vs. HGGs differentiation task, while the prediction region of grade 3 vs. grade 4 resulted to be larger and more oriented toward superior FPRs. This suggests that although we have not witnessed any advancements in performance thus far, we have reason to believe that we may observe progress in the upcoming years in distinguishing between LGGs and HGGs. This is shown by the SROC curve, which illustrates the potential for future improvement.
Given the blossoming of AI models over the last decade, we expected the year of publication to significantly impact the performance of the models. Yet, we did not find a linear correlation between time of development and performance.
Textural features, along with deep and voxel intensities features were the most performing. Feature extraction is driven by algorithms to select ones appropriate for a precise task [9]. For glioma grading, textural features were reported as the best-performing. These common features, quantify the spatial variation of grey-level intensity inferring image heterogeneity [17, 69]. Frequent textural features are energy, entropy, inertia, correlation, and others [47, 50]. Zhou et al. [56] identified gray-level co-occurrence matrix (GLCM)-homogeneity as the main texture feature to predict histological grade from CE-T1W images. Likewise, Liu et al. [37] found two GLCM texture features to reflect the glioma grade. The multiparametric texture analysis of Hashido et al. [31] also included the GLCM features. Their study found that GLCM-based entropy effectively differentiated between LGGs and HGGs in PWI. Skogen et al. [17] used MRI texture analysis (MRTA) to assess tumor heterogeneity. Extraction of texture features at fine anatomical scales best discriminated LGG and HGG.
The most reported best-performing MRI sequences were the CE-T1W, T2-FLAIR, and DWI. The CE-T1W and T2-FLAIR modalities were particularly regarded for the HGGs vs. LGGs differentiation task, while the DWI has mostly been reported as the best-performing sequence for the WHO grade 3 vs. grade 4 discrimination task. Apparent diffusion coefficient (ADC) maps calculated from DWI, along with dynamic contrast-enhanced (DCE), dynamic susceptibility contrast (DSC), and arterial spin labeling (ASL) PWI have been increasingly reported as promising alternatives and will perhaps be the sequences most chosen for feature extraction over the next years [27, 30, 43, 45].
Future models should not disregard external validation to ascertain a sufficient level of reproducibility and reliability. The lack of standardization and the customizability of radiomics reduce the applicability of AI in daily practice. Clear routes of development should be delineated to overcome the diversities in radiomics laboratories. Hopefully, this meta-analysis will orient forthcoming models to more defined and shared processes that will be possibly implemented in clinical practice.
Gliomas, a collection of primary brain tumors, result from the abnormal proliferation of glial cells, including ependymal cells, oligodendrocytes, and astrocytes. These tumors display distinct cellular characteristics based on their histological subtype. For example, astrocytomas, which make up a significant portion of gliomas, primarily consist of proliferating astrocytes. Oligodendrogliomas, on the other hand, stem from oligodendrocyte precursor cells and are identified by their “fried-egg” appearance, characterized by round nuclei and clear cytoplasm. Ependymomas, the third major subtype, develop from ependymal cells that line the brain and spinal cord ventricles. Each glioma subtype exhibits unique cellular features crucial for precise diagnosis and classification.
The classification and comprehension of gliomas heavily rely on their cellular characteristics. However, the 2016 WHO classification ushered in a transformative era of understanding these tumors through their molecular traits, carrying substantial implications for diagnosis, prognosis, and therapy selection [70]. A pivotal molecular anomaly identified in gliomas is the isocitrate dehydrogenase (IDH) mutation, especially the IDH1 and IDH2 mutations, which manifest in a subset of gliomas. These mutations are linked to distinct clinical and histological attributes and wield a critical role in glioma classification and management. Another crucial molecular alteration in glioma involves the loss of the tumor suppressor gene TP53, contributing to the pathogenesis of high-grade gliomas. Furthermore, the methylation status of the O-6-methylguanine-DNA methyltransferase (MGMT) gene promoter stands as a noteworthy molecular marker that impacts the response to alkylating chemotherapy agents. Molecular profiling has empowered the refinement of glioma categorization into more precise groups, providing guidance for treatment decisions and enhancing prognostic accuracy [71, 72, 73].
The 2021 WHO classification of gliomas brought significant innovations to the molecular classification of these brain tumors, offering a more detailed and comprehensive approach. It emphasizes the importance of integrating both histological and molecular data to establish an integrated diagnosis, leading to a more precise glioma classification. These molecular markers play a pivotal role in refining glioma classification, offering valuable insights for prognosis, treatment planning, and personalized therapeutic strategies. The inclusion of telomerase reverse transcriptase (TERT) promoter mutation, H3K27M mutation, v-raf murine sarcoma viral oncogene homolog B1 (BRAF)-fusion mutation, and the increased focus on O-6-methylguanine-DNA methyltransferase (MGMT) promoter methylation reflects the evolving understanding of glioma biology and the growing need for more accurate diagnosis and management [2].
In this context, radiomics can assume a central role in unveiling the cellular and molecular patterns of gliomas. Utilizing advanced radiomic analyses, which are based on quantitative characteristics derived from medical images, offers a non-invasive approach to assessing the tumor’s heterogeneity, microenvironment, and genetic attributes. For instance, research such as Kickingereder et al. [74] demonstrates that radiomic features can capture variations in cell density, vascularity, and necrosis within gliomas, effectively reflecting their histological and cellular diversity. Furthermore, radiomics can aid in the identification of crucial genetic and molecular markers, like IDH mutations and 1p/19q co-deletion [75]. Additionally, radiomic features may also mirror the molecular diversity of gliomas, contributing to more precise diagnoses, treatment planning, and patient stratification, as highlighted by Lambin et al. [6]. However, it is worth noting that at present, there is still a scarcity of radiomic studies that indicate predictive features for the molecular sub-classifications proposed in the WHO 2021 classification.
The meta-analysis limiting in nature since it was mostly based on retrospective cohort studies, even if the number of patients was considerable. Due to the limited data available, we could not ascertain the performance of radiomics in other discrimination tasks, such as WHO grade 1 vs. grade 2, and grade 3 vs. grade 4. Given the bivariate model of the meta-analysis, we did not calculate the overall accuracy for the differentiation tasks. Nonetheless, to the best of our knowledge, this is the first meta-analysis to picture the current performance of radiomics for glioma grading, providing cutting-edge conclusions to pilot future models.
Moreover, a limitation possibly affecting the generalizability of radiomics studies for classification tasks consists of the data drift phenomenon. Data drift in radiomics can occur due to changes over time in the classification used in radiomics analysis, in this specific case represented by the CNS WHO Classification of Tumors [1]. This can lead to a loss of accuracy and reliability in radiomics models trained on the old classification criteria, which may no longer apply to the new classification [76]. It is worth noting that when it comes to radiomics models, the classification criteria used can also have an impact on their effectiveness. A recent study by Moodi et al. [77] found that ML algorithms delivered superior results in grading gliomas based on WHO 2021 criteria, as compared to the WHO 2016 classification criteria. To mitigate this type of data drift, it is important to update radiomics models and retrain them on the new classification criteria. It is also important to carefully track any changes in classification criteria and ensure that they are well documented so that radiomics analysis can be properly adjusted and validated accordingly [78].
This meta-analysis suggests that the current radiomics models perform better in distinguishing between LGGs and HGGs than between WHO grade 3 and WHO grade 4 gliomas, in terms of both SPE and SEN. Enhanced future models with increased accuracy can prove to be of clinical use for categorizing HGGs and LGGs.
MRI is the most preferred imaging method, and the CE-T1W sequence is found to be the most effective for current radiomics models. Textural features are commonly used, and modern non-linear classifiers show a promising trend.
1H-MRS, proton magnetic resonance spectroscopy; ADC, apparent diffusion coefficient; AI, artificial intelligence; ASL, arterial spin labeling; CE-T1W, contrast-enhanced T1W; CIs, confidence intervals; CNN, convolutional neural network; CNS, central nervous system; DCE, dynamic contrast-enhanced; DL, deep learning; DSC, dynamic susceptibility contrast; DWI, diffusion-weighted imaging; ENR, elastic net regression; FN, false negative; FNR, false negative rate; FP, false positive; FPR, false positive rate; GAN, generative adversarial network; GB, gradient boost; GLCM, gray-level co-occurrence matrix; HGG, high-grade glioma; IDH, isocitrate dehydrogenase; LDA, linear discriminant analysis; LGG, low-grade glioma; LR, logistic regression; MGMT, O-6-methylguanine-DNA methyltransferase; ML, machine learning; MLP, multilayer perceptron; MRTA, MRI texture analysis; NB, naïve bayes; NN, nearest neighbors; NOS, Newcastle-Ottawa scale; PRISMA, preferred reporting items for systematic reviews and meta-analysis; PWI, perfusion-weighted imaging; RF, random forest; SE N, sensitivity; SPE, specificity; SROC, summary receiver operating characteristic; SVM, support vector machine; T2-FLAIR, T2W-fluid-attenuated inversion recover; TN, true negative; TP, true positive; WHO, world health organization.
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conceptualization, EA, MZ, LDM, MMF and PPP; methodology, EA, MZ, LDM, FP, HC, KS, IT, MG, LU and WB; validation, LDM, EA, FP, LU, MG and MMF; formal analysis, LDM, EA, PPP and FP; investigation, LDM, EA, PPP and FP; resources, EA, MZ, LDM, FP, HC, KS, IT, MG, PPP and LU; data curation, LDM, EA, PPP and FP; writing—original draft preparation, LDM, EA, PPP and FP; writing—review and editing, EA, LDM, FP, MG, MMF, WB and LU; visualization, EA, LDM, FP, MG, MMF, WB, TI, MZ, PPP and LU; supervision, EA, LDM, PPP, MMF and WB; project administration, LDM, EA, PPP and MMF. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Not applicable.
Not applicable.
This research received no external funding.
The authors declare no conflict of interest.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
