- Academic Editor
A prevalent condition during pregnancy, gestational diabetes mellitus (GDM) affects a significant proportion of pregnancies worldwide and poses substantial risks to maternal as well as fetal health. Polymorphisms in the glucokinase (GCK) and glucokinase regulatory protein (GCKR) genes, which are crucial for glucose homeostasis, may modulate susceptibility to GDM. Hence, this meta-analysis aimed to assess the relationship between GDM and polymorphisms in GCK (rs1799884, rs4607517) and GCKR (rs780094, rs1260326).
In this systematic review, we retrieved data from PubMed, EMBASE, Medline, EBSCO, Cochrane Library, and Chinese National Knowledge Infrastructure (CNKI) databases. Studies were critically appraised using the Newcastle-Ottawa Scale, and meta-analyses were performed using STATA 12.0. The odds ratios (ORs) were calculated with 95% confidence intervals (CIs) and heterogeneity was assessed with Cochran’s Q test as well as I2 statistical tests, respectively. Moreover, Begg’s test helped in evaluating publication bias.
We included 20 studies, comprising 9745 GDM women and 15,830 controls. All genetic models showed a strong correlation between the GCK rs1799884 polymorphism and GDM, with carriers of the A allele exhibiting an increased risk. Conversely, GCK rs4607517, GCKR rs780094, and rs1260326 were not significantly associated. However, heterogeneity was influenced by ethnicity and diagnostic criteria.
The GCK rs1799884 polymorphism can be a potential predictive marker because it is significantly associated with an increased risk of GDM.
A significant public health concern, gestational diabetes mellitus (GDM) affects approximately 7%–18% of pregnancies worldwide [1, 2, 3]. GDM is characterized by the development of pregnancy-related glucose intolerance [4] and is substantially risky for maternal and fetal health. GDM increases the risk of cesarean delivery, hypertensive disorders, and long-term metabolic complications for both the mother and child [5, 6, 7]. Increased obesity and sedentary lifestyles exacerbated the incidence of GDM [8, 9], necessitating a comprehensive understanding of its pathophysiology and genetic underpinnings to enhance prevention and treatment strategies. The glucokinase (GCK) and glucokinase regulatory protein (GCKR) genes, essential for glucose metabolism and homeostasis, are linked to GDM [10, 11]. Primarily expressed in pancreatic beta cells, GCK acts as a glucose sensor that regulates insulin secretion [12, 13]. Conversely, GCKR modulates GCK activity, thereby influencing glucose metabolism and insulin sensitivity [14, 15, 16]. Thus, variations in these genes can disrupt glucose regulation and lead to GDM [17, 18].
Several studies have implicated GCK and GCKR gene
polymorphisms in GDM susceptibility [19, 20]. These genetic variants may affect
glucose metabolism and insulin response. She et al. [19] suggested that
the GCK gene rs1799884 (–30G
This meta-analysis aimed to systematically evaluate the association between GCK (rs1799884 and rs4607517) and GCKR (rs780094 and rs1260326) polymorphisms as well as GDM susceptibility. By integrating data from multiple studies, we sought to clarify the genetic components of GDM and provide relavant insights for future research and clinical practice.
We used databases like PubMed (https://pubmed.ncbi.nlm.nih.gov/), EMBASE (https://www.embase.com/), Medline (https://www.nlm.nih.gov/medline/medline_home.html), EBSCO (http://search.ebscohost.com/), Cochrane Library (https://www.cochranelibrary.com/), and the Chinese National Knowledge Infrastructure (CNKI) (https://www.cnki.net/) for literature search. Keywords like “glucokinase” or “GCK”, “glucokinase regulatory protein” or “GCKR”, “gestational diabetes mellitus” or “gestational diabetes” or “GDM”, along with terms for genetic variations such as “polymorphisms”, “mutations”, or “variants”, as well as “risk” and “susceptibility” were used. Reference lists from identified articles were screened to ensure a comprehensive collection of relevant studies.
Our inclusion criteria were: (1) case-control studies comprising both a GDM group and a control group of pregnant women without GDM, and (2) those with adequate data on the genotypes of the GCK gene variants rs1799884 and rs4607517, as well as the GCKR gene variants rs780094 and rs1260326. Our exclusion criteria were: (1) studies lacking publication details; (2) case reports, reviews, abstracts, or meta-analyses; (3) those not on GCK or GCKR polymorphisms or GDM susceptibility; (4) research without case-control design; (5) those with incomplete odds ratio (OR) calculation data, and (6) those with genotype distributions that deviated from the control group’s Hardy-Weinberg equilibrium (HWE).
Using eligible publications, we meticulously extracted data like the first author’s name, publication year, the participants’ ethnicity, the genotype methodology, sample size, gestational age, and the number of genotypes as well as alleles in both study and control groups, respectively.
Each study was rigorously assessed using the Newcastle-Ottawa Scale (NOS) score
[21]. Our analysis only included studies that met the NOS score of
Our literature search yielded 368 articles from Embase, Medline, EBSCO, PubMed,
CNKI, and Wanfang databases. After removing 53 duplicates, we excluded 69
articles that did not fit our inclusion criteria, like being non-human studies,
lacking full texts, or being meta-analyses. Additionally, 203 articles were
disqualified because they were irrelevant to GDM, did not employ a case-control design, or did not investigate the specified GCK or GCKR
polymorphisms. This resulted in 43 full-text articles for further analysis. Our
final analysis included 20 articles [19, 20, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43] comprising 9745 GDM patients
and 15,830 healthy controls after excluding those without investigations on
polymorphisms of interest (rs1799884, rs4607517, rs780094, or rs1260326) or
lacked sufficient data (Fig. 1; Table 1, Ref. [19, 20, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43]). All included
trials had NOS scores
 Fig. 1.
Fig. 1.
Flow diagram for eligible article selection. CNKI, China National Knowledge Infrastructure; GDM, gestational diabetes mellitus; GCK, glucokinase; GCKR, glucokinase regulatory protein.
| Author | Year | Country | Ethnicity | Diagnosis | GDM group | Control group | p HWE | Genotype method | SNPs | |||||
| Procedure | Criteria | Size | Gestational age | Age | Size | Gestational age | Age | |||||||
| (mean |
(mean | |||||||||||||
| Chiu et al. [26] | 1994 | USA | African | OGTT | WHO | 174 | / | 28.2 |
99 | / | 22.1 |
0.286 | PCR-SSCP | GCK rs1799884 |
| Zaidi et al. [30] | 1997 | UK | Caucasian | 75 g OGTT | WHO | 92 | 28–32 | 31 |
45 | 28–32 | / | 0.414 | PCR-SSCP | GCK rs1799884 |
| Shaat et al. [29] | 2006 | Sweden | Caucasian | 75 g OGTT | DPSG-EASD | 642 | 27–28 | 32.3 |
1229 | 27–28 | 30.5 |
0.504 | PCR-RFLP | GCK rs1799884 |
| Freathy et al. [27] | 2010 | UK, Australia, Thailand | Caucasian, Asian | 75 g OGTT | IADPSG | 998 | 24–32 | / | 5587 | 24–32 | / | 0.91 | Illumina | GCK rs1799884 |
| Santos et al. [28] | 2010 | Brazil | Caucasian | 75 g OGTT | ADA | 150 | 24–28, 32–36 | 31.9 |
600 | 24–28, 32–36 | 25.2 |
0.371 | PCR-RFLP | GCK rs1799884 |
| Li W [31] | 2011 | China | Asian | 100 g OGTT | ADA | 668 | 28.2 |
32.5 |
758 | 27.7 |
31.2 |
0.558 | TaqMan | GCK rs1799884, GCKR rs780094 |
| Han et al. [34] | 2015 | China | Asian | 75 g OGTT | / | 948 | 2–32 | / | 975 | / | / | 0.985 | PCR-based invader assay | GCK rs1799884 |
| Tarnowski et al. [36] | 2017 | Poland | Caucasian | 75 g OGTT | IADPSG | 207 | 24–28 | 31.7 |
204 | / | 29.2 |
0.875 | TaqMan | GCK rs1799884, GCKR rs780094 |
| Zhou [43] | 2020 | China | Asian | 75 g OGTT | National | 835 | 24–28 | 30.97 |
870 | / | 28.84 |
0.246 | MassARRAY | GCK rs1799884 and rs4607517, GCKR rs780094 and rs1260326 |
| Popova et al. [41] | 2021 | Russia | Caucasian | 75 g OGTT | IADPSG | 688 | 24–28 | 31.9 |
454 | 24–28 | 29.5 |
0.141 | TaqMan | GCK rs1799884 |
| She et al. [19] | 2022 | China | Asian | 75 g OGTT | IADPSG | 835 | 24–28 | 30.97 |
870 | / | 28.84 |
0.246 | MassARRAY | GCK rs1799884 and rs4607517, GCKR rs780094 and rs1260326 |
| Wang et al. [32] | 2011 | China | Asian | 100 g OGTT | ADA | 1701 | / | 30 (30, 35) | 1023 | / | 32 (28, 33) | 0.804 | Taqman | GCK rs4607517 |
| Stuebe et al. [33] | 2014 | USA | Caucasian, African–American | 100 g OGTT | / | 80 | 24–29 | 28.3 |
1138 | 24–29 | / | 0.324 | Sequenom iPLEX | GCK rs4607517, GCKR rs780094 and rs1260326 |
| Ao et al. [40] | 2021 | China | Asian | 75 g OGTT | National | 562 | / | 30.18 |
452 | / | 29.50 |
0.28 | MassARRAY | GCK rs4607517 |
| Anghebem-Oliveira et al. [35] | 2017 | Brazil | Caucasian | / | / | 127 | / | 31.9 |
125 | / | 30.6 |
0.094 | RT-PCR | GCKR rs780094 |
| Jamalpour et al. [37] | 2018 | Malaysia | Asian | OGTT | / | 186 | / | / | 588 | / | 29.9 |
0.512 | Sequenom iPLEX | GCKR rs780094 |
| Li et al. [42] | 2018 | China | Asian | 75 g OGTT | ADA | 127 | 24–32 | 31.9 |
125 | / | 30.6 |
0.094 | RT-PCR | GCKR rs780094 |
| Franzago et al. [38] | 2017 | Italy | Caucasian | 75 g OGTT | IADPSG | 102 | 24–28 | 34.6 |
66 | 24–28 | 31.9 |
0.46 | High resolution melting | GCKR rs1260326 |
| Franzago et al. [39] | 2018 | Italy | Caucasian | 75 g OGTT | IADPSG | 104 | 24–28 | 34.6 |
124 | 24–28 | 32.5 |
0.276 | RT-PCR | GCKR rs1260326 |
| Zhu et al. [20] | 2023 | China | Asian | 75 g OGTT | IADPSG | 519 | 24–28 | 31 (28–34) | 498 | 24–28 | 29 (27–32) | 0.737 | Illumina | GCKR rs1260326 |
Notes: GDM, gestational diabetes mellitus; OGTT, oral glucose tolerance test; WHO, World Health Organization; DPSG-EASD, Diabetic Pregnancy Study Group of the European Association for the Study of Diabetes; IADPSG, new International Association of Diabetes and Pregnancy Study Groups; ADA, American Diabetes Association; PCR-SSCP, polymerase chain reaction–single strand conformation polymorphism; PCR-RFLP, polymerase chain reaction restriction fragment-length polymorphism; RT-PCR, real time-polymerase chain reaction; SD, standard deviation; p HWE, p value of Hardy-Weinberg Equilibrium in control group; SNPs, single nucleotide polymorphisms; GCK, glucokinase; GCKR, glucokinase regulatory protein; MassARRAY, Sequenom MassARRAY iPLEX system.
Eleven articles (encompassing 12 studies) focused on the GCK rs1799884 polymorphism with a power of 1.0, while five articles on GCK rs4607517 had a power of 0.812. Eight articles (including 11 studies) and six articles (including 7 studies) concentrated on GCKR rs780094 with a power of 0.829 and GCKR rs1260326 with a power of 1.0, respectively. Table 2 (Ref. [19, 20, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43]) shows the genotype distributions for these SNPs.
| Author | Year | Country | Ethnicity | GDM group | Control group | |||||
| 11 | 12 | 22 | 11 | 12 | 22 | |||||
| rs1799884 | GG | GA | AA | GG | GA | AA | ||||
| Chiu et al. [26] | 1994 | USA | African | 56 | 37 | 4 | 63 | 34 | 2 | |
| Zaidi et al. [30] | 1997 | UK | Caucasian | 47 | 42 | 3 | 25 | 20 | 2 | |
| Shaat et al. [29] | 2006 | Sweden | Caucasian | 435 | 181 | 26 | 889 | 316 | 24 | |
| Freathy et al. [27] | 2010 | UK, Australia | Caucasian | 388 | 194 | 32 | 2575 | 1114 | 122 | |
| Freathy et al. [27] | 2010 | Thailand | Asian | 288 | 91 | 5 | 1375 | 311 | 20 | |
| Santos et al. [28] | 2010 | Brazil | Caucasian | 86 | 56 | 8 | 387 | 186 | 27 | |
| Li W [31] | 2011 | China | Asian | 632 | 349 | 42 | 552 | 315 | 40 | |
| Han et al. [34] | 2015 | China | Asian | 705 | 226 | 17 | 787 | 178 | 10 | |
| Tarnowski et al. [36] | 2017 | Poland | Caucasian | 163 | 42 | 2 | 147 | 52 | 5 | |
| Zhou [43] | 2020 | China | Asian | 506 | 277 | 47 | 556 | 280 | 27 | |
| Popova et al. [41] | 2021 | Russia | Caucasian | 488 | 173 | 27 | 343 | 99 | 12 | |
| She et al. [19] | 2022 | China | Asian | 506 | 277 | 47 | 556 | 280 | 27 | |
| rs4607517 | GG | GA | AA | GG | GA | AA | ||||
| Wang et al. [32] | 2011 | China | Asian | 1420 | 244 | 37 | 618 | 356 | 49 | |
| Stuebe et al. [33] | 2014 | USA | Caucasian | 49 | 3 | 0 | 731 | 53 | 2 | |
| Ao et al. [40] | 2021 | China | Asian | 316 | 200 | 46 | 283 | 154 | 15 | |
| Zhou [43] | 2020 | China | Asian | 702 | 112 | 18 | 735 | 124 | 7 | |
| She et al. [19] | 2022 | China | Asian | 702 | 112 | 18 | 735 | 124 | 7 | |
| rs780094 | TT | TC | CC | TT | TC | CC | ||||
| Li W [31] | 2011 | China | Asian | 275 | 502 | 247 | 265 | 453 | 225 | |
| Stuebe et al. [33] | 2014 | USA | Caucasian | 24 | 23 | 5 | 266 | 376 | 150 | |
| Stuebe et al. [33] | 2014 | USA | African–American | 16 | 6 | 0 | 255 | 87 | 4 | |
| Anghebem-Oliveira et al. [35] | 2017 | Brazil | Caucasian | 15 | 48 | 64 | 14 | 68 | 43 | |
| Jamalpour et al. [37] | 2018 | Malaysia | Asian | 18 | 69 | 95 | 84 | 284 | 214 | |
| Jamalpour et al. [37] | 2018 | Malaysia | Asian | 5 | 30 | 13 | 23 | 76 | 64 | |
| Jamalpour et al. [37] | 2018 | Malaysia | Asian | 3 | 13 | 16 | 16 | 47 | 39 | |
| Tarnowski et al. [36] | 2017 | Poland | Caucasian | 33 | 101 | 73 | 28 | 99 | 77 | |
| Li et al. [42] | 2018 | China | Asian | 64 | 48 | 15 | 43 | 68 | 14 | |
| Zhou [43] | 2020 | China | Asian | 200 | 371 | 213 | 227 | 401 | 212 | |
| She et al. [19] | 2022 | China | Asian | 200 | 371 | 213 | 227 | 401 | 212 | |
| rs1260326 | TT | TC | CC | TT | TC | CC | ||||
| Stuebe et al. [33] | 2014 | USA | Caucasian | 5 | 26 | 25 | 154 | 395 | 291 | |
| Stuebe et al. [33] | 2014 | USA | African–American | 1 | 7 | 16 | 7 | 107 | 248 | |
| Franzago et al. [38] | 2017 | Italy | Caucasian | 21 | 58 | 23 | 15 | 36 | 15 | |
| Franzago et al. [39] | 2018 | Italy | Caucasian | 21 | 58 | 25 | 30 | 68 | 26 | |
| Zhou [43] | 2020 | China | Asian | 220 | 404 | 182 | 238 | 424 | 195 | |
| She et al. [19] | 2022 | China | Asian | 220 | 404 | 182 | 238 | 424 | 195 | |
| Zhu et al. [20] | 2023 | China | Asian | 142 | 241 | 122 | 164 | 245 | 86 | |
Notes: GDM, gestational diabetes mellitus; GCK, glucokinase; GCKR, glucokinase regulatory protein.
For these SNPs, 2 referes risk allele and 1 defiene as reference allele. The combined analyses for GCK (rs1799884 and rs4607517) and GCKR (rs780094 and rs1260326) polymorphisms indicated that these SNPs were significantly correlated with elevated GDM susceptibility under 22 vs. 11 (OR = 1.28, 95% CI = 1.08–1.51, Fig. 2A); decreased GDM susceptibility under 12 vs. 11 (OR = 0.65, 95% CI = 0.52–0.81, Fig. 2B) and 22 vs. 11 + 12 (OR = 0.80, 95% CI = 0.70–0.92, Fig. 2C) genetic models. However, no significant association was discovered in 22 + 12 vs. 11 (OR = 1.05, 95% CI = 0.91–1.21, Fig. 2D) and 2 vs. 1 (OR = 1.07, 95% CI = 0.96–1.20, Fig. 2E) genetic models, respectively.
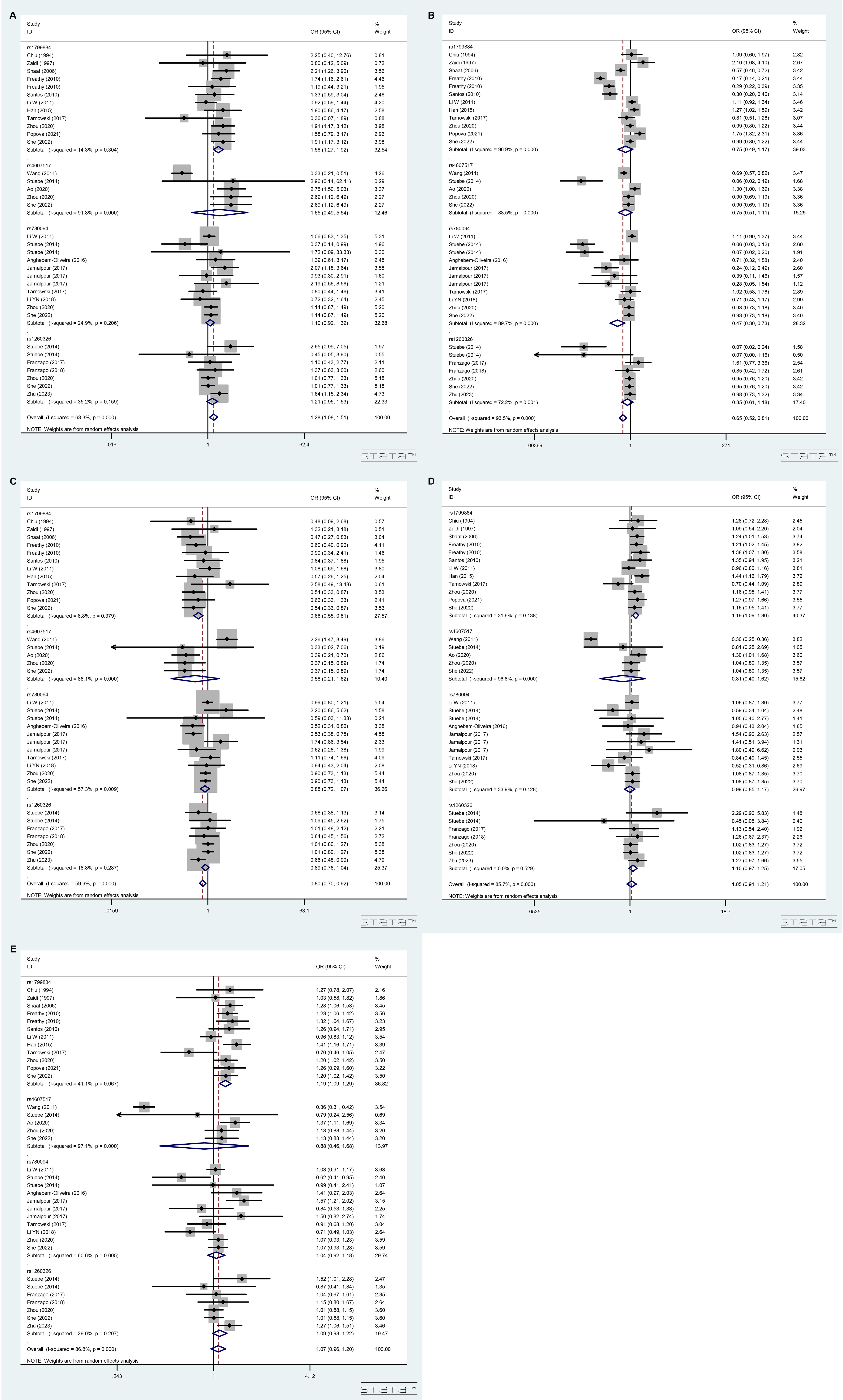 Fig. 2.
Fig. 2.
Forest plot for merged odds ratios (ORs) with 95% confidence intervals (CIs) for GCK and GCKR polymorphisms. (A) Forest plot under 22 vs. 11 model. (B) Forest plot under 12 vs. 11 model. (C) Forest plot under 22 vs. 11 + 12 model. (D) Forest plot under 22 + 12 vs. 11 model. (E) Forest plot under 2 vs. 1 model.
Regarding the rs1799884 polymorphism, significant heterogeneity was observed in
the GA vs. GG model (p
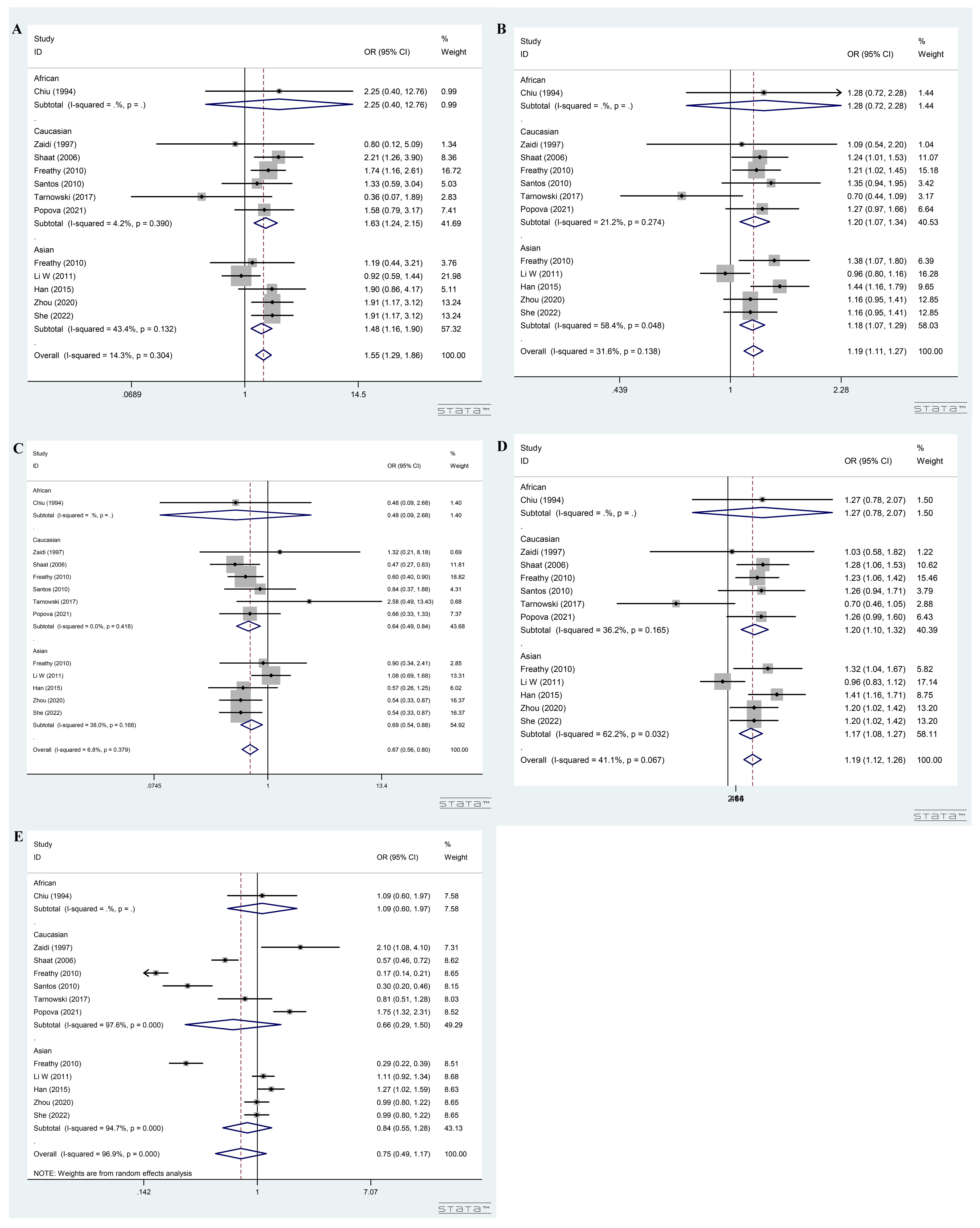 Fig. 3.
Fig. 3.
Forest plot for merged odds ratios (ORs) with 95% confidence intervals (CIs) for GCK rs1799884 polymorphism. (A) Forest plot for subgroup analysis based on ethnicity under AA vs. GG model. (B) Forest plot for subgroup analysis based on ethnicity under AA + GA vs. GG model. (C) Forest plot for subgroup analysis based on ethnicity under AA vs. GG + GA model. (D) Forest plot for subgroup analysis based on ethnicity under A vs. G model. (E) Forest plot for subgroup analysis based on ethnicity under AG vs. GG model.
The rs780094 polymorphism showed significant heterogeneity in the TC
vs. TT, CC vs. TT+TC, and C vs. T
genetic models (p
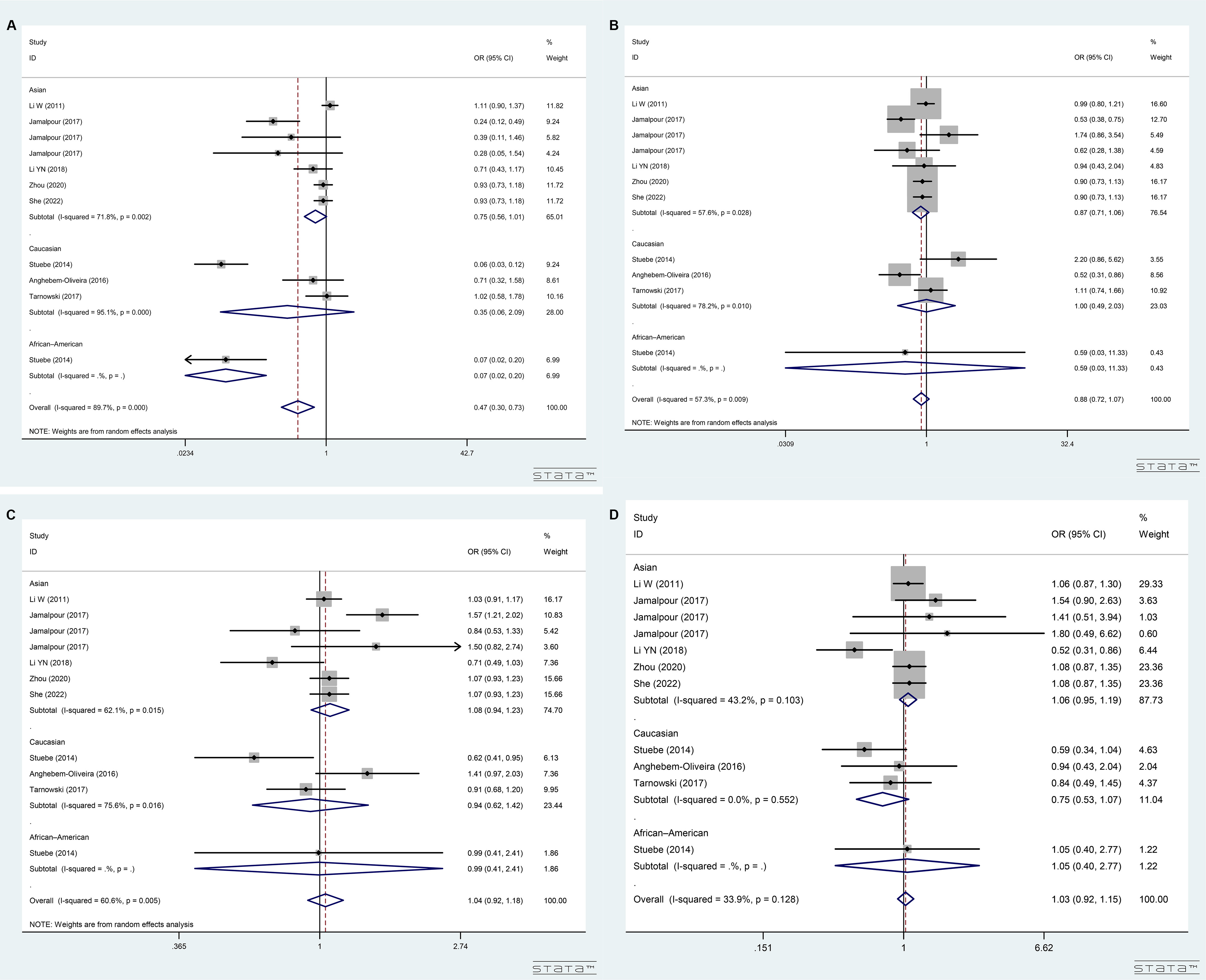 Fig. 4.
Fig. 4.
Forest plot for merged odds ratios (ORs) with 95% confidence intervals (CIs) for GCKR rs780094 polymorphism. (A) Forest plot for subgroup analysis based on ethnicity under TC vs. TT model. (B) Forest plot for subgroup analysis based on ethnicity under CC vs. TT + TC model. (C) Forest plot for subgroup analysis based on ethnicity under C vs. T model. (D) Forest plot for subgroup analysis based on ethnicity under CC + TC vs. TT model.
Stratified by ethnicity and diagnostic criteria, subgroup analyses were conducted to fine heterogeneity sources. The heterogeneity observed in the rs1799884 polymorphism’s GA vs. GG model might have stemmed from variable ethnic and diagnostic criteria. Due to fewer studies for rs4607517, rs780094, and rs1260326 polymorphisms and multiple diagnostic criteria, diagnostic criteria-based subgroup analysis was not feasible. The heterogeneity in these polymorphisms was attributed to varying ethnicity.
In the ethnicity-based subgroup analysis, a positive association between rs1799884 polymorphism and GDM susceptibility was evident in the AA vs. GG model in both Asian (OR = 1.48, 95% CI = 1.16–1.90) and Caucasian (OR = 1.63, 95% CI = 1.24–2.15) subgroups, respectively (Fig. 3A). Analysis by diagnostic criteria revealed a positive association in the AA vs. GG model in subgroups following the Diabetic Pregnancy Study Group of the European Association for the Study of Diabetes (DPSG-EASD) (OR = 2.21, 95% CI = 1.26–3.90), International Association of Diabetes and Pregnancy Study Groups (IADPSG) (OR = 1.63, 95% CI = 1.25–2.13), and National criteria (OR = 1.91, 95% CI = 1.17–3.12) (Supplementary Fig. 3A). We also observed positive associations in the AA+GA vs. GG (Fig. 3B), AA vs. GG+GA (Fig. 3C), and A vs. G (Fig. 3D) models in the specified ethnic and diagnostic subgroups (Supplementary Fig. 3B–D). Conversely, the ethnicity-based subgroup analysis revealed a negative correlation between the rs780094 polymorphism and GDM susceptibility in the African-American subgroup under the TC vs. TT model (OR = 0.07, 95% CI = 0.02–0.20, Fig. 4A), but not in other for genetic models (Fig. 4B–D). Only one Caucasian study was conducted on the rs4607517 (Supplementary Fig. 1) and one African-American study was conducted on rs1260326 (Supplementary Fig. 2) polymorphism. Small number of studies, especially only one study in the subgroup, usually lead to unreliable estimation of heterogeneity, such as false positive result. Therefore, the ethnicity-based subgroup analysis was not performed in five genetic models of rs4607517 and rs1260326.
Hence, these findings suggest a significant association between the rs1799884 as well as rs780094 polymorphisms and GDM susceptibility. However, the rs4607517 and rs1260326 polymorphisms were not be significantly associated with the overall population.
Begg’s test helped to assess publication bias across the combined analysis of GCK and GCKR genetic polymorphisms. None of the publications showed any significant publication bias (Begg’s test: p = 0.063 for 22 vs. 11, Fig. 5A; p = 0.386 for 12 vs. 11; p = 0.806 for 22 vs. 11+12, Fig. 5B; p = 0.088 for 22+12 vs. 11; p = 0.629 for 2 vs. 1).
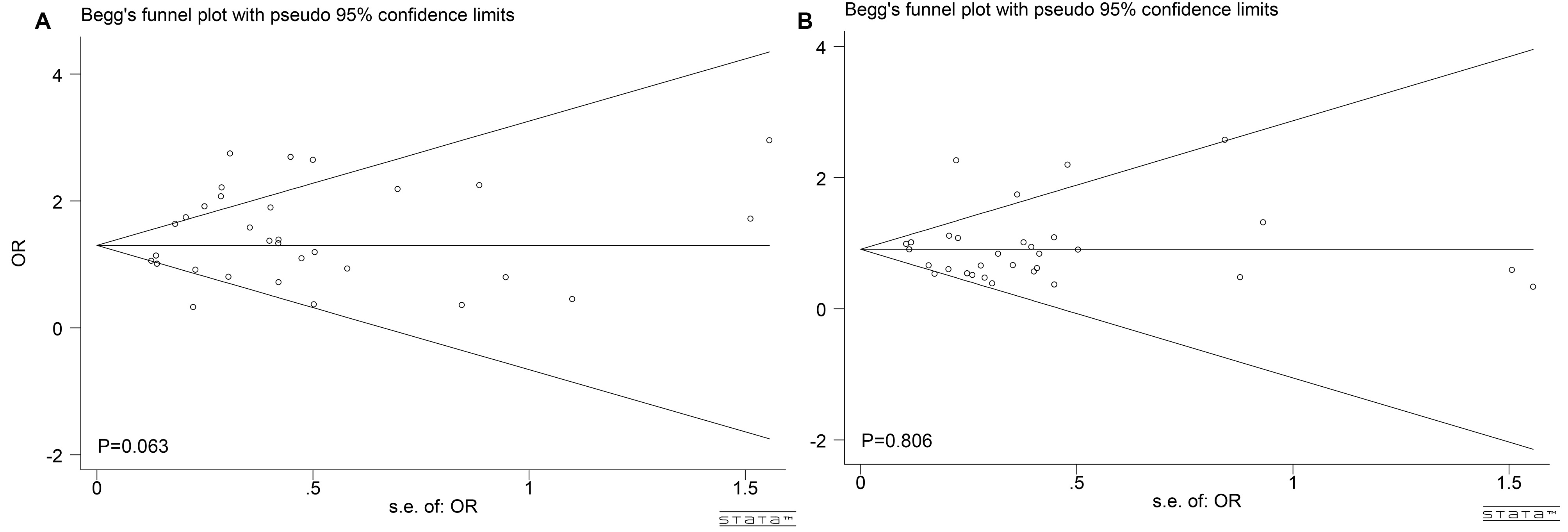 Fig. 5.
Fig. 5.
Publication bias for eligible studies of GCK and GCKR polymorphisms. (A) Publication bias under 22 vs. 11 model. (B) Publication bias under 22 vs. 11 + 12 model. 22 is risk genotype, 12 is heterozygote genotype and 11 is the reference genotype.
We systematically reviewed 20 eligible studies in this extensive meta-analysis that examined the relationship between GDM susceptibility and GCK (rs1799884 and rs4607517) and GCKR genes (rs780094 and rs1260326). Our results indicated that these SNPs were significantly correlated with GDM susceptibility. Meanwhile, GCK rs1799884 polymorphism was significantly associated with GDM susceptibility in five genetic models; the A allele carriers displayed a higher risk for GDM. This is consistent with previous meta-analytical results that identified a positive association between the rs1799884 polymorphism and GDM susceptibility [20, 34, 44]. Moreover, a positive association between rs1799884 polymorphism and GDM susceptibility was also noted. We found that GCK rs4607517 was a risk factor for GDM susceptibility in the AA vs. GG genetic model. Mao et al. [45] also reported a positive correlation between the GCK rs4607517 A allele and GDM risk, suggesting that GCK polymorphisms might lead to GDM.
However, we could not find a significant association between GDM risk and the GCKR genes rs780094 as well as rs1260326. This was in contrast with a few meta-analyses that suggested a high risk of GDM for rs780094 G allele carriers [37, 46, 47]. This discrepancy may be attributed to variable ethnicities among the study populations.
Enhanced heterogeneity was discovered in the rs1799884 polymorphism’s GA vs. GG genetic model. We examined the rs1799884 polymorphism’s heterogeneity origin across different ethnicities and diagnostic criteria. The possible causes of heterogeneity include various ethnicities and diagnostic criteria. Due to limited studies on rs4607517, rs780094, and rs1260326 polymorphisms, we discussed their heterogeneities in different ethnic groups. Subsequently, we found that the GCK rs4607517 heterogeneities in five genetic models stemmed from different ethnicities. Heterogeneities were also discovered in the TC vs. TT model of GCKR rs780094 as well as the TC vs. TT model of rs1260326 polymorphism and were derived from different ethnicities. However, no significant heterogeneity was discovered in previous meta-analyses [45, 46, 47, 48]. Different diagnosis standards and ethnicities might be the cause of this discrepancy.
Ethnicity-based subgroup analysis revealed a positive correlation between rs1799884 polymorphism and GDM susceptibility in Asian and Caucasian populations. This is in line with the findings of Yang and Du [44] that A allele of rs1799884 is a risk factor for GDM in White and African populations. Notably, we also observed a negative correlation between the rs780094 polymorphism and GDM susceptibility in the African-American subgroup’s TC vs. TT genetic model. Conversely, Jamalpour et al. [37] observed that the C allele of rs780094 was positively correlated with GDM susceptibility in the Asian population. The rs1260326 polymorphism’s subgroup analysis was consistent with previous meta-analyses, indicating no significant association with GDM susceptibility across different populations [46, 47].
Furthermore, diagnostic criteria-based subgroup analysis confirmed their influence on the association between the rs1799884 polymorphism and GDM susceptibility. The DPSG-EASD and IADPSG subgroups were positively correlated. Yang and Du [44] demonstrated that the A allele of rs1799884 was not significantly associated with GDM susceptibility under World Health Organization (WHO) and American Diabetes Association (ADA) subgroups. This underscores the importance of standardized criteria for future studies and the possible effects of varying GDM diagnostic criteria on research outcomes.
Begg’s test revealed no significant publication bias, suggesting the reliability of observed associations. However, selection and information biases could potentially affect our results’ generalizability and statistical power. Our findings highlight the importance of targeted screening guidelines for Asian and Caucasian populations as well as focusing on nutritional and lifestyle interventions for pregnant women. Additional research with larger sample sizes and multicenter studies should be conducted to validate our findings and explore the biological mechanisms influencing the associations between GCK and GCKR polymorphisms as well as GDM susceptibility.
According to our meta-analysis, the GCK rs1799884 polymorphism’s A allele is associated with an increased risk of GDM. Although certain individuals with GCK rs4607517 and GCKR rs780094 polymorphisms may also be at higher risk for GDM, the overall association is less clear and warrants further investigation. These insights can help in identifying high-risk pregnant women early as well as emphasizing the necessity of genetic counseling and interdisciplinary collaboration for preventing and managing GDM.
Corresponding authors may provide data and materials.
HXT and MYW designed the research study. YYZ performed the research. HXT, YYZ and MYW analyzed the data. HXT and MYW wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Not applicable.
Not applicable.
This research received no external funding.
The authors declare no conflict of interest.
Supplementary material associated with this article can be found, in the online version, at https://doi.org/10.31083/CEOG26710.
Publisher’s Note: IMR Press stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
