† These authors contributed equally.
Background: To investigate the effects of GSDMB
polymorphism on sensitivity to chemoradiation. Methods: 108 cervical
cancer patients were selected and treated with a combination of radiotherapy and
chemotherapy. After 2 cycles, patients were grouped into sensitive group and
non-sensitive group based on the chemoradiation therapy outcomes. GSDMB
polymorphism was assessed by high-resolution melting (HRM) analysis, and the
GSDMB gene expression was detected using RT-qPCR. Results: Our results
indicate that the allele and genotype distribution of GSDMB in patients from
sensitivity group were significantly different as compared to non-sensitive
group. Experimental analysis showed a close correlation between GSDMB
polymorphism and sensitivity to chemoradiation therapy for cervical cancer.
Moreover, ATT, GCC, GCT and GTC halotype of GSDMB gene was significantly
different among sensitive and non-sensitive groups (p
Cervical cancer is regarded as the most common gynecologic cancer in developing
countries which ranks third among all cancer types [1, 2, 3]. Recent studies
demonstrated that treatment of cervical epithelial atypical hyperplasia, a
precancerous of cervical cancer, displays 1% and 0.5% decrease in the incidence
and mortality, respectively [4]. The rise in the occurrence of cervical cancer
in developing countries is associated with sexual behavior, such as multiple
partners, multiple pregnancy with Chlamydia psittaci infection, and
HIV-associated immunosuppression, which could increase the risk of HPV infection,
therefore promote the occurrence of cervical cancer [5, 6]. The main therapy to
treat cervical cancer is surgical operation and radiotherapy combined with
chemotherapy [7, 8, 9]. The efficacy of chemoradiation therapy determines their
prognosis and survival time among the patients who cannot receive radical
operation [10, 11]. It has been established that the sensitivity to
chemoradiation therapy for cervical cancer is associated with numerous factors,
including immune state, neoplasm staging and genovariation [12, 13]. Among these,
gene polymorphism has a close relationship with the efficacy of chemoradiation
therapy for cervical cancer [14, 15]. However, the relationship between GSDMB
polymorphism and sensitivity to chemoradiation therapy for cervical cancer has
been unclarified yet. Therefore, we designed this study to investigate the effect
of GSDMB polymorphism on sensitivity to chemoradiation therapy for cervical
cancer. Briefly, this study analyzed the GSDMB rs8067378 A
108 patients with cervical cancer registered in Chenzhou First People’s Hospital
(Chenzhou, China) from January 2019 till now were selected as participants. All
of them were confirmed for cervical cancer, and the stage and grade of
differentiation was assessed by an experienced histopathologist based on the
International Federation of Gynecology and Obstetrics classification system and
the World Health Organization. The possibility of other serious
cancerous diseases, such as primary hepatic carcinoma and lung cancer, was ruled
out. All patients had no previous history of any chemoradiation therapy, while
they were also unsuitable for radical surgery. Based on the
outcomes of radiotherapy and chemotherapy, patients were grouped into sensitive
group (62 cases) and non-sensitive group (46 cases). The median age was 58.32
Radiation regimen: All participants were irradiate using a linear accelerator
(Primus, Siemens Corporation, Beijing, China). Firstly, whole pelvic radiotherapy
was given (36 Gy, four times per week), and then four weeks irradiation was
performed (56 Gy, 30 times), and eventually brachytherapy was given (90 Gy).
Chemotherapeutic regimen: Cisplatin was given at a dose of 30 mg/m
The therapeutic efficacy of chemoradiation therapy which is classified into complete response (CR), partial response (PR), stable disease (SD) and progression of disease (PD), was evaluated after 2 cycles of treatment according to RECIST1.1. Patients were grouped into sensitive group and non-sensitive group based on the therapeutic effect of chemoradiation therapy, i.e., CR and PR were grouped into sensitive group, while SD and PD were grouped into non-sensitive group.
4–5 mL of peripheral blood was collected in tubes containing EDTA anticoagulant from all participants before chemoradiation therapy. Genomic DNA from whole blood was extracted by using DNA extraction kit (Tiangen, Nobleride Corporation, Beijing, China) following the manufacturer’s protocol. Quality and quantity of extracted DNA was determined by using a spectrophotometer (Spectrum Instrument Co., Ltd., Shanghai, China).
High-resolution melting (HRM) analysis was used to analyze GSDMB rs8067378
A
| Primer | Sequence | |
| rs8067378 | Sense | TGTGAGTGGAAAGCTTGACAG |
| Anti-sense | AGCTTGCTACAGTGAGACCC | |
| rs7216389 | Sense | AGTTCTGTCGCTGTTGTTTGT |
| Anti-sense | ACACATCCTCCACGAACCTG | |
| rs11650680 | Sense | CTTCCTTCCCTGCTAGTCCC |
| Anti-sense | TCAAAAGACTTGAGGAGGTTCA | |
| GSDMB | Sense | TGATTGCCGTTAGAAGCCTTG |
| Anti-sense | TCCCGTTGAGTCTACATTATCCA | |
| GAPDH | Sense | AGGTCGGTGTGAACGGATTTG |
| Anti-sense | GGGGTCGTTGATGGCAACA |
RNA from peripheral blood cells was extracted by using Trizol, cDNA was synthesized via reverse transcription of total RNA, and the gene expression was detected by qPCR. GAPDH was used as housekeeping gene. Primer sequences used for RT-qPCR analysis are shown in Table 1.
SHEsis online program
(http://analysis.bio-x.cn/)
was used for polymorphism, haplotype and linkage disequilibrium analysis. The
distinction in genotypic prevalence between the patients and controls, and their
genotype deviation using the Hardy–Weinberg equilibrium was evaluated using a
The ratio of CR, PR, SD and PD patients among all 108 patients with cervical cancer after chemoradiation therapy was 24% (26/108), 33.3% (36/108), 29.6% (32/108) and 12.9% (14/108) respectively, which showed an effective rate of 57.3%. Sensitive group comprising of CR and PR patients, which were sensitive to chemoradiation therapy, accounted for 54.7% (62/108). While SD and PD patients accounted for 42.6% (46/108), belong to non-sensitive group as they were not sensitive to chemoradiation therapy.
No significant difference was observed among the age (p = 0.236), pathology results (p = 0.168), stage distribution (p = 0.078), and histological grade (p = 0.069). However, the tumor size (p = 0.011) and CEA (p = 0.000) level showed a statistically significant difference between sensitive and non-sensitive group. This suggested that tumor size and CEA level contribute to the sensitivity of patients to chemoradiation therapy rather than age, pathology results, stage and histological grades (Table 2).
| Parameters | Cases | Sensitive group | Non-sensitive group | p | |
| Age (years) | 0.236 | ||||
| 50 | 26 | 24 | |||
| 58 | 36 | 22 | |||
| Pathology results | 0.168 | ||||
| Squamous carcinoma | 90 | 51 | 49 | ||
| Adenocarcinoma | 18 | 11 | 7 | ||
| Tumor size (cm) | 0.011 | ||||
| 70 | 52 | 18 | |||
| 38 | 10 | 28 | |||
| CEA (ng/mL) | 0.000 | ||||
| 66 | 50 | 16 | |||
| 42 | 12 | 30 | |||
| Tumor stage | 0.078 | ||||
| Stage IA | 5 | 3 | 2 | ||
| Stage IB | 7 | 4 | 3 | ||
| Stage IIA | 30 | 13 | 17 | ||
| Stage IIB | 31 | 26 | 5 | ||
| Stage IIIA | 30 | 15 | 15 | ||
| Stage IIIB | 5 | 1 | 4 | ||
| Stage IV | 0 | 0 | 0 | ||
| Histological grade | 0.069 | ||||
| G1 | 47 | 25 | 22 | ||
| G2 | 35 | 26 | 9 | ||
| G3 | 21 | 10 | 11 | ||
| G |
5 | 1 | 4 | ||
There was no significant association between age (p = 0.439), pathology results (p = 0.573), stage (p = 0.736) and sensitivity to chemoradiation therapy. However, tumor size (r = 0.263, p = 0.005) and CEA level (r = –0.548, p = 0.000) showed statistically significant association with chemoradiation therapy (Table 3).
| Parameter | r | p |
| Age | 0.017 | 0.439 |
| Pathology results | 0.105 | 0.573 |
| Tumor size | 0.263 | 0.005 |
| CEA | –0.548 | 0.000 |
| Stage | 0.021 | 0.736 |
| Histological grade | 0.015 | 0.532 |
The values for the
| Site | Allele | Sensitive group | Non-sensitive group | OR value | 95% CI | p | |
| rs8067378 | A | 95 (0.766) | 41 (0.446) | 4.07 | 2.27–7.31 | 23.26 | 0.000 |
| G | 29 (0.234) | 51 (0.554) | |||||
| rs7216389 | C | 36 (0.290) | 56 (0.609) | 0.26 | 0.14–0.46 | 21.89 | 0.000 |
| T | 88 (0.710) | 36 (0.391) | |||||
| rs11650680 | C | 47 (0.379) | 46 (0.500) | 0.61 | 0.35–1.05 | 3.15 | 0.075 |
| T | 77 (0.621) | 46 (0.500) |
The genotypic distribution of GSDMB rs8067378 A
| Site | Genotype | Sensitive group | Non-sensitive group | p | |
| rs8067378 | AA | 39 (0.629) | 8 (0.174) | 22.67 | 0.000 |
| AG | 17 (0.274) | 25 (0.543) | |||
| GG | 6 (0.097) | 13 (0.283) | |||
| rs7216389 | CC | 8 (0.129) | 17 (0.370) | 19.15 | 0.000 |
| CT | 20 (0.323) | 22 (0.478) | |||
| TT | 34 (0.548) | 7 (0.152) | |||
| rs11650680 | CC | 4 (0.065) | 13 (0.283) | 9.83 | 0.007 |
| CT | 39 (0.629) | 20 (0.435) | |||
| TT | 19 (0.306) | 13 (0.283) |
Our further investigation revealed that there was a dramatic correlation between
GSDMB rs8067378 A
| Polymorphism site | r | p |
| rs8067378 A |
0.437 | 0.000 |
| rs7216389 C |
–0.274 | 0.003 |
| rs11650680 C |
0.178 | 0.064 |
The distribution of rs8067378 A
| Site | Genotype | Sensitive group | Non-sensitive group | p | ||
| Dominant model | ||||||
| rs8067378 | AA + AG | 56 (0.903) | 33 (0.717) | 4.38 | 0.112 | |
| GG | 6 (0.097) | 13 (0.283) | ||||
| rs7216389 | CC + CT | 28 (0.452) | 39 (0.848) | 15.34 | 0.000 | |
| TT | 34 (0.548) | 7 (0.152) | ||||
| rs11650680 | CC + CT | 43 (0.694) | 31 (0.717) | 3.57 | 0.168 | |
| TT | 19 (0.306) | 13 (0.283) | ||||
| Recessive model | ||||||
| rs8067378 | AA | 39 (0.629) | 8 (0.174) | 14.38 | 0.001 | |
| AG + GG | 23 (0.371) | 38 (0.826) | ||||
| rs7216389 | CC | 8 (0.129) | 17 (0.370) | 4.32 | 0.115 | |
| CT + TT | 54 (0.871) | 29 (0.630) | ||||
| rs11650680 | CC | 4 (0.065) | 13 (0.283) | 5.34 | 0.069 | |
| CT + TT | 58 (0.935) | 33 (0.717) | ||||
| Hybrid model | ||||||
| rs8067378 | AA | 39 (0.629) | 8 (0.174) | 3.25 | 0.197 | |
| AG | 17 (0.274) | 25 (0.543) | ||||
| rs7216389 | CC | 8 (0.129) | 17 (0.370) | 4.28 | 0.118 | |
| CT | 20 (0.323) | 22 (0.478) | ||||
| rs11650680 | CC | 4 (0.065) | 13 (0.283) | 2.34 | 0.312 | |
| CT | 39 (0.629) | 20 (0.435) | ||||
| Homozygous model | ||||||
| rs8067378 | AA | 39 (0.629) | 8 (0.174) | 3.85 | 0.146 | |
| GG | 6 (0.097) | 13 (0.283) | ||||
| rs7216389 | CC | 8 (0.129) | 17 (0.370) | 3.17 | 0.205 | |
| TT | 34 (0.548) | 7 (0.152) | ||||
| rs11650680 | CC | 4 (0.065) | 13 (0.283) | 2.84 | 0.242 | |
| TT | 19 (0.306) | 13 (0.283) |
Our data showed ATT, GCC, GCT and GTC of GSDMB rs8067378 A
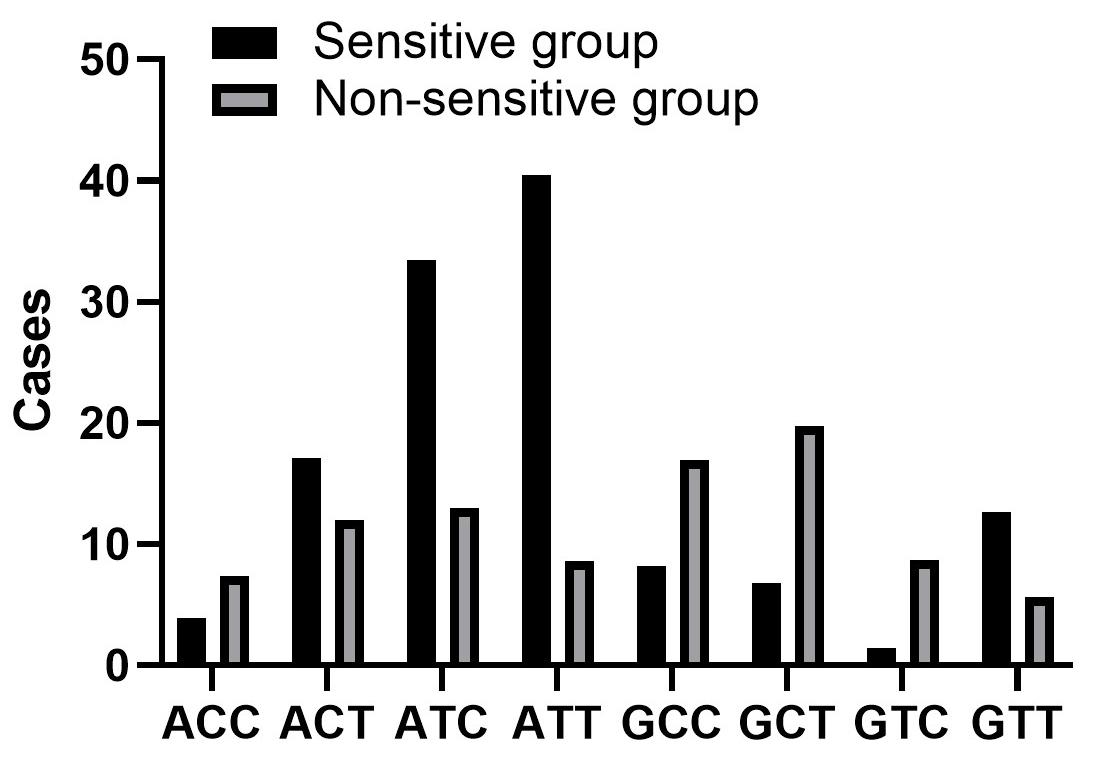 Fig. 1.
Fig. 1.
Haplotype distribution of GSDMB rs8067378 A
We further revealed that linkage disequilibrium was higher in GSDMB rs8067378
A
| D’ | rs8067378 | rs7216389 | rs11650680 |
| rs8067378 | - | 0.415 | 0.002 |
| rs7216389 | 0.415 | - | 0.004 |
| rs11650680 | 0.002 | 0.004 | - |
| r |
rs8067378 | rs7216389 | rs11650680 |
| rs8067378 | - | 0.137 | 0.001 |
| rs7216389 | 0.137 | - | 0.001 |
| rs11650680 | 0.001 | 0.001 | - |
Finally, we found that GSDMB rs8067378 A
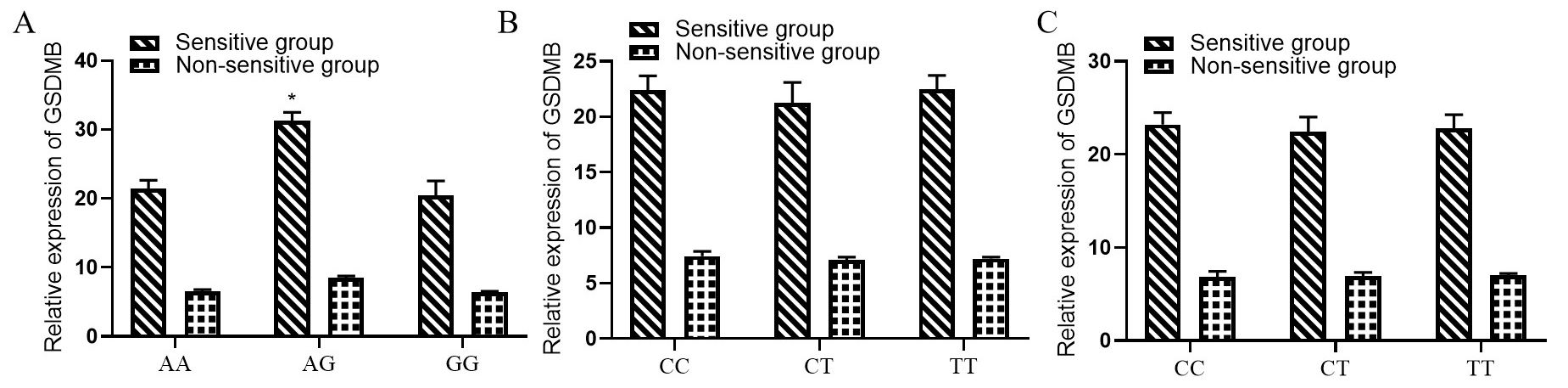 Fig. 2.
Fig. 2.
Correlation between GSDMB rs8067378 A
Cervical cancer is one of the most common malignant tumors in women. It has been estimated that the five-year survival rate of localized cervical cancer is 91.5%, while metastatic cervical cancer is 16.5% [16]. Cisplatin-based chemotherapy has long been the major treatment for intermediate and advanced cervical cancer. However, the survival rate shows no significant improvement as patients respond differently to chemotherapy. Adding of new therapeutical drugs, such as antiangiogenic agent, can greatly improve the outcome of curative capability [17]. However, the sensitivity of patients to chemoradiation varies because of tumor heterogeneity and individual difference, which directly impact the tumor prognosis and survival of patients [18, 19]. Therefore, identifying factors which are associated with the sensitivity to cervical cancer chemoradiation therapy could contribute to prolong the overall survival of patients. In this study, we analyzed a total 108 cases of cervical cancer after chemoradiation therapy. We discovered that 24% of total patients were CR, while the percentages of PR, SD and PD patients were 33.3%, 29.6% and 12.9% respectively, with an effective ratio of 57.3%. Tumor size and CEA level were significantly different, meanwhile, age, pathology results, stage and histological grade distribution were not changed significantly between sensitive and non-sensitive group. Moreover, no significant correlation was found between age, pathology results, stage, histological grade, and sensitivity to chemoradiation therapy. Instead, tumor size and CEA were correlated with chemoradiation sensitivity. These results suggested that tumor size and CEA level contribute to the sensitivity of patients to chemoradiation therapy rather than age, pathology results, stage and histological grades.
GSDMB belongs to the Gasdermin protein family, and these gene family members
share conserved N- and C-terminal structural domain with a sequence homology of
45% [20, 21]. The Gasdermin protein family has five members in humans, which are
termed as GSDMA, GSDMB, GSDMC, GSDMD and GSDME [22, 23]. GSDMB displays broader
expression pattern as compared to other GSDM family members by showing higher
tissue-specific expression in skin epithelium, gastrointestinal tract and immune
cells [24, 25]. Genome-wide association study (GWAS) revealed a close
relationship between GSDMB and susceptibility to inflammatory diseases such as
Crohn’s disease, asthma, and type I diabetes [26, 27]. A study on GSDMB
transgenic mice indicated that GSDMB promotes the development of asthma, and
increases airway responsiveness and remodeling [28, 29]. Besides, researchers
have found that GSDMB was highly expressed not only in healthy tissues, but also
in cancerous tissues from gastric cancer, metrocarcinoma, cervical cancer and
breast cancer. GSDMB was located in amplicons which often amplified during the
progression of cancer [30]. Therefore, GSDMB might be involved in the occurrence,
development and metastasis of cancer. However, the functions of GSDMB are still
not studied well. Meanwhile, the effect of GSDMB on sensitivity to chemoradiation
therapy in cervical cancer also remains undocumented. This study compared the
GSDMB rs8067378 A
By haplotype analysis and linkage disequilibrium analysis, we found that ATT
(p = 0.000), GCC (p = 0.008), GCT (p = 0.000) and GTC
(p = 0.004) of GSDMB rs8067378 A
We further concluded that GSDMB rs8067378 A
The limitations of this study is that the sample size is not large enough to
fully confirm the effect of GSDMB rs8067378 A
This analysis provides new insights into the genetic causes of cervical cancer and is of great significance for the prevention and treatment of cervical cancer.
NZ, YY and HL designed the research study. NZ performed the research. GHP, HC and QYT provided help and advice on the ELISA experiments. NZ, YY, HKL, QP, HZZ and YBZ analyzed the data. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Chenzhou First People’s Hospital (Approval number: QZ6C11 ).
We would like to express our gratitude to all those who helped us during the writing of this manuscript. And thanks to all the peer reviewers for their opinions and suggestions at the same time.
Chenzhou Science and Technology Bureau 2019 (No.zdyf201945).
The authors declare no conflict of interest.
